Abstract
Background and Aims
Brassinosteroids (BRs) are plant hormones involved in many developmental processes as well as in plant–environment interactions. Their role was investigated in this study through the analysis of lilliputian1-1 (lil1-1), a dwarf mutant impaired in BR biosynthesis in maize (Zea mays).
Methods
We isolated lil1-1 through transposon tagging in maize. The action of lil1 was investigated through morphological and genetic analysis. Moreover, by comparing lil1-1 mutant and wild-type individuals grown under drought stress, the effect of BR reduction on the response to drought stress was examined.
Key Results
lil1-1 is a novel allele of the brassinosteroid-deficient dwarf1 (brd1) gene, encoding a brassinosteroid C-6 oxidase. We show in this study that lil1 is epistatic to nana plant1 (na1), a BR gene involved in earlier steps of the pathway. The lill-1 mutation causes alteration in the root gravitropic response, leaf epidermal cell density, epicuticular wax deposition and seedling adaptation to water scarcity conditions.
Conclusions
Lack of active BR molecules in maize causes a pleiotropic effect on plant development and improves seedling tolerance of drought. BR-deficient maize mutants can thus be instrumental in unravelling novel mechanisms on which plant adaptations to abiotic stress are based.
Keywords: Brassinosteroids, brassinosteroid-deficient dwarf1 (brd1), cell density, cuticular waxes, CYP85A, drought stress, dwarf mutant, gravitropic response, lilliputian1 (lil1), nana plant1 (na1), P450 C-6 oxidase, Zea mays
INTRODUCTION
Brassinosteroids (BRs) are a class of steroid hormones essential for plant growth and development (Clouse, 2011). They control cell elongation, division and differentiation and are also involved in the control of many developmental traits of agronomic importance, such as seed germination, leaf curvature, flowering time and seed yield (Divi and Krishna, 2009; Vriet et al., 2012).
Several BR biosynthetic steps have been unravelled through the study of mutants, the main features of which are reduced stature and the presence of round and dark green leaves. Castasterone (CS) and brassinolide (BL), the two active BR molecules, are produced in the final steps of the BR pathway by members of the CYP85A family of cytochrome P450 monooxygenases. The isolation of their corresponding genes was first achieved in tomato (Lycopersicon esculentum) through the molecular characterization of two extreme dwarf mutants. The tomato Dwarf gene, encoding LsCYP85A1, which catalyses the C-6 oxidation of 6-deoxo castasterone (6-deoxoCS) to CS, is expressed in the vegetative part of the plant (Bishop et al., 1996, 1999). LsCYP85A3, involved in the conversion of CS to BL, is preferentially expressed in fruits (Nomura et al., 2005). Analogously, two AtCYP85A genes involved in the final reactions of BR biosynthesis were detected in Arabidopsis thaliana. AtCYP85A2 is specifically involved in the CS-to-BL conversion (Nomura et al., 2005) while AtCYP85A1 catalyses the C-6 oxidation reactions of multiple substrates (Shimada et al., 2001). Mutations in the AtCYP85A2 gene result in severe dwarfism, whereas Atcyp85a1 mutations do not affect plant development, but cause failure of female gametogenesis (Pérez-España et al., 2011). Unlike what has been reported in dicot species, studies in monocots reported a single CYP85A gene. The OsCYP85A1/OsBRD1 and Zmbrd1 genes, both encoding a BR C6-oxidase, were isolated in rice (Oryza sativa) (Kim et al., 2008) and maize (Zea mays) (Makarevitch et al., 2012), respectively.
In maize, two additional genes involved in BR biosynthesis, i.e. nana plant1 (na1) and nana plant2 (na2), have been isolated. The na1 gene, encoding a 5α-reductase enzyme (Hartwig et al., 2011), is the homologue of the A. thaliana gene DEETIOLATED2 (DET2) (Li et al., 1996) , while na2, encoding a D24-sterol reductase (Best et al., 2016), corresponds to DWARF1 in A. thaliana (Choe et al., 1999). Moreover, the maize BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) has been characterized by using a transgenic RNA interference (RNAi) approach that knocked down the expression of all five maize BRI1 homologous genes (Kir et al., 2015). The resulting mutant phenotype includes dwarf stature, due to shortened internodes, dark green, upright, and twisted leaves with decreased auricle formation, and feminized male flowers.
So far, mutant studies conducted in maize have focused on the role of BRs in developmental processes. However, there is growing evidence that mutants with deficiencies either in the synthesis or the perception of BR are instrumental for unravelling the role of these hormones in the interactions between plant and environment (Feng et al., 2015; Gruszka et al., 2016). Data obtained in this context will have important implications in crop breeding since genetic manipulation of the BR level could improve plant tolerance to environmental stresses.
Here we report the characterization of lil1-1, a novel allele at the maize brd1 locus. A mutant allele at this locus, referred to as brd1-m1, was previously isolated in a population mutagenized with ethyl methanesulfonate (EMS). Molecular characterization of brd1-m1 revealed the presence of single base-pair substitution resulting in the creation of a premature stop codon and thus generating a truncated protein (Makarevitch et al., 2012).
We detected some peculiar traits in the lil1-1 mutant, such as altered root gravitropic response and epicuticular wax deposition. We also analysed the genetic interaction between lil1-1 and the na1-1 mutant, which is impaired in earlier steps of biosynthesis and exhibits a less severe phenotype. Moreover, by comparing lil1-1 mutant and wild-type individuals, we examined the effect of BR reduction on the response to drought stress during early phases of plant development.
MATERIALS AND METHODS
Plant materials and growth conditions
The maize (Zea mays) lil1-1 mutant was originally isolated from the selfed progeny of a Mutator stock outcrossed to an unrelated stock (Dolfini et al., 1999). The lil1-1 mutant allele was introgressed into B73, A188, H99 and Rsc-m2 inbred lines. The brd1-m1 (Makarevitch et al., 2012) and na1-1 (Hartwig et al., 2011) mutant lines were obtained from the Maize Genetics Cooperation Stock Center. Due to impaired inflorescence development, mutant plants were maintained as heterozygotes. In all the experiments performed, homozygous mutants and their wild-type control plants were from the same F2 segregating progeny.
Plants were grown in a growth chamber with controlled temperature (25 °C night, 30 °C day) and under a long-day photoperiod (16 h of light/8 h of dark) with photon fluence of 70 µmol m−2 s−1. For phenotypic analysis at the seedling level, seeds were germinated and grown for 10 days on wet filter paper; for analysis at later developmental stages, plants were grown on soil.
Cloning of lil1 and transcript analysis
Co-segregation analysis was performed on DNA samples extracted from single-mutant (lil1-1/lil1-1) and wild-type plants, whose homozygous (+/+) or heterozygous (+/lil-1) constitution was ascertained through self-pollination. Genomic DNA extraction and Southern analysis were performed as previously described (Gutiérrez-Marcos et al., 2007). Hybridization probes were as follows: the Mu1-specific probe corresponded to the AvaI/BstNI 650-bp central fragment of the Mu1 element; the lil1-specific probe was the EcoRV/SalI 443-bp fragment obtained from the genomic region flanking the Mu1 insertion. The genomic fragment co-segregating with the lil1 mutant phenotype was cloned by the screening of a size-fractionated EcoRI library of genomic DNA with the lil1 probe. For PCR-based co-segregation analysis, genomic DNA was extracted as previously described (Arthur et al., 2003).
For reverse-transcription (RT) PCR, total RNA was extracted with TRIzol reagent following the manufacturer’s instructions (Thermo Fisher Scientific). DNase treatment was performed with the reagent RQ1 RNase-Free DNase (Promega) and first-strand cDNA was synthesized from 750 ng of RNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific).
Genomic DNA and cDNA were subjected to amplification with GoTaq® Flexi DNA Polymerase (Promega), and PCRs were conducted with different sets of primers (Supplementary Data Table S6).
Allelism test
Pollen of a given +/brd1-m1 plant, whose heterozygous condition was ascertained by selfing, was applied to the silks of +/lil1-1 plants, whose heterozygous condition was determined by PCR analysis. F1 seeds were germinated to score the seedling phenotypes. About one-quarter of seedlings are expected to be mutant if there is lack of complementation (allelism). Statistical analysis was conducted using the χ2 test to confirm the observed segregation.
Microscopy analyses
To obtain histological images of the shoot apical meristem (SAM), 2-week-old homozygous lil1-1 and wild-type plants were sampled from the same segregating progeny grown under standard conditions in the growth chamber. Shoot tissues were immediately fixed in 3 % glutaraldehyde in phosphate-buffered saline (PBS) (130 mm NaCl, 7 mm Na2HPO4, 3 mm NaH2PO4) for 24 h. The fixed material was placed in 70 % ethanol and embedded in LR White resin. Semi-thin sections (2 μm) were applied to polylysine-coated slides, stained with Toluidine Blue O [1 % toluidine blue 1 % sodium tetraborate (1:1, v/v)] and imaged under a light microscope (Ortholux, Leitz, Germany). Leaf-blade samples were fixed with 3 % glutaraldehyde in PBS and then embedded in 5 % agar. Sections of 30 µm were cut with a microtome.
For the analysis of epicuticular waxes, leaf pieces of wild-type and lil1-1 mutant plants were dried and processed according to La Rocca et al. (2015). The specimen surfaces were examined with a SEMLEO 1430 (Zeiss) microscope.
Phenotypic analysis
Mesocotyl, coleoptile, primary root length and seedling height, measured from the scutellar node to the tip of the last emerging leaf, were determined on a sample of 20 plants per genotype. Data are reported as the mean ± standard error. Pairwise comparisons between means of the two genotypes were performed using a two-tailed Student’s t-test.
Plant height analysis in the progeny segregating for lil1-1 and na1-1 alleles was performed for a sample of 102 wild-type, 31 homozygous na1-1, 26 homozygous lil1-1 and 12 double-homozygous na1-1 lil1-1 seedlings. A one-way ANOVA test was performed with the statistical package SPSS 21.0.
Cell density and stomatalindex analysis
To measure stomatal density and stomatal index, a leaf surface imprint method was used. We analysed the fourth and sixth fully expanded leaves of ~50-day old mutant and wild-type plants. Briefly, a thin film of vinyl glue (SuperVinil) was applied with a small paintbrush on both the adaxial and the abaxial surface of the median portion of the leaf still attached to the plant, in order to prevent wilting. One imprint per leaf was taken from a minimum of eight independent plants for wild-type, na1-1 or lil1-1 and four independent plants for lil1-1 na1-1 genotypes. The imprints were then put on glass microscope slides and observed under a light microscope (DM RA2 optical microscope, Leica, Wetzlar, Germany) at 10× magnification. Two viewable leaf areas (0.807 mm2) between vascular tissues were randomly selected per imprint and photographed. The total numbers of stomata and pavement cells were determined by counting all the cells in each area and mean parameters were calculated. The stomatal index was determined as [number of stomata/(number of epidermal cells + number of stomata)] × 100. A one-way ANOVA test was performed with the statistical package SPSS 21.0.
Chlorophyll leaching analysis
Completely expanded sixth leaves were taken from five independent plants per genotype and dissected into pieces of 8-cm length measured from the apex. Leaf sectors were weighed, immersed in 80 % ethanol and incubated at room temperature. Every vial was carefully covered to protect the samples from light. A series of 1-mL aliquots were taken at 12, 24, 36, 48, 60 and 72 h. Absorbance was measured at 647 and 664 nm with a spectrophotometer (Agilent Technologies Cary 60 UV-Vis) to quantify the chlorophyll released in the solution. The micromolar concentration of total chlorophyll per gram of fresh weight (FW) of tissue was calculated using the equation described by Lolle et al. (1997): total micromoles of chlorophyll = 7.93 (A664) + 19.53 (A647).
Plant growth conditions in drought stress and recovery experiments
For the drought stress experiment, seedlings at the coleoptile developmental stage were transferred to pots (8 × 8 cm) containing SER V8-14L substrate (Vigorplant, Piacenza, Italy). Each pot contained one seedling and the same amount of soil. To monitor the drought stress level, the relative soil water content (RSWC) was measured throughout the experiment. First, the maximum soil water content (RSWC = 100 %) was determined (Janeczko et at. 2016). The pots were watered until complete imbibition, left to lose the water excess and then weighed (wet weight). Subsequently, the soil was completely dried out at 60 °C for 5 days (dry weight). The 100 % RSWC was calculated as wet weight minus dry weight. All plants were grown under the well-watered condition (RSWC = 70 %) until the fourth leaf was emerging, and on the next day (Day 0) drought stress was initiated by withholding irrigation. The RSWC were measured at 0, 3, 6 and 9 days from the start of the drought treatment. At each time point there were 20 pots per genotype.
For the recovery assay, 9 days after the beginning of the drought treatment, 40 plants of each genotype were re-watered and after 3 and 7 days from re-watering were classified on a visual scale into three categories: 1 (severely wilted); 2 (slightly wilted); and 3 (recovered). The data collected for each category were expressed as percentage of the total number of tested plants.
Determination of leaf relative water content
To determine the leaf relative water content (RWC), discs 7 mm in diameter were taken from the widest portion of the fourth leaf at 0 and 9 days after the cessation of irrigation.
After fresh weight determination, the discs were floated in Petri dishes with distilled water for 24 h at room temperature. Following surface drying with absorbent paper towels and turgid weight determination, the discs were oven-dried at 37 °C for 4 days and reweighed to determine the dried weight. The leaf RWC was calculated as (fresh weight − dry weight)/(turgid weight − dry weight) × 100 (Pieczynski et al., 2013). Leaves were from ten individual plants per genotype and two discs were taken from each leaf. The average was calculated, and pairwise comparison between the two genotypes was performed using a two-tailed Student’s t-test.
Leaf gas exchange analysis
Measurements of gas exchange were made at 0, 3 and 4 days after the cessation of irrigation, with a portable open-system infra-red gas analyser (CIRAS-2, PP Systems, Hitchin, UK). Measurements were made on portions of leaf lamina of 1.7 cm2, with an air flow rate of 300 mL min−1, at a photosynthetic photon flux density (PPFD) of 1000 μmol m−2 s−1. The ambient temperature was 35–38 °C and the vapour pressure deficit (VPD) was similar during the whole experiment. Measurements were taken three times for each plant. Stomatal conductance (gs), transpiration rate (E), intercellular CO2 (Ci) and net photosynthesis rate (A) were directly retrieved from CIRAS-2 measurements. Water use efficiency (WUE) was calculated as the ratio between A and E. Physiological parameters were measured on well-developed fourth leaves in five replicates/genotype, where one replicate represented one leaf from an individual plant. Pairwise comparisons between means of different genotypes were performed using a two-tailed Student’s t-test.
RESULTS
The maize lil1-1 mutant is a novel allele of the brd1 gene
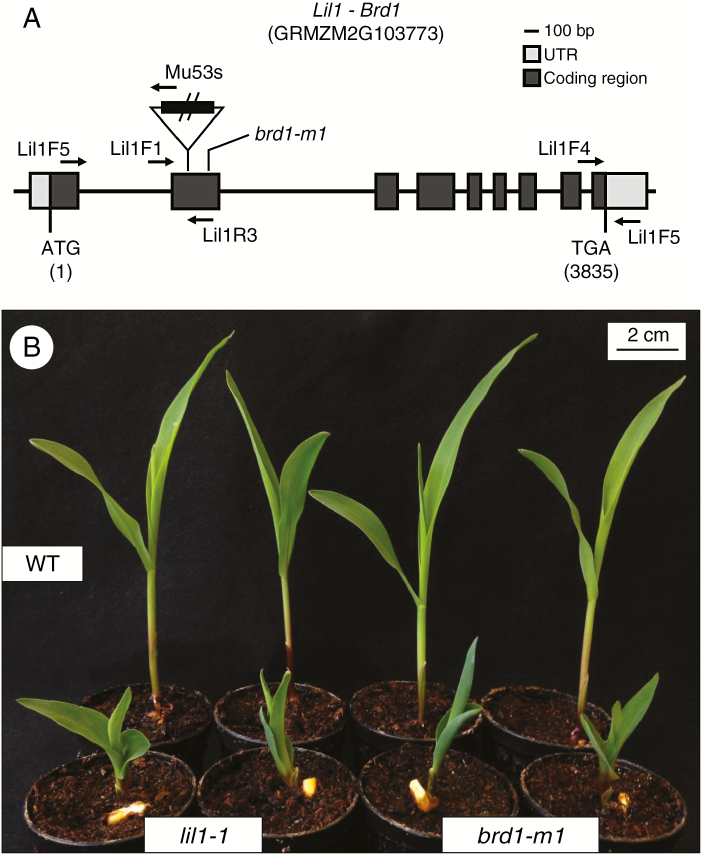
The lil1-1 recessive mutant was isolated from an active Mutator (Mu) stock (Dolfini et al., 1999). Genomic sequences associated with the lil1 mutant phenotype were identified through a Southern-based co-segregation analysis. In this analysis, genomic DNA was extracted from 40 wild-type (+/lil1-1 and +/+) and 20 mutant (lil1-1/lil1-1) single individuals and cut with the EcoRI restriction enzyme. Hybridization with a Mu1-specific probe revealed the presence of a polymorphic restriction fragment, of ~9.5 kb, co-segregating with the mutant allele (Supplementary Data Fig. S1A). Cloning and subsequent sequencing of the fragment confirmed the presence of a Mu1 element and disclosed the presence of a genomic region flanking the transposon insertion. To verify the identity of the cloned fragment, the same EcoRI-digested DNA blot was hybridized with a probe obtained from the flanking sequence. Individuals carrying the lil1-1 allele showed the same 9.5 Kb fragment (Supplementary Data Fig. S1B). A PCR-based co-segregation analysis was also performed and showed absence of recombination between the lil1 phenotype and the Mu1 insertion on a sample of a total of 100 F2 individuals, thus indicating a correspondence between this insertion and the lil1-1 mutant allele (Supplementary Data Fig. S1C). BLAST analysis of the cloned region flanking the Mu1 insertion showed complete homology with the brd1 gene (GRMZM2G103773) encoding a cytochrome P450 CYP85A1 C-6 oxidase (CYP85A1) (Makarevitch et al., 2012). In the lil1-1 allele the Mu1 element was inserted in the second exon of the brd1 gene (Fig. 1A). RT–PCR analysis was also performed to compare wild-type and mutant transcripts (Supplementary Data Fig. S1C). Results obtained with the Lil1F4-Lil1R5 primer set showed that the CYP85A1 transcript accumulates in wild-type as well as in lil1-1 and brd1-1 mutants. An amplification product of the expected size was specifically detected with the Lil1F5-Mu53 primer set in the lil1-1 mutant, while the Lil1F5-Lil1R3 set of primers spanning the Mu1 insertion gave an amplification product only in wild-type and brd1-1 mutant DNAs. This indicates that the lil1-1 mutant transcript retained the Mu1 element. This aberrant transcript is thus predicted to encode a non-functional protein.
Fig. 1.
The lil1-1 mutant is an allele of brd1. (A) Schematic representation of the maize cytochrome P450 CYP85A1 C-6 oxidase gene structure (Maize B73 RefGen_v3). The lil1-1 mutant allele carries a Mutator1 (Mu1) insertion, indicated as a triangle, in the second exon at 877 bp from the ATG start codon. Positions of the brd1-m1 mutation at 1145 bp (Makarevitch et al., 2012) and of primers (arrows) used in this work are also indicated. (B) Representative phenotypes of 10-day old wild-type, homozygous lil1-1 and homozygous brd1-m1 seedlings. Scale bar = 2 cm.
The lil1-1 allele was indistinguishable from the previously isolated brd1-m1 allele (Makarevitch et al., 2012), since both exhibited an obvious and similar dwarf seedling phenotype, which was clearly distinguishable from that of wild-type seedlings (Fig. 1B, Supplementary Data Fig. S2). To confirm that lil1-1 and brd1-m1 mutants were ascribable to the same gene an allelism test was done. Heterozygous +/lil1-1 female plants, the genotype of which had been determined by PCR analysis, were crossed to +/brd1-m1 plants, whose genotype was ascertained by self-pollination. The mutant segregation value observed in the progeny confirmed that the two mutants are allelic (Supplementary Data Table S1), thus providing a further proof of lil1 gene identity.
The lil1-1 mutation has a pleiotropic effect on plant growth
Involvement of BRs in seed production has been shown in studies conducted in arabidopsis and rice (Vriet et al., 2012). However, lack of lil1 activity does not appear to have an effect on the maize kernel. No visible defects were present at morphological level and no difference in weight was detected between lil1-1 mutant and wild-type seeds, whose genotypes were ascertained through germination (Supplementary Data Table S2). We also excluded the possibility that the lil1 constitution of the mother plant affects seed production. Seed weight and number were compared in F2 progeny ears obtained from selfing either F1 heterozygous or homozygous wild-type plants. In most of the lines, segregating and non-segregating ears did not differ in seed weight. Similarly, seed number per ear did not differ between segregating and non-segregating ears (Supplementary Data Table S3).
The effects of the lil1-1 mutation on plant growth were analysed in more detail. Segregating F2 progenies were obtained by introgressing the mutation in different genetic backgrounds. The lil1-1 mutant allele was introgressed three times in B73 and once in R-scm2, H99 and A188 genetic backgrounds. At 10 days after sowing, homozygous lil1-1 mutant seedlings showed the same phenotype in all backgrounds tested. Mutant mesocotyl elongation was blocked, and mutant coleoptile elongation was ~25 % of that of the wild type (Table 1).
Table 1.
Morphometric analysis of homozygous lil1-1 mutant and wild-type (WT) seedlings in four different genetic backgrounds. Standard error is reported (±). All measured differences between homozygous lil1-1 mutant and wild-type plants were statistically significant (P < 0.001, two-tailed Student’s t-test)
| Genetic background | Seedling height (cm) | Mesocotyl length (cm) | Coleoptile length (cm) | Root length (cm) | ||||
|---|---|---|---|---|---|---|---|---|
| lil1-1 | WT | lil1-1 | WT | lil1-1 | WT | lil1-1 | WT | |
| B73 | 3.70 ± 0.39 | 13.02 ± 0.73 | 0.04 ± 0.04 | 0.94 ± 0.05 | 0.66 ± 0.11 | 2.59 ± 0.08 | 11.52 ± 0.89 | 16.07 ± 1.12 |
| R-scm2 | 3.40 ± 0.29 | 11.84 ± 0.85 | 0.00 ± 0.00 | 0.99 ± 0.15 | 0.77 ± 0.12 | 2.83 ± 0.08 | 8.75 ± 0.80 | 13.72 ± 1.29 |
| H99 | 5.34 ± 0.39 | 18.93 ± 0.66 | 0.00 ± 0.00 | 0.59 ± 0.05 | 0.59 ± 0.05 | 2.57 ± 0.11 | 12.60 ± 1.50 | 23.80 ± 0.99 |
| A188 | 4.27 ± 0.33 | 15.20 ± 0.60 | 0.00 ± 0.00 | 0.44 ± 0.04 | 0.62 ± 0.06 | 2.08 ± 0.11 | 12.71 ± 0.97 | 22.14 ± 0.93 |
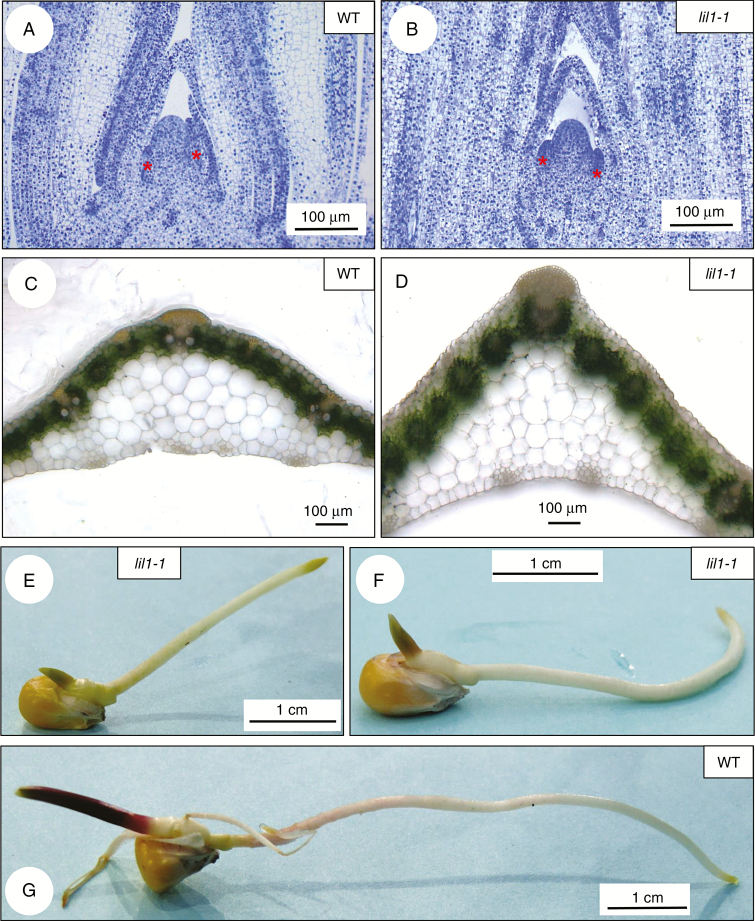
Reduced elongation of mutant compared with wild-type leaf primordia was also observed in histological analysis of shoot apices (Fig. 2A, B). Mutant leaves, at later developmental stages, exhibited an altered shape, visible in blade transverse sections. They also appeared thicker (Fig. 2D) than wild-type leaves (Fig. 2C) and presented supernumerary cell layers in the mesophyll region between leaf vessels and the leaf epidermis (Fig. 2D).
Fig. 2.
Morphological features of lil1-1 mutants. (A, B) Longitudinal sections of shoot apices of 2-week-old wild-type (A) and homozygous lil1-1 mutant (B) plants. Samples were stained with Toluidine Blue O. Red asterisks mark leaves primordia. (C, D) Transverse section of leaf blades of 2-week-old wild-type (C) and homozygous lil1-1 mutant (D) plants. (E–G) Primary root of 4-day old seedlings. Altered gravitropic response in homozygous lil1-1 mutant roots growing upwards (E) or parallel to the surface (F) compared with wild-type (WT) root, which shows a positive gravitropic response (G).
Primary root elongation was also significantly reduced in comparison with wild type (Supplementary Data Fig. S2A, C) and this was observed in all the different lines tested (Table 1). Interestingly, seedlings germinated on wet filter paper showed altered root gravitropic curvature. Mutant roots grew either upwards or parallel to the surface (Fig. 2E, F, Supplementary Data Fig. S2A). A positive gravitropic response (Fig. 2G) was instead observed in all wild-type seedlings analysed (Supplementary Data Table S4). The mutant-specific root phenotypes were detected in all genetic backgrounds tested (Supplementary Data Table S4) and was also observed in brd1-m1 mutants (Supplementary Data Fig. S2A). We thus conclude that lack of active BR molecules does not interfere with seed formation in maize but causes a pleiotropic effect on plant development.
The lil1-1 mutant is epistatic to the na1-1 mutant
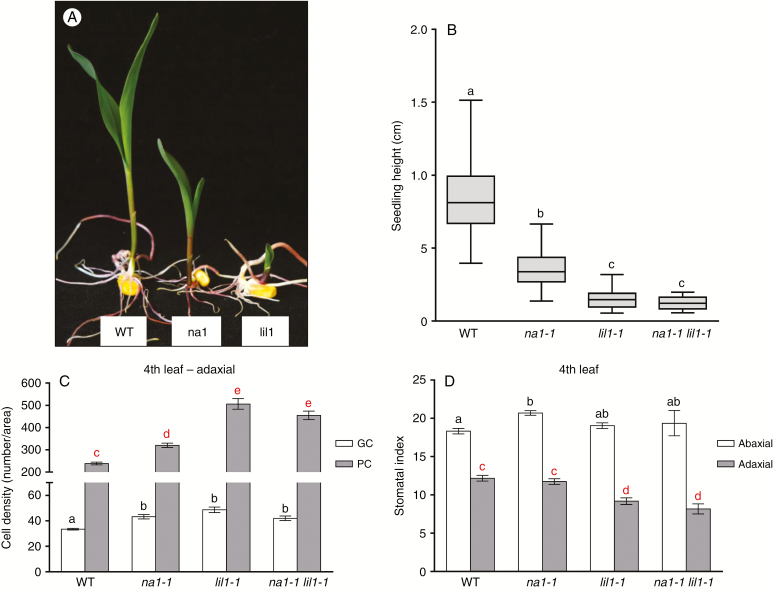
The na1-1 (Hartwig et al., 2011) and lil1-1 mutants are impaired in the earlier and the final steps of the BR biosynthesis, respectively. We observed that in homozygous na1-1 (na1-1/na1-1) plants growth defects appeared less severe than in homozygous lil1-1 (lil1-1/lil1-1) plants. Therefore, to compare their phenotypes as well as to analyse the genetic relationship between the two mutants, F2 progenies were produced from selfing heterozygous na1-1/+ lil1-1/+ F1 plants. A first analysis was performed at the seedling developmental stage through visual scoring of F2 plant phenotypes. Three phenotypic classes were detected, comprising the wild-type class and two distinct mutant classes, referred to as ‘na1-1 like’ and ‘lil1-1 like’, respectively (Fig. 3A). The latter exhibited the most severe phenotype and the more severe reduction in shoot elongation (Fig. 3A). When tested for fit to a 9:3:4 segregation ratio, the calculated χ2 value (0.9142) confirmed the segregation hypothesis (Supplementary Data Table S5). The genotype of single individuals was then determined through PCR analysis (see the Materials and methods section). The analysis confirmed the presence of four genotypic classes along with the 9:3:3:1 segregation ratio expected (χ2 = 1.6939). It also showed that individuals homozygous for na1-1 (na1-1/na1-1 Lil1/−) were all included in the na1-like mutant class while both homozygous lil1-1 (Na1/− lil1-1/lil1-1) and double-homozygous na1-1 lil1-1 (na1-1/na1-1 lil1-1/lil1-1) mutant seedlings belonged to the lil1-like category (Supplementary Data Table S5). This classification was maintained at later developmental stages (Supplementary Data Fig. S3A–D). Wild-type plants (Supplementary Data Fig. S3A) were clearly distinguishable from mutant plants (Supplementary Data Fig. S3B–D). Homozygous na1-1 plants (Supplementary Data Fig. S3B) were recognizable for their erect leaves (Hartwig et al., 2011). Homozygous lil1-1 (Supplementary Data Fig. S3C) and double-homozygous na1-1 lil1-1 (Supplementary Data Fig. S3D) mutant plants were phenotypically identical and showed a drastic reduction in growth compared with both wild-type and homozygous na1-1 plants. Analysis of the four genotypic classes for seedling height showed that their mean values were statistically different except for lil1-1 and na1-1 lil1-1 plants, which did not differ from each other (Fig. 3B). Overall these data confirmed that the lil1 phenotype is more severe than the na1 phenotype and showed that lil1 is epistatic to na1.
Fig. 3.
The lil1-1 mutant is epistatic to the na1-1 mutant . (A) Representative wild-type (WT), na1-like (na1) and lil1-like (lil1) phenotypes of 10-day old F2 plants obtained from selfing F1 heterozygous na1-1/+ lil1-1/+ plants. (B–D) Characterization of WT, homozygous na1-1, homozygous lil1-1 and double-homozygous na1-1 lil1-1 genotypic classes. (B) Whisker plot representing seedling height of 10-day old plants. (C) Numbers of guard cells (GC) and pavement cells (PC). (D) Stomatal index of fourth fully expanded leaves of 50-day old plants. Error bars show the standard error. In (B), (C) and (D) different letters denote differences between values that are statistically significant (P < 0.05) as calculated by one-way ANOVA. In (C) GC (black letters) or PC (red letters) values were compared. In (D) abaxial (black letters) or adaxial (red letters) values were compared.
The genetic relationship between na1 and lil1 was also investigated for epidermal traits. The cell density and stomatal index were analysed in the median portion of fully expanded fourth and sixth leaves (Fig. 3, Supplementary Data Fig. S3).
Comparison of the cell density in the fourth leaf adaxial side (Fig. 3C) among wild-type, single and double mutant plants revealed that both single homozygous na1-1 and lil1-1 mutants have a higher density of pavement cells (PCs) compared with wild type. Analogously to seedling height (Fig. 3B), single homozygous lil1-1 and double homozygous lil1-1 na1-1 mutants showed a higher PC density compared with na1-1 (Fig. 3C). With regard to guard cell (GC) density, that of the wild type was lower and significantly different from that of all single and double mutants (Fig. 3C). Moreover, in plants carrying the lil1-1 mutation in homozygous state the stomata appeared unevenly distributed (Supplementary Data Fig. S3G, H) compared with wild-type (Supplementary Data Fig. S3E) and na1-1 plants (Supplementary Data Fig. S3F).
The adaxial side (Fig. 3C) and the abaxial side (Supplementary Data Fig. S3I) of the fourth leaf presented the same trend, but in the first the differences in PC density between the mutant and wild-type plants was higher. The resulting stomatal index (Fig. 3D) revealed statistically significant differences only in the adaxial side (grey); wild-type and na1-1 plants were identical, while lil1-1 and double lil1-1 na1-1 mutants had similar values to each other but differed from the first two.
The stomatal index (Supplementary Data Fig. S3J) and cell density (Supplementary Data Fig. S3K, L) were also analysed for the adaxial and abaxial sides of the sixth leaf. The results were consistent for the fourth and the sixth leaves and both surfaces.
The lil1-1 plants showed altered leaf permeability and cuticular wax deposition
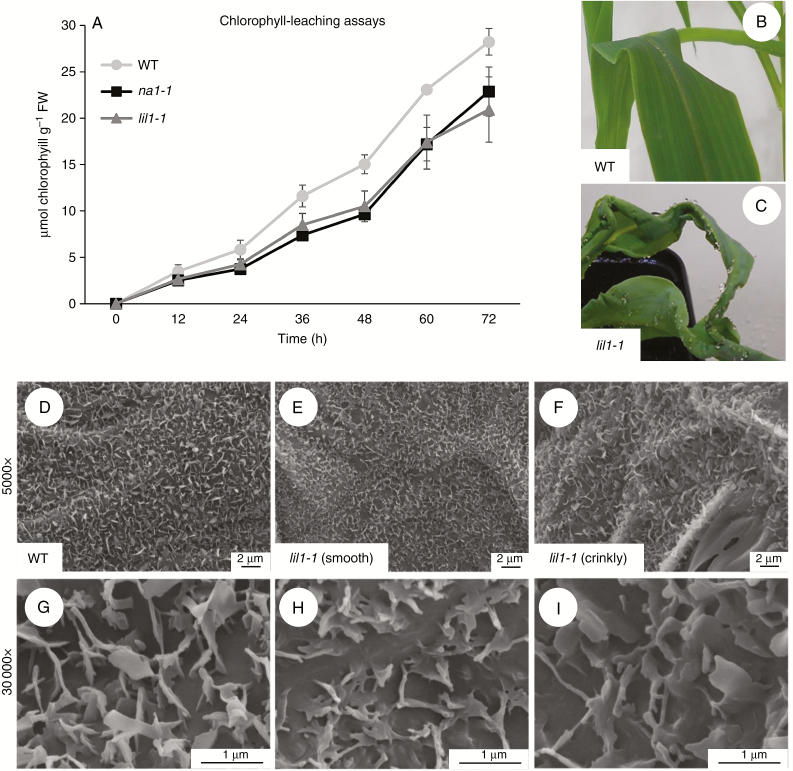
We observed that lil1-1 seedling leaves were thicker than wild-type leaves (Fig. 2C) and had crinkly sectors on their surfaces. To assess the effect of the mutations on leaf permeability a chlorophyll-leaching assay was performed and showed that chlorophyll was more readily released from wild-type leaves than from mutant ones, thus indicating that BR reduction leads to a decrease in leaf permeability. Similar behaviour was observed for both lil1-1 and na1-1 mutants during the assay (Fig. 4A).
Fig. 4.
Permeability and cuticular wax analysis. (A) Chlorophyll leaching assay on the sixth fully expanded leaf of 40-day old wild-type (WT), homozygous na1-1 and homozygous lil1-1 mutant plants. Values represent the mean of five leaves per genotype. Error bars show the standard error. FW, fresh weight. (B, C) Representative adult leaf phenotype of WT (B) and homozygous lil1-1 mutant (C) plants misted with water. (D–I) Scanning electron microscope images of epicuticular waxes of the fourth adaxial leaf surface of WT (D, G) and homozygous lil1-1 mutant plants (E, F, H, I). Mutant samples were taken from a smooth (E, H) and a crinkly sector (F, I) of the leaf.
Further analyses were then conducted for the lil1-1 mutant to test the involvement of cuticle changes in the altered morphology and permeability. We first observed that mutant leaves, unlike wild-types leaves, retained water beads on their surface when they were misted with water (Fig. 4B, C). This was indicative of an alteration in epicuticular waxes, which was indeed highlighted by scanning electron microscopic analysis of the abaxial surface of wild-type and mutant leaves (Fig. 4D–I). Mutant crystalloids appeared less dense, slightly smaller and more embedded in the subtending cuticular material (Fig. 4E, F, H, I) if compared with wild-type ones (Fig. 4D, G).
Homozygous lil1-1 plants are more tolerant to drought stress
Cuticular waxes provide a continuous waterproof barrier and protect plant organs against different environmental stresses. They have been shown to be actively involved in reducing leaf passive water loss (Post-Beittenmiller, 1996). On this basis, we compared wild-type and lil1-1 seedlings for their response to drought stress conditions.
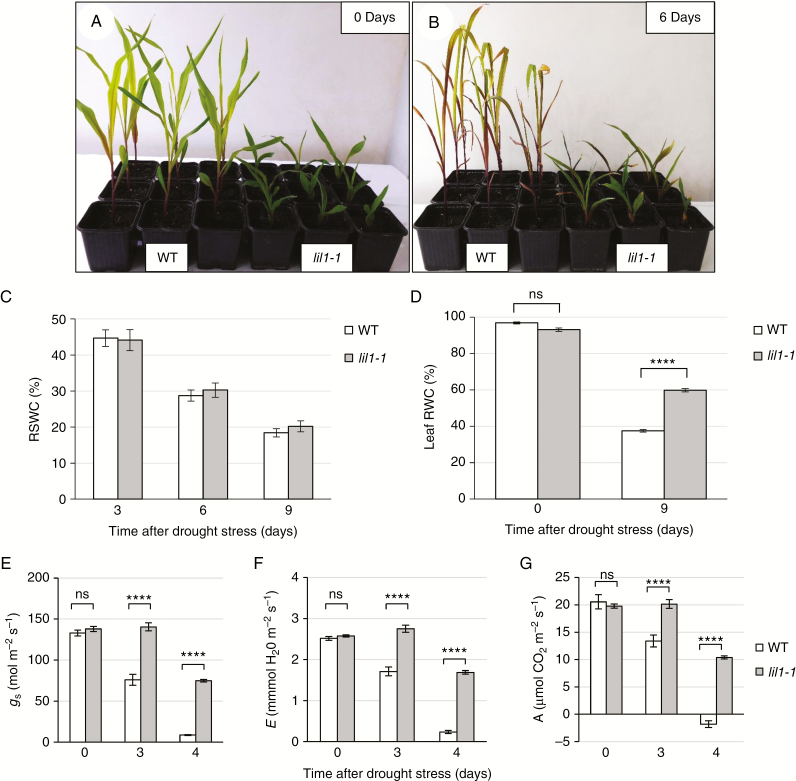
Plants at the fourth-leaf stage grown in pots in well-watered conditions (Fig. 5A) were subjected to drought stress by terminating irrigation. From this time, which was defined as Day 0, soil was allowed to dry progressively. The RSWC, as determined after 3, 6 and 9 days (Fig. 5C), was the same in the two genotypes, thus indicating that the drought stress treatment was identical.
Fig. 5.
The lil1-1 mutant plants are more tolerant to drought stress. (A) Representative phenotype of 14-day old wild-type (WT) and homozygous lil1-1 mutant plants grown under well-watered conditions (0 Days) and (B) after 6 days of drought stress initiation by withholding irrigation (6 Days). (C) Relative soil water content (RWSC) measured at 0, 3, 6 and 9 days after drought stress indicates that the water loss from the soil of pots in which WT plants were grown was identical to that of pots in which mutant (lil1-1) plants were grown. (D) Leaf relative water content (RWC) of WT and homozygous lil1-1 mutant plants at 0 and 9 days of drought treatment. (E, F) Physiological parameters were measured at 0, 3 and 4 days. (E) Stomatal conductance (gs). (F) Transpiration rate (E). (G) Net photosynthesis rate (A). Error bars in (C–G) show standard error. Comparison was made at each time point between wild-type and homozygous lil1-1 genotypes. ****P < 0.0001 (two tailed Student’s t-test); ns, not significant.
After 6 days without water, we observed that lil1-1 plants were less damaged than wild types and their leaves remained greener, whereas wild-type plants showed more severe wilting and leaf chlorosis (Fig. 5B). In addition, measurement of leaf RWC, a marker indicating the balance between intrinsic water supply and transpiration rate, revealed similar values in well-watered conditions, but higher values in mutant than in wild-type plants, 9 days after the cessation of irrigation (Fig. 5D). Taken together, these observations indicated that lil1-1 mutant plants have better tolerance to water stress.
Physiological parameters were assayed at Day 0 (well-watered plants) and after 3 and 4 days of drought treatment and comparisons were made between wild-type and lil1-1 values at each time point. Measurements of stomatal conductance (gs) and transpiration rate (E) showed higher mean values in mutant than in wild-type plants at the same developmental stage under drought stress (Fig. 5E, F). Similarly, net photosynthesis rate (A), which had the same values in the two genotypes at Day 0, showed a higher level in mutants under drought stress. Wild types showed low and negative values at 3 and 4 days of drought, respectively (Fig. 5G). In wild types, the high intercellular CO2 concentration (Ci) measured at 4 days probably indicated almost complete stomatal closure, whereas in mutants the constant Ci values suggested that they maintain their stomata slightly open and functional even during drought treatment (Supplementary Data Fig. S4A). Despite this, mutants deprived of water for 4 days could maintain water use efficiency (WUE) at a constant level, whereas wild types had a negative value for this parameter (Supplementary Data Fig. S4B).
A subset of plants (40 plants per genotype) subjected to water stress was then re-watered. After 3 and 7 days from the starting of re-watering, plants were classified in three categories: (1) severely wilted; (2) slightly wilted; and (3) recovered (Supplementary Data Fig. S4C–I). The percentage of plants showing complete recovery (category 3) was higher in mutant than wild-type plants (Supplementary Data Fig. S4I). The recovery assay indicated that, upon rehydration, mutant plants have a more rapid response, and this confirms their dehydration tolerance.
DISCUSSION
The lil1-1 mutant, isolated from an active Mu population, shows a drastic reduction in plant growth (Dolfini et al., 1999). The molecular and genetic analysis carried out in this study disclosed that the lil1 phenotype is due to a Mu1 insertion in the second exon of the maize cytochrome P450 CYP85A1 C-6 oxidase gene, which is involved in the last steps of the BR pathway. lil1-1 is allelic to the previously isolated brd1-m1 (Makarevitch et al., 2012; Fig. 1; Supplementary Data Table S1).
The phenotype of lil1-1 plants appears more severe than that of the other BR-related mutants so far isolated in maize, such as na1-1 (Hartwig et al., 2011). Analysis of F2 progenies segregating for both lil1-1 and na1-1 mutants revealed that lil1 is epistatic to na1. The product of na1, which is the maize orthologue of A. thaliana DET2, is a 5α-reductase located further upstream in the BR pathway, which catalyses multiple steps in different branches of the pathway. Our data suggest the existence in the maize BR pathway of an additional na1-independent branch leading to the production of CS precursors. Accordingly, CS, although detected at a reduced concentration, was not completely absent in na1 mutants (Hartwig et al., 2011).
We show in this study that lil1 gene action is involved in different aspects of plant development but is not required for seed development. Many reports have highlighted the importance of BRs in seed development in different species (Vriet et al., 2012; Jiang and Lin, 2013). In arabidopsis, seeds of the BR-deficient mutant det2 and the BR-insensitive mutant bri1-5 are smaller compared with wild-type seeds (Jiang et al., 2013). Similarly, the rice BR-deficient mutants brd2 (Hong et al., 2005) and dwarf mutant d61 (Morinaka et al., 2006) have shortened and smaller grains. In contrast, we did not find any obvious phenotypic variations related to seed development and seed weight in homozygous lil1-1 mutant compared with wild-type kernels (Supplementary Data Table S2). However, in the aforementioned studies, mutant seeds were produced by homozygous mutant plants. The observed variations in seed size and production could therefore be attributed to alterations in the BR level of the mother plant. This was observed in transgenic rice plants, in which the BR level was altered only in the stems, leaves and roots and not in seeds. Changes in plant architecture had an impact on seed production, which was higher in transgenic than in the control wild-type plants (Wu et al., 2008). In our work, comparisons were among seeds produced by wild-type plants. We assumed that mother plants had a normal BR level and concluded that the lil1-1 mutation does not have an effect on seed size and production.
Comparison of lil1-1 mutant and wild-type seedlings soon after germination highlighted defective primary root growth. In germinating seeds, wild-type primary roots exhibited a downward curvature, whereas homozygous lil1-1 roots grew either parallel to the surface or with an upward orientation (Fig. 2E, F). Brassinosteroid hormones were detected in root tissues in maize (Kim et al., 2000) as well as in arabidopsis and tomato (Yokota et al., 2001; Bancos et al., 2002). In maize, the work of Kim et al. (2000) also showed that exogenous application of different BRs, including CS and BL, stimulated the gravitropic response. A positive influence of BL on root curvature was also observed in Pisum sativum (Amzallag and Vaisman, 2006), and in arabidopsis, in which BR signalling mutants exhibited a reduced gravitropic curvature, whereas transgenic plants overexpressing BRI1 displayed a greater gravitropic curvature than wild-type plants (Kim et al., 2007).
The present work provides the first genetic evidence of BR involvement in gravitropic curvature in maize. Positive gravitropic movement normally occurs in response to the stimulus of gravity and causes a downward curvature of the root. Beside BRs, the plant hormones auxin and ethylene exert a stimulatory role on the root gravitropic response in maize (Kim et al., 2000; Chang et al., 2004). A model has been proposed to explain their roles, in which BL promotes the gravitropic response by stimulating auxin-induced ethylene production (Chang et al., 2004). The higher level of ethylene would lead to an increase in gravitropic curvature. A different pathway might also be implicated in this response, since gravity perception and signal transduction are also stimulated by BL in a way independent of ethylene (Chang et al., 2004). A deeper characterization of BR-deficient mutants like lil1-1 will allow a better comprehension of the process.
Many studies have shown that plant tolerance to drought stress is increased by exogenous application of BRs (Krishna, 2003; Bajguz and Hayat, 2009). However, the analyses in various species of mutants with reduced endogenous levels and/or perception of these hormones seems to lead to contrasting conclusions. In A. thaliana, reduced BR accumulation, due to either loss of CYP85A2 activity or CYP85A farnesylation, increased drought tolerance (Northey et al., 2016). Higher tolerance to both salt and drought stresses was also observed in the loss-of-function gsk1 mutant carrying a T-DNA insertion in the rice orthologue of the A. thaliana BIN2 (BR-INSENSITIVE 2) gene (Koh et al., 2007). In the same species, the d1 mutant, carrying a mutation in the RGA1 gene involved in both Gibberellin (GA) and BR signalling, showed reduced sensitivity to drought (Ferrero-Serrano and Assman, 2016). RNAi mutants of the Brachypodium distachyon BdBRI1 gene, encoding the BR receptor, exhibited enhanced drought tolerance, accompanied by high expression of drought-responsive genes (Feng et al., 2015). Similarly, in our study we show that lil1-1 mutant plants were better adapted to drought stress conditions. Moreover, recovery from stress conditions occurred more rapidly in mutant than in wild-type plants (Supplementary Data Fig. S4), an observation that was also reported for the mutant in the BdBRI1 gene (Feng et al., 2015).
This behaviour might be due to a better capacity of mutant plants to retain water, as shown by the analysis of leaf RWC, which was significantly higher in mutants after the cessation of irrigation (Fig. 5D; Feng et al., 2015). We propose that morphological changes, such as leaf thickness (Fig. 2C), lower stomatal index (Fig. 3D, Supplementary Data Fig. S3J) and altered epicuticular waxes (Fig. 4) may improve water retention by mutant leaves. In agreement with this, the chlorophyll leaching assay showed that permeability was lower in mutant than in wild-type leaves.
It has been shown in barley that drought causes a significant increase in the accumulation of biologically active BR molecules (Gruszka et al., 2016). In addition, an interesting hypothesis suggests that alteration in BR level induces the presence, by default, of a physiological stress condition. The activity of oxidative stress-related genes, including cold and drought stress response genes and heat stress-related genes, was shown to be enhanced in BR-deficient plants. In the arabidopsis bri1-9 mutant the transcript level of stress-inducible genes and of transcription factors that regulate their expression was constitutively higher in comparison with wild types (Kim et al., 2010). This condition may confer a higher capacity to respond to and tolerate stresses. Of interest in this context is the study of the det2 mutant showing enhanced resistance to general oxidative stress, in which the ATPA2 and ATP24a genes, encoding peroxidases, were constitutively upregulated (Goda et al., 2002; Shuqing et al., 2005). Analogies between lil1-1 and det2 phenotypic traits, such as thicker leaves, thicker cuticle and increased guard cell density, suggest that the two mutants share the same type of response. Further investigation is needed to unravel the molecular mechanisms by which BRs influence drought stress response in maize. It is conceivable that new information arising from these studies will allow us to establish novel manipulation approaches aimed at improving crop adaptability to environmental stress.
CONCLUSIONS
We show in this study that the characterization of mutants with deficiency in BR accumulation and the analysis of their interactions are instrumental in improving our knowledge of the architecture of the pathway in maize. Detailed mutant analysis has also been instrumental in highlighting novel developmental processes in which these hormones are involved. In addition, we provide the first evidence in maize that BRs play a role in the plant response to drought. Although further studies are required to clarify the molecular mechanisms at the basis of the BR-mediated response, this may offer a possible direction to future breeding programmes aimed at improving crop adaptation to changes in climate and environmental conditions.
Supplementary Data
Supplementary Data are available online at www.aob.oxfordjournals.org and consist of the following.
Figure S1: the Mu1 insertion within the maize CYP85A1 gene co-segregates with the lil1 mutant phenotype. Figure S2: both lil1-1 and brd1-m1 mutants showed reduced plant size. Figure S3: phenotypic and morphological analysis of F2 progeny plants obtained from selfing double-heterozygous na1-1/+ lil1-1/+ plants. Figure S4: recovery from drought stress. Table S1: allelism test. Table S2: seed weight is not affected by the lil1-1 mutation. Table S3: mother plant genotype does not affect seed production. Table S4: gravitropic response is altered in the lil1-1 mutant seedling primary root. Table S5: segregation of seedling phenotypes and genotypes in F2 progeny obtained from selfing double-heterozygous plants. Table S6: PCR primers used in this study.
ACKNOWLEDGEMENTS
We wish to thank the Maize Genetics Cooperation Stock Center, Urbana, IL, USA for sending the na1-1 and brd1-m1 stocks, Dr Nadia Santo at the Biosciences Imaging Facility (University of Milan) for the analysis with the scanning electron microscope and Dr Lesley Currah for editing the manuscript.
LITERATURE CITED
- Amzallag GN, Vaisman J. 2006. Influence of brassinosteroids on initiation of the root gravitropic response in Pisum sativum seedlings. Biologia Plantarum 50: 283–286. [Google Scholar]
- Arthur K, Vejlupkova Z, Meeley R, Fowler J. 2003. Maize ROP2 GTPase provides a competitive advantage to the male gametophyte. Genetics 165: 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz A, Hayat S. 2009. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiology and Biochemistry 47: 1–8. [DOI] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, et al. 2002. Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiology 130: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best NB, Hartwig T, Budka J, et al. 2016. nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis protein Dwarf1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiology 171: 00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Harrison K, Jones JD. 1996. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, et al. 1999. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proceedings of the National Academy of Sciences of the USA 96: 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Kim YS, Lee JY, et al. 2004. Brassinolide interacts with auxin and ethylene in the root gravitropic response of maize (Zea mays). Physiologia Plantarum 121: 666–673. [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, et al. 1999. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiology 119: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. 2011. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Krishna P. 2009. Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnology 26: 131–136. [DOI] [PubMed] [Google Scholar]
- Dolfini S, Landoni M, Consonni G, Rascio N, Dalla Vecchia F, Gavazzi G. 1999. The maize lilliputian mutation is responsible for disrupted morphogenesis and minute stature. Plant Journal 17: 11–17. [Google Scholar]
- Feng Y, Yin Y, Feia S. 2015. Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Science 234: 163–173. [DOI] [PubMed] [Google Scholar]
- Ferrero-Serrano Á, Assmann SM, 2016. The α-subunit of the rice heterotrimeric G protein, RGA1, regulates drought tolerance during the vegetative phase in the dwarf rice mutant d1. Journal of Experimental Botany 67: 3433–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. 2002. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiology 130: 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszka D, Janeczko A, Dziurka M, Pociecha E, Oklestkova J, Szarejko I. 2016. Barley brassinosteroid mutants provide an insight into phytohormonal homeostasis in plant reaction to drought stress. Frontiers in Plant Science 7: 1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos JF, Dal Prà M, Giulini A, et al. 2007. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, et al. 2011. Brassinosteroid control of sex determination in maize. Proceedings of the National Academy of Sciences of the USA 108: 19814–19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, et al. 2005. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17: 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeczko A, Gruszka D, Pociecha E, et al. 2016. Physiological and biochemical characterisation of watered and drought-stressed barley mutants in the HvDWARF gene encoding C6-oxidase involved in brassinosteroid biosynthesis. Plant Physiology and Biochemistry 99: 126–141. [DOI] [PubMed] [Google Scholar]
- Jiang WB, Lin WH. 2013. Brassinosteroid functions in Arabidopsis seed development. Plant Signaling & Behavior 8: e25928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH. 2013. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiology 162: 1965–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BK, Fujioka S, Takatsuto S, Tsujimoto M, Choe S. 2008. Castasterone is a likely end product of brassinosteroid biosynthetic pathway in rice. Biochemical and Biophysical Research Communications 374: 614–619. [DOI] [PubMed] [Google Scholar]
- Kim S-K, Chang SC, Lee EJ, et al. 2000. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiology 123: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Kim BH, Lim CJ, Lim CO, Nam KH. 2010. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Plant Physiology 138: 191–204. [DOI] [PubMed] [Google Scholar]
- Kim TW, Lee SM, Joo SH, et al. 2007. Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant, Cell & Environment 30: 679–689. [DOI] [PubMed] [Google Scholar]
- Kir GYH, Nelissen H, Neelakandan AK, et al. 2015. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiology 169: 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, Lee SC, Kim MK, et al. 2007. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Molecular Biology 65: 453–466. [DOI] [PubMed] [Google Scholar]
- Krishna P. 2003. Brassinosteroid-mediated stress responses. Journal of Plant Growth Regulation 22: 289–297. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. 1996. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–402. [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Berlyn GP, Engstrom EM, Krolikowski KA, Reiter W. 1997. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: a role for the epidermal cell wall and cuticle. Developmental Biology 321: 311–321. [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM. 2012. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PloS ONE 7: e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, et al. 2006. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiology 141: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northey JGB, Liang S, Jamshed M, et al. 2016. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nature Plants 2: 1–7. [DOI] [PubMed] [Google Scholar]
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S. 2005. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. Journal of Biology Chemistry 280: 17873–17879. [DOI] [PubMed] [Google Scholar]
- Pérez-España VH, Sánchez-León N, Vielle-Calzada JP. 2011. CYP85A1 is required for the initiation of female gametogenesis in Arabidopsis thaliana. Plant Signaling & Behavior 6: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczynski M, Marczewski W, Hennig J, et al. 2013. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnology Journal 11: 459–469. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D. 1996. Biochemistry and molecular biology of wax production in plants. Annual Review of Plant Biology 47: 405–430. [DOI] [PubMed] [Google Scholar]
- La Rocca N, Manzotti PS, Cavaiuolo M, et al. 2015. The maize fused leaves1 (fdl1) gene controls organ separation in the embryo and seedling shoot and promotes coleoptile opening. Journal of Experimental Botany 66: 5753–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, et al. 2001. Brassinosteroid-6- oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiology 126: 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuqing C, Qiaoting X, Yajun C, et al. 2005. Loss‐of‐function mutations in DET2 gene lead to an enhanced resistance to oxidative stress in Arabidopsis. Physiologia Plantarum 123: 57–66. [Google Scholar]
- Vriet C, Russinova E, Reuzeau C. 2012. Boosting crop yields with plant steroids. Plant Cell 3: 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Trieu A, Radhakrishnan P, et al. 2008. Brassinosteroids regulate grain filling in rice. Plant Cell Online 20: 2130–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Sato T, Takeuchi Y, et al. 2001. Roots and shoots of tomato produce 6-deoxo-28-norcathasterone, 6-deoxo-28-nortyphasterol and 6-deoxo-28-norcastasterone, possible precursors of 28-norcastasterone. Phytochemistry 58: 233–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.