Gut microbiomes abundant with Proteobacteria, Enterococcaceae, or Streptococcaceae predicted infection during therapy for acute lymphoblastic leukemia. A gut microbiome biomarker may be used to stratify the infection risk in children with acute lymphoblastic leukemia before and during chemotherapy.
Keywords: microbiome, cancer, children, acute lymphoblastic leukemia, infection
Abstract
Background
Myelosuppression-related infections remain important causes of morbidity and mortality in children with acute lymphoblastic leukemia (ALL).
Methods
By analyzing fecal samples collected at diagnosis and after each of the initial 3 phases of chemotherapy, we evaluated the role of gut microbiota in predicting infections in 199 children with newly diagnosed ALL. The bacterial 16S rRNA gene was analyzed by high-depth sequencing to determine the diversity and composition of the microbiome.
Results
After the induction and reinduction I phases of chemotherapy, microbial diversity decreased significantly relative to the prechemotherapy value. After chemotherapy, the relative abundance of certain bacterial taxa (eg, Bacteroidetes) decreased significantly, whereas that of other taxa (eg, Clostridiaceae and Streptococcaceae) increased. A baseline gut microbiome characterized by Proteobacteria predicted febrile neutropenia. Adjusting for the chemotherapy phase and ALL risk level, Enterococcaceae dominance (relative abundance ≥30%) predicted significantly greater risk of subsequent febrile neutropenia and diarrheal illness, whereas Streptococcaceae dominance predicted significantly greater risk of subsequent diarrheal illness.
Conclusions
In children undergoing therapy for newly diagnosed ALL, the relative abundance of Proteobacteria before chemotherapy initiation predicts development of febrile neutropenia, and domination of the gut microbiota by Enterococcaceae or Streptococcaceae at any time during chemotherapy predicts infection in subsequent phases of chemotherapy.
Clinical Trial Registration
Emerging epidemiologic evidence shows associations between gut microbiota and overall health [1–3]. Alterations in gut microbiota affect the immune system and resistance to infection in healthy individuals, adults with acute myeloblastic leukemia or non-Hodgkin lymphoma, and hematopoietic stem cell transplant (HSCT) recipients [4–7]. Disruption of the gut microbiome by chemotherapy and broad-spectrum antibiotics may facilitate domination by a single pathogen that can cross the intestinal mucosa to the bloodstream [8, 9]. The diversity and composition of gut microbiome predict infection in adults with cancer [5, 6, 8–10]. Although one study [11] showed that children with cancer had less-diverse gut microbiota than their healthy siblings, the relevance of the microbiome to the outcome of antineoplastic therapy in children is unknown.
We hypothesized that changes in the gut microbiome are associated with infection risk in children with ALL. This study examined changes in fecal microbiota in children with ALL before and during chemotherapy and aimed to identify characteristics of gut microbiome that predicted outcomes such as diarrhea, bloodstream infections, or febrile neutropenia (FN).
METHODS
Study Design and Participants
Patients were enrolled at the time of ALL diagnosis at St Jude Children’s Research Hospital between January 2012 and August 2015. The protocol was approved by the institutional review board, and informed consent was obtained. The ALL treatment regimen and risk classification (ClinicalTrials.gov identifier NCT00549848) were described previously [12, 13]. Therapy consisted of a 6-week remission induction, 8-week consolidation, and 120-week continuation phase that included two 3-week periods of intensive chemotherapy (reinduction I in weeks 7–9 and reinduction II in weeks 17–20). Demographic, antibiotic and probiotic exposure, treatment, and outcomes were abstracted from the study database and electronic medical records.
Bloodstream infection was defined as detection of bacteria or fungi in blood cultures obtained for clinical care; diarrhea as ≥3 loose or watery stools in a 24-hour period or a documented clinical diagnosis; fever as an oral temperature >38.0°C persisting for >1 hour; and neutropenia as an absolute neutrophil count ≤500 cells/µL. Patients were followed from the date of ALL diagnosis until censoring at the first of: end of reinduction II, death, or removal from the Total XVI study because of ALL relapse, HSCT, or other factors.
Sample Collection
Stool specimens were obtained at 4 predefined time points for microbiome analysis: before or within 72 hours after induction chemotherapy initiation (baseline); within 2 weeks after induction completion (postinduction); and at the initiation of reinduction I (postconsolidation) and reinduction II (postreinduction) phases. Samples were frozen at −80°C until DNA extraction.
DNA Extraction, Library Construction, and Sequence Analysis
DNA was extracted by bead beating on a FastPrep instrument (MP Biomedicals, Santa Ana, California) followed by genomic DNA extraction with a FastDNA kit (MP Biomedicals). The V1–V3 region of the bacterial 16S rRNA gene was amplified using a NEXTflex 16S Amplicon-Seq Library Prep kit (Bioo Scientific, Austin, Texas). The multiplexed products were used for high-depth sequencing on the Illumina Mi-seq platform. The quality of raw 16S ribosomal RNA (rRNA) pair-ended reads (300 bp ×2) was examined by FastQC, and low-quality read ends (quality score <20) were trimmed with Trim Galore [14, 15]. The reads were subsequently merged with PANDAseq and processed with QIIME [16, 17]. The average read depth is 133290 (median, 147411). The closed-reference mapping protocol of QIIME was used for operational taxonomic unit (OTU) assignment at 97% sequence similarity, using the UCLUST algorithm [18]. A representative sequence was selected from each OTU for taxonomic assignment with the Greengenes database [19]. Using the OTU table, alpha diversity estimates (Shannon, Simpson, and Chao 1 indices) were calculated using R phyloseq [20]. Rarefaction curves were generated for number of sequences per sample (Supplementary Figure 1).
All sequence read files are accessible through the National Center for Biotechnology Information (project accession number PRJNA449103).
Statistical Analysis
OTU Assignment
All samples were included in the analysis. OTUs were grouped in 14 taxa based on frequency (Supplementary Figure 2).
Changes in Microbiota Diversity and Composition During Chemotherapy
A linear mixed-effects model examined changes in the diversity indices using SAS version 9.4 software (SAS Institute, Cary, North Carolina). For microbiome compositional change, a mixed-effects negative binomial regression model was used [21]. The P value was adjusted for the false discovery rate with the Benjamini-Hochberg method [22] and reported as Q value.
Effect of Antibiotics and Probiotics on Microbiome
Antibiotic prophylaxis against pneumocystis pneumonia was excluded from analysis because of ubiquitous exposure. Because all patients received other antibiotics during the study, exposure was calculated as the proportion of days receiving antibiotics for each chemotherapy phase. Correlation between antibiotic exposure and change in diversity was evaluated using Spearman rank test, and change in dominance of Bacteroidetes was tested using Fisher exact test. Probiotic use was analyzed using a Swimmer plot.
Impact of Baseline Microbiota on Infection Outcomes
For initial risk stratification at the time of ALL diagnosis, the Wilcoxon-Mann-Whitney test and logistic regression were used to assess potential risk factors for subsequent infection occurring at any time during the study period, including the baseline microbiome. For each taxon whose relative abundance (RA) significantly differed among patients who had or did not have the event, the RA was dichotomized at the optimal cutoff value based on the Youden index. Factors with a P value of <.1 in univariate analysis were examined by multiple logistic regression. The cumulative incidence of outcomes was compared among risk factors with Gray test.
Microbiota Composition Type Assignment
The “community state” refers to the RA of taxa in a stool sample at a specific time point. The “microbiota composition type” is a cluster of community states with similar microbiota composition. The community states were grouped using hierarchical clustering of the Jensen-Shannon divergence matrix, which assesses beta diversity and the optimal number of clusters [23], and ward linkage and presented as a heat map [21].
Modeling Association Between Microbiota and Infection Outcomes
To evaluate the effect of the time-dependent microbiome on the recurrent infection outcomes throughout all courses of chemotherapy, Anderson-Gill proportional hazards models were adopted [24, 25]. ALL risk level, Shannon diversity index and microbiome composition at the collection time point, and the chemotherapy phase were treated as time-dependent predictors. To assess sensitivity of the results, a bootstrap method (with 10000 replicates) was used to examine the association between Enterococcaceae or Streptococcaceae dominance and subsequent infections. Dominance was defined as an RA of ≥30% [9].
RESULTS
Patient Characteristics and Infection Outcomes
A total of 199 children provided 406 stool samples (Table 1). Twenty-six patients (13%) had 31 episodes of bloodstream infection, 122 (61%) had 248 FN episodes, and 73 (37%) had 112 episodes of diarrheal illness (Table 1). Characteristics of patients with samples at each time point are shown in Supplementary Table 1.
Table 1.
Patient Characteristics (N = 199)
| Characteristic | No. (%) |
|---|---|
| Age at diagnosis, y, median (range) | 6.4 (0.5–18.4) |
| Age at diagnosis, y, No. (%) | |
| <1.5 | 9 (5) |
| 1.5–4 | 72 (36) |
| 5–9 | 65 (33) |
| ≥10 | 53 (27) |
| Sex, No. (%) | |
| Male | 118 (59) |
| Female | 81 (41) |
| Race, No. (%) | |
| Black | 28 (14) |
| White | 158 (79) |
| Other | 13 (7) |
| Acute lymphoblastic leukemia risk level at induction | |
| Low | 106 (53) |
| Standard | 81 (41) |
| High | 12 (6) |
| Stool samples, No. (%) | 406 |
| Baseline before induction | 112 (28) |
| After induction completion | 97 (24) |
| Postconsolidation at reinduction I initiation | 107 (26) |
| Postreinduction at reinduction II initiation | 90 (22) |
| Patients with infection outcomes, No. (%) | |
| Bloodstream infectionsa | 26 (13) |
| Febrile neutropenia | 122 (61) |
| Diarrheal illnessb | 73 (37) |
| No infection outcomec | 42 (21) |
aEtiologic agents of bloodstream infections included coagulase-negative staphylococci (7 episodes), Escherichia coli (3), Enterococcus species (3, all susceptible to vancomycin), Staphylococcus aureus (3), Streptococcus pneumoniae (3), Rothia mucilaginosa (2), Neisseria polysaccharea (2), Stenotrophomonas maltophilia (1), Enterobacter cloacae (1), Micrococcus species (1), Bacillus cereus (1), Pseudomonas aeruginosa (1), Streptococcus salivarius (1), Capnocytophaga sputigena (1), and Burkholderia cepacia (1).
bEtiologies included Clostridium difficile (45 episodes), rotavirus (5), norovirus (5), adenovirus (4), and Cryptosporidium (2).
cNo fever, bacteremia, febrile neutropenia, or diarrheal illness.
Changes in Gut Microbiota Diversity
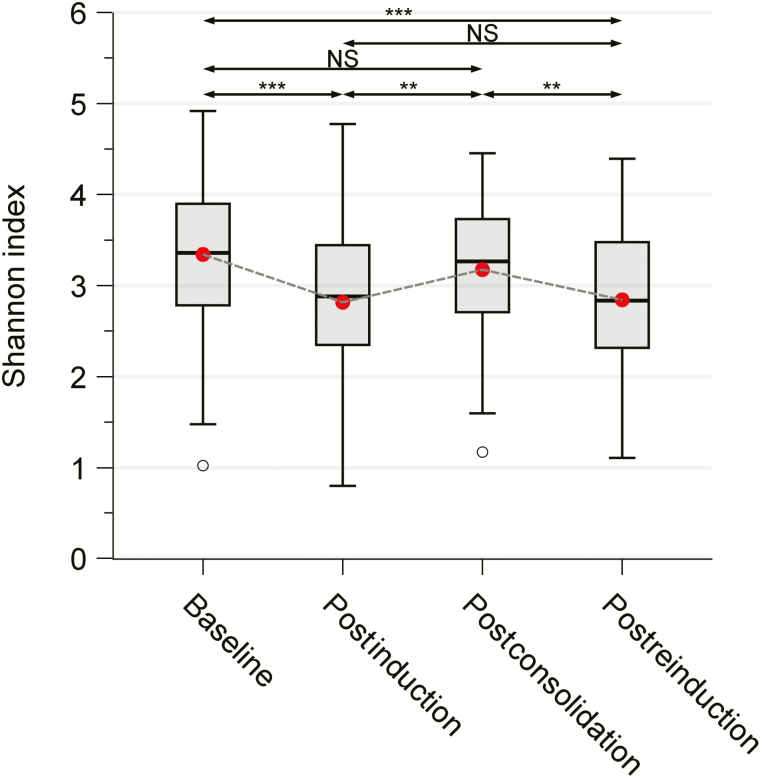
Microbiome diversity significantly decreased from diagnosis to the end of induction (3.34 [standard deviation {SD}, 0.72] vs 2.82 [SD, 0.88]; P < .001; Figure 1 and Supplementary Table 2). However, after less-intensive consolidation and continuation treatment, diversity reverted to the baseline level (3.18 [SD, 0.69] postconsolidation vs 2.82 [SD, 0.88] postinduction; P < .01). Gut microbiota diversity again decreased significantly after reinduction I (2.84 [SD, 0.76] postreinduction vs 3.18 [SD, 0.69] postconsolidation; P < .01). Although there was no significant difference between the diversity indices at baseline and postconsolidation or between those at postinduction and postreinduction, the index at postreinduction was significantly lower than that at baseline (P < .001; Figure 1), suggesting that induction and reinduction chemotherapy reduced the gut microbial diversity. Similar results were shown across all alpha diversity indices (Supplementary Figure 3).
Figure 1.
Change in gut microbiota diversity in 199 children during chemotherapy for acute lymphoblastic leukemia. Fecal samples were obtained at 4 time points (baseline, n = 112; postinduction, n = 97; postconsolidation, n = 107; and postreinduction, n = 90). A mixed-effects linear model was used to examine the change in Shannon indices over time. Patients were included as a random effect. **P < .01; ***P < .001. Abbreviation: NS, not significant.
Changes in Gut Microbiota Composition
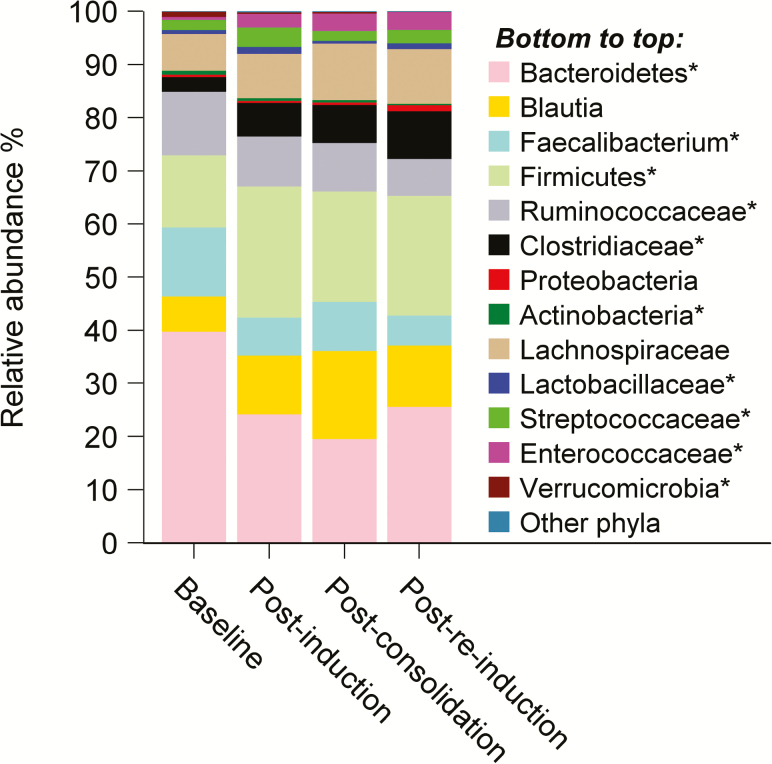
Compared to baseline, the mean log RA of Bacteroidetes, Faecalibacterium species, Ruminococcaceae, Actinobacteria, and Verrucomicrobia significantly decreased in at least one of the postchemotherapy samples (Figure 2 and Supplementary Tables 2 and 3). In contrast, the mean log RA of Clostridiaceae, Streptococcaceae, Lactobacillaceae, Enterococcaceae, and other Firmicutes significantly increased after chemotherapy (Figure 2 and Supplementary Tables 2 and 3). Although mean diversity of the postconsolidation samples did not differ significantly from that of the baseline samples nor did the diversity of the postinduction samples compared to postreinduction samples, the gut microbiome composition changed significantly. The diversity at postconsolidation reverted back to baseline level, but the composition was different.
Figure 2.
Change in gut microbiota composition during chemotherapy at the same time points as in Figure 1. *Significant change.
Effect of Antibiotics and Probiotics on Microbiome
Impact of antibiotics on microbiome changes was examined for patients who had stool specimens collected before and after each phase of chemotherapy. Antibiotic exposure was not significantly associated with changes in diversity for any chemotherapy phase (Supplementary Table 4 and Supplementary Figure 4). Similarly, no significant association was found between cumulative antibiotic exposure and change in dominance of Bacteroidetes (Supplementary Table 5). Due to the small number (n = 10) of patients who received probiotics at any time, the findings regarding microbiome changes are unlikely to be affected by probiotics (Supplementary Figure 5).
Using the Baseline Microbiome to Predict Any Infection Throughout Therapy
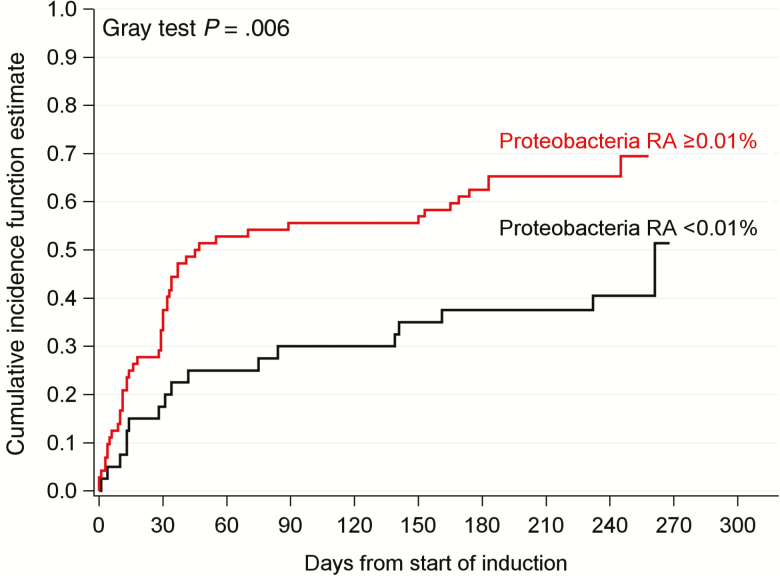
The relationship between baseline microbiome at the time of ALL diagnosis and subsequent development of infection during chemotherapy was characterized. Of the 112 patients with a baseline sample, 65 developed at least one FN episode. There was no significant difference between diversity of baseline samples from patients who did or did not develop FN (median, 3.38 [interquartile range {IQR}, 2.83–4.04] vs 3.34 [IQR, 2.72–3.87]; Supplementary Table 6). However, Proteobacteria were significantly more abundant at baseline in patients who subsequently developed FN (median, 0.03 [IQR, 0.01–0.14] vs 0.01 [IQR, 0–0.06]; P = 0.027; Supplementary Table 5). Using an optimal RA cutoff of 0.01%, Proteobacteria RA in baseline microbiome was the only independent predictor of subsequent FN (Supplementary Table 7). After adjusting for sex and the competing risk of death and ALL relapse, the cumulative incidence of first FN episodes in patients with Proteobacteria RA ≥0.01% was significantly higher than in patients with RA <0.01% (hazard ratio [HR], 2.12 [95% confidence interval {CI}, 1.22–3.69]; P = .008; Figure 3 and Supplementary Table 7). In summary, the composition, rather than diversity, of baseline gut microbiome was an independent predictor of FN during chemotherapy. Patients presenting at ALL diagnosis with Proteobacteria RA ≥0.01% had a 67% chance of developing FN. The differences found in similar analyses of bloodstream infections and diarrheal illness were not significant (data not shown).
Figure 3.
The relative abundance (RA) of Proteobacteria in the baseline gut microbiota at acute lymphoblastic leukemia diagnosis is associated with increased cumulative incidence of first episodes of febrile neutropenia. The estimated hazard ratio was 2.12 (95% confidence interval, 1.22–3.69; P = .008) for a Proteobacteria RA of ≥0.01 vs an RA of <0.01%, adjusting for gender.
Infections During a Course of Chemotherapy Associated With Gut Microbiome at the Start of That Course
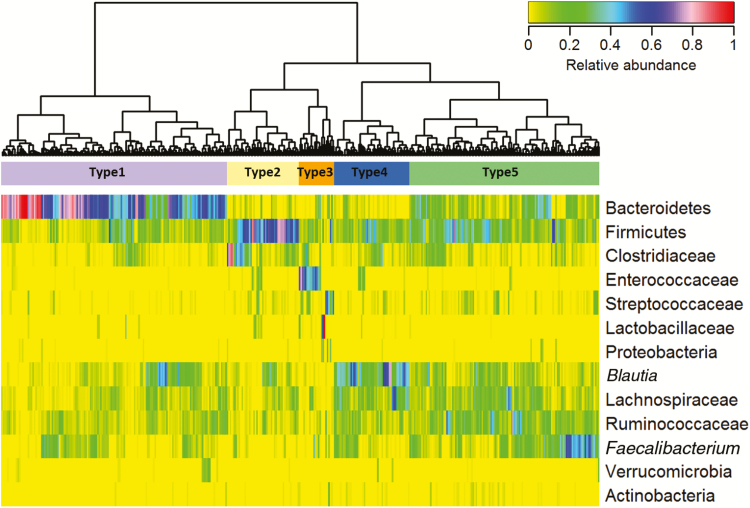
Using all stool samples collected at all 4 time points to generate a heat map and hierarchical clustering, 5 meaningful clusters of composition type were identified: Bacteroidetes-dominant (type 1); dominance of other Firmicutes or Clostridiaceae (type 2); dominance of Enterococcaceae, Streptococcaceae, Lactobacillaceae, or Proteobacteria (type 3); Blautia species dominant (type 4); and even distribution of taxa (type 5) (Figure 4). The distribution of the 4 stool-sample time points and the infection outcomes in each cluster are shown in Supplementary Tables 7 and 8. Forty percent of baseline samples were dominated by type 5 cluster and 49% by Bacteroidetes (type 1 cluster). This dominance by Bacteroidetes has previously been shown in the gut microbiome of healthy children and adults [26, 27].
Figure 4.
Heat map of taxa relative abundance in microbiota of all 406 fecal samples collected at any of the 4 time points. Five clusters (types 1–5) were identified using unsupervised hierarchical clustering of the Jensen-Shannon divergence matrix and ward linkage. Type 1 is dominated by Bacteroidetes; type 2 by other Firmicutes or Clostridiaceae; type 3 by Enterococcaceae, Streptococcaceae, Lactobacillaceae, or Proteobacteria; and type 4 by Blautia species. Type 5 has evenly distributed relative abundances of taxa.
These 5 microbiota composition types at any of the 4 time points, along with diversity, chemotherapy phase, ALL risk level at the time of sample collection, age at ALL diagnosis, sex, and race, were tested to identify risk factors for infection outcomes during chemotherapy phase subsequent to the stool collection time point. Supplementary Table 10 shows that ALL risk level, chemotherapy phase, and time-dependent microbiome composition type were all associated with infection. FN episodes and diarrheal illness were more likely to occur during induction. Similarly, high-risk ALL was significantly associated with an increased risk for diarrheal illness, as compared to standard-risk (HR, 2.16 [95% CI, 1.01–4.59]; P = .046) and low-risk ALL (HR, 2.74 [95% CI, 1.26–5.95]; P = .011). We found no significant association between ALL risk level and FN. Microbiome diversity, age, sex, and race were not associated with FN or diarrheal illness.
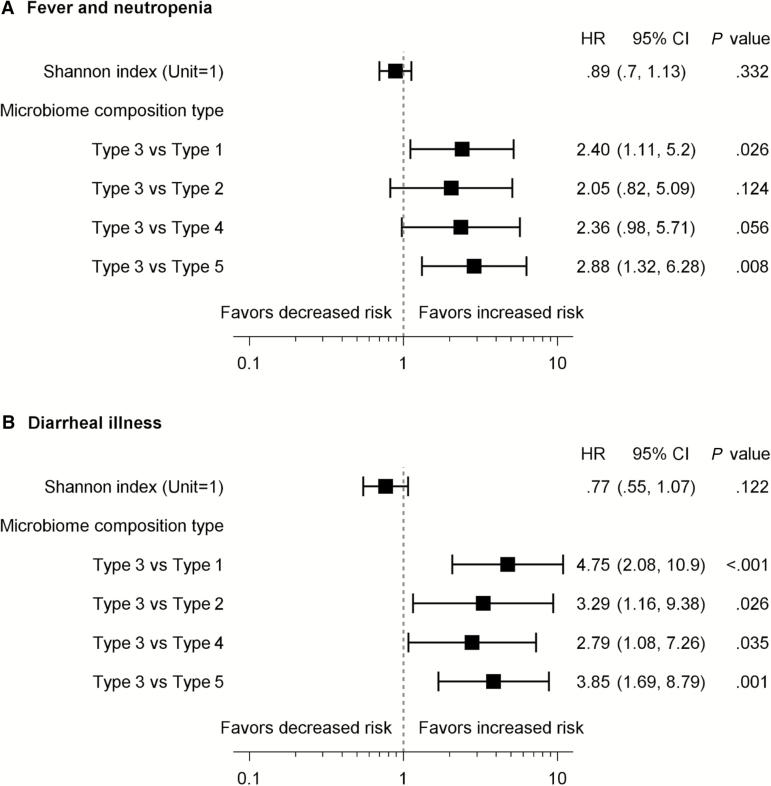
Adjusting for chemotherapy phase and ALL risk level, a type 3 gut microbiome composition at any time point was a significant risk factor for FN, diarrheal illness, and any infection outcome (Figure 5). Compared with type 1 and type 5, type 3 composition conferred a higher risk for FN in subsequent chemotherapy phase (HR, 2.40 [95% CI, 1.11–5.20] and 2.88 [95% CI, 1.32–6.28], respectively; Figure 5A). Similarly, type 3 composition was significantly associated with higher risk for diarrheal illness compared with type 1, type 2, type 4, and type 5 (Figure 5B). Diversity was not significantly associated with infection outcomes. We also found no significant association with the time-independent factors, age, sex, and race (data not shown).
Figure 5.
Forest plot of hazard ratios (HRs) with 95% confidence intervals (CIs) for febrile neutropenia (A) and diarrheal illness (B) associated with gut microbiome composition and diversity at any of the 4 time points. The Anderson-Gill model was used to compare the risk of each outcome among the microbiome Shannon indices and composition types over time. The reported HRs were adjusted for subsequent chemotherapy phase and acute lymphoblastic leukemia risk level, and the 95% CI and P values were estimated using the robust sandwich estimator.
Characterization of Type 3 Microbiome Composition
Most (79%) of the fecal samples in the type 3 composition cluster had dominance of Enterococcaceae or Streptococcaceae. Of 14 patients with dominant Enterococcaceae, 7 (50%) developed subsequent infection, as did 4 of 5 (80%) patients with dominant Streptococcaceae (Supplementary Table 11). Adjusting for chemotherapy phase and ALL risk level, Enterococcaceae dominance was significantly associated with increased risk of subsequent FN (HR, 2.97 [95% CI, 1.35–6.53]) and diarrheal illness (HR, 4.23 [95% CI, 1.77–10.1]) compared to nondominance of both Streptococcaceae and Enterococcaceae, while Streptococcaceae dominance carried a greater risk (HR, 7.94 [95% CI, 3.27–19.3]) of subsequent diarrheal illness (Table 2). Thus, a gut microbiota dominated by Enterococcaceae or Streptococcaceae is associated with subsequent infections.
Table 2.
Domination of Gut Microbiome at Any Time Point by Streptococcaceae or Enterococcaceae Was Associated With Diarrheal Illness and Febrile Neutropenia in the Subsequent Phase of Chemotherapy
| Dominance of Microbiome | Associated With FN | Associated With Diarrheal Illness | ||||
|---|---|---|---|---|---|---|
| HR | (95% CI) | P Value | HR | (95% CI) | P Value | |
| Streptococcaceae-dominanta vs Enterococcaceae-dominantb | 0.63 | (.1–4.11) | .628 | 1.88 | (.59–5.96) | .285 |
| Streptococcaceae-dominant vs othersc | 1.87 | (.34–10.4) | .475 | 7.94 | (3.27–19.3) | <.001 |
| Enterococcaceae-dominant vs others | 2.97 | (1.35–6.53) | .007 | 4.23 | (1.77–10.1) | .001 |
Anderson-Gill model was used to compare the risk of each infection outcome in different groups over time. The reported HRs were adjusted for the phase of chemotherapy and acute lymphoblastic leukemia risk level at the time of stool sample collection as appropriate.
Abbreviations: CI, confidence interval; FN, febrile neutropenia; HR, hazard ratio.
aStreptococcaceae-dominant: ≥30% relative abundance (RA) of Streptococcaceae.
bEnterococcaceae-dominant: ≥30% RA of Enterococcaceae.
cOthers: RA <30% for both Streptococcaceae and Enterococcaceae.
Validation of Enterococcaceae or Streptococcaceae Dominance as Infection Predictor
A bootstrap method confirmed that patients with a gut microbiome dominated by Enterococcaceae or Streptococcaceae had a significantly higher risk of diarrheal illness and any infection outcome (Supplementary Figure 6A and 6B). Patients with Enterococcaceae or Streptococcaceae dominance also had a higher but nonsignificant risk of FN (Supplementary Figure 6C). In summary, the performance of Enterococcaceae or Streptococcaceae dominance as a predictor was validated for diarrheal illness and any infection outcome, and showed a strong trend toward predicting FN.
DISCUSSION
Our study is the first to evaluate changes in the diversity and composition of gut microbiome and its role in predicting infections in a large cohort of children with ALL.
Diversity decreased significantly after intensive induction and reinduction chemotherapy, despite a rebound after recovery from induction. The decreased microbial diversity after immunosuppressive and myelosuppressive treatment is consistent with studies in adult HSCT recipients and acute myeloblastic leukemia or non-Hodgkin lymphoma [5, 9, 28, 29]; however, the only other pediatric study to date [11] detected increases in gut microbial diversity during chemotherapy. Although both studies sequenced the V1–V3 regions of the 16S rRNA gene, their divergent findings might be explained by differences in the chemotherapy regimens, antibiotic use, methods of sample preparation and sequencing, data analysis methods, or time points for stool collection.
Although the empirical diversity index recovered to levels similar to those of baseline, this renewed diversity was characterized by a different microbiotal composition. In contrast to the studies in adults, we found that microbiota composition, but not diversity, was independently predictive of infections during chemotherapy. As previously reported [26], the baseline microbiome was dominated by Bacteroidetes; which decreased after chemotherapy. A reduction in Bacteroidetes has been linked to human disease [30]. The metabolic function of some of these bacterial taxa was explored in pathogenesis studies [28, 31]. For example, Ruminococcaceae and Faecalibacterium species have been implicated in anti-inflammatory effects and may maintain intestinal epithelial integrity [31]. The loss of the protective effect of these taxa after chemotherapy might compromise gut integrity and increase the risk of bacterial translocation into the bloodstream.
On the other hand, the RA of bacterial taxa that commonly infect immunocompromised patients, such as Streptococcaceae and Enterococcaceae, increased significantly after chemotherapy. This is consistent with a study evaluating 94 adult HSCT recipients whose gut microbiota became dominated by Enterococcus species, Streptococcus species, and Proteobacteria [9]. Studies have demonstrated that gut microbial communities recover after resolution of perturbing factors [10, 32, 33]. We saw no such recovery of gut microbiota composition at the 3 time points of stool sampling during chemotherapy. We found no association between antibiotic exposure and microbiome changes. As shown in Supplementary Figure 4, all patients received antibiotics during induction chemotherapy, and the majority did during consolidation and/or reinduction. It is not clear whether the change in microbiome is due to antibiotics or the chemotherapy itself. We believe that the confounding between antibiotic administration and the chemotherapy makes it difficult to isolate an effect of antibiotics alone on the microbiota. It would be interesting to examine these patients’ microbiomes after completion of chemotherapy and resolution of their immunosuppressed state.
We have shown that the composition, but not diversity, of gut microbiota is a significant predictor of infections. Presentation with a gut characterized by Proteobacteria at ALL diagnosis significantly predicted FN during chemotherapy. The Proteobacteria phylum of gram-negative bacteria includes Enterobacteriaceae, Pseudomonas species, and other bacteria commonly causing infections in immunocompromised patients. The importance of Proteobacteria as markers of imbalanced gut microbiota and increased risk for disease has been previously documented in immunocompetent and immunocompromised patients [9, 10, 34].
In addition to the baseline gut microbiome, characterization of the microbiome composition before each phase of chemotherapy independently predicted infections during the upcoming phase. Adjusting for chemotherapy phase and ALL risk level, domination of the gut by Enterococcaceae or Streptococcaceae predicted subsequent FN or diarrheal illness. This is consistent with adult studies showing that an increased abundance of certain taxa (Proteobacteria, Enterococcus species, and Streptococcus species) was associated with subsequent bloodstream infections and mortality [9, 35].
Our study included the largest pediatric ALL cohort with gut microbiome evaluation, and it represents the most comprehensive analysis of the microbiome’s relation to infections. However, it did not adjust for additional factors that might contribute to changes in gut microbiota, including diet, ethnicity, body mass index, residence geographic location, and pet exposure. Also, not all patients provided stool samples at all 4 time points.
In conclusion, we have shown that microbiome composition can be used as an infection risk stratification tool at the time of presentation with ALL diagnosis. If validated, evaluations of the gut microbiome at the onset of chemotherapy could be used to determine biomarkers of the clinical course during chemotherapy. Our findings emphasize the need for enhanced understanding of microbiome in children with cancer during chemotherapy to design targeted microbiome-based diagnostic and biomarker tools. Therefore, once a patient is identified at risk for infections, customized treatment regimens could be designed to mitigate this risk by selectively manipulating the gut microbiota composition using customized probiotics, fecal microbiota transplant, modified diet, or other innovative approaches. However, more studies are needed. Our findings can guide the design of future studies to evaluate the performance of microbiota biomarkers and the proposed treatment regimens that modify gut dysbiosis.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. H. H. designed the study, performed sample and clinical data collection, interpreted the analysis, and wrote the manuscript. R. D. and V. D. contributed to the clinical data abstraction and the sample collection and tracking. S. J. and C.-H. P. contributed to patient recruitment and reviewed the findings and the manuscript. J. W. R., S. S.-C., C. J., and E. A. K. designed and validated the 16S sample-analysis pipeline. T. C. completed the computational analysis. L. T., Y. S., and S. P. designed and performed the statistical analysis. J. W., S. S.-C., R. T. H., E. T., and J. W. R. assisted with results interpretation and with writing and reviewing the manuscript.
Acknowledgments. We thank Melissa Shenep for help in data abstraction; Keith A. Laycock, PhD, ELS, for scientific editing of the manuscript; and the St Jude Hartwell Center for assistance with sequencing.
Financial support. This work was supported by the Children’s Infection Defense Center at St Jude Children’s Research Hospital; American Lebanese Syrian Associated Charities; and the National Institutes of Health (grant number CA21765).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 15th Annual St Jude/Pediatric Infectious Diseases Society (PIDS) Research Meeting, Memphis, Tennessee, March 2016; 16th Annual St Jude/PIDS Research Conference, Memphis, Tennessee, March 2017; and 2017 Pediatric Academic Societies Conference, San Francisco, California, May 2017.
References
- 1. Martin R, Nauta AJ, Ben Amor K, Knippels LM, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes 2010; 1:367–82. [DOI] [PubMed] [Google Scholar]
- 2. Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer 2016; 114:237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012; 486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buffie CG, Bucci V, Stein RR et al. . Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galloway-Peña JR, Smith DP, Sahasrabhojane P et al. . The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer 2016; 122:2186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montassier E, Al-Ghalith GA, Ward T et al. . Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med 2016; 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taur Y, Pamer EG. Microbiome mediation of infections in the cancer setting. Genome Med 2016; 8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taur Y, Xavier JB, Lipuma L et al. . Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singh P, Teal TK, Marsh TL et al. . Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 2015; 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rajagopala SV, Yooseph S, Harkins DM et al. . Gastrointestinal microbial populations can distinguish pediatric and adolescent acute lymphoblastic leukemia (ALL) at the time of disease diagnosis. BMC Genomics 2016; 17:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. ClinicalTrials.gov. Total therapy study XVI for newly diagnosed patients with acute lymphoblastic leukemia 2015. Available at: https://clinicaltrials.gov/ct2/show/NCT00549848. Accessed 24 July 2017.
- 13. Wolf J, Tang L, Flynn PM et al. . Levofloxacin prophylaxis during induction therapy for pediatric acute lymphoblastic leukemia. Clin Infect Dis 2017; 65:1790–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krueger F. “Trim Galore.” A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files 2015. Available at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 24 July 2017.
- 15. Andrews S. FastQC: a quality control tool for high throughput sequence data 2010. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 24 July 2017.
- 16. Caporaso JG, Kuczynski J, Stombaugh J et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 2012; 13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460–1. [DOI] [PubMed] [Google Scholar]
- 19. DeSantis TZ, Hugenholtz P, Larsen N et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romero R, Hassan SS, Gajer P et al. . The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995; 57:289–300. [Google Scholar]
- 23. Kaufman L, Rousseeuw PJ.. Finding groups in data: an introduction to cluster analysis. New York: Wiley, 1990. [Google Scholar]
- 24. Amorim LD, Cai J. Modelling recurrent events: a tutorial for analysis in epidemiology. Int J Epidemiol 2015; 44:324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersen PK, Gill RD. Cox’s regression model for counting processes: a large sample study. Ann Stat 1982; 10:1100–20. [Google Scholar]
- 26. Hollister EB, Riehle K, Luna RA et al. . Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 2015; 3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Human Microbiome Project. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Montassier E, Gastinne T, Vangay P et al. . Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther 2015; 42:515–28. [DOI] [PubMed] [Google Scholar]
- 29. Taur Y, Jenq RR, Ubeda C, van den Brink M, Pamer EG. Role of intestinal microbiota in transplantation outcomes. Best Pract Res Clin Haematol 2015; 28:155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gevers D, Kugathasan S, Denson LA et al. . The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014; 15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Vliet MJ, Harmsen HJ, de Bont ES, Tissing WJ. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog 2010; 6:e1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 2009; 77:2367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol 2004; 42:1203–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 2015; 33:496–503. [DOI] [PubMed] [Google Scholar]
- 35. Taur Y, Jenq RR, Perales MA et al. . The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.