Abstract
Loss of function mutations in CTNNB1 have been reported in individuals with intellectual disability [MIM #615075] associated with peripheral spasticity, microcephaly and central hypotonia, suggesting a recognisable phenotype associated with haploinsufficiency for this gene. Trio based whole exome sequencing via the Deciphering Developmental Disorders (DDD) study has identified eleven further individuals with de novo loss of function mutations in CTNNB1. Here we report detailed phenotypic information on ten of these. We confirm the features that have been previously described and further delineate the skin and hair findings, including fair skin and fair and sparse hair with unusual patterning.
Keywords: CTNNB1, intellectual disability, microcephaly
Introduction
Whole exome sequencing is increasingly being utilised in the investigation of intellectual disability (ID), with more genes being implicated as a result. A gene, recently linked to a syndromic form of ID, is CTNNB1, which encodes for beta 1 catenin. This is a component of the cadherin adhesion complex which mediates cell-cell adhesion, and is also part of the Wnt signalling pathway. This pathway is important in controlling cell growth and differentiation, both in normal and tumour tissue. Beta catenin knockout mice display behaviours consistent with autistic spectrum disorder (Dong et al, 2016), as well as defects in neural development (Brault et al, 2001) and in hair follicle formation (Huelsken et al, 2001). Somatic mutations in CTNNB1 have been detected in certain tumours, and more recently germline, loss of function mutations have been linked to intellectual disability [MIM #615075].
Mutations in CTNNB1 were first linked to intellectual disability when it was identified as a candidate gene during a trio exome sequencing study of 100 patients with intellectual disability (de Ligt et al, 2012). One patient was found to have a frameshift mutation (p.Ser425Thrfs*11), with a further two patients found to have nonsense mutations (p.Arg515* and p.Gln309*) on sequencing of an additional cohort of 765 patients with intellectual disability. The phenotypes of these three patients were investigated in detail, and features included mild to severe intellectual disability, craniofacial anomalies, severe speech delay, microcephaly and childhood hypotonia with progressive peripheral spasticity and delayed motor development (Tucci et al, 2014). A report providing details of a patient with a whole gene deletion of CTNNB1, along with a further case series of 16 individuals with loss of function mutations in CTNNB1, confirmed these phenotypic features, as well as providing more detail regarding associated dysmorphic features, including thin upper lip vermillion and small alae nasi (Dubruc et al, 2014; Kuechler et al, 2015). One further case of a patient with a de novo nonsense mutation in CTNNB1 has been reported, presenting with characteristic features such as microcephaly, peripheral spasticity and delayed speech, along with atypical features of hyperekplexia and apraxia of upward gaze (Winczewska-Wiktor et al, 2016). This case series aims to further delineate the phenotype associated with loss of function mutations in CTNNB1, reviewing ten patients ascertained through the Deciphering Developmental Disorders study.
Methods
The Deciphering Developmental Disorders (DDD) study is a project which recruited patients through genetics centres in the UK and Ireland, with the aim to utilise genome-wide microarray and whole exome sequencing to increase diagnostic rates for children and adults with severe undiagnosed developmental disorders. Specifically congenital or early onset severe phenotypes were the focus for recruitment, with family trios sampled in the majority of cases. 13,632 families were recruited, and analysis of 4,293 has been completed so far. DNA samples from patients and their parents were analysed at the Wellcome Trust Sanger Institute with microarray analysis (Agilent 2x1M array CGH [Santa Clara, CA, USA] and Illumina 800K SNP genotyping [San Diego, CA, USA]) to identify copy number variants (CNVs) in the child, and exome sequencing (Agilent SureSelect 55MB Exome Plus with Illumina HiSeq) to investigate single nucleotide variants (SNVs), small insertion-deletions (indels), and CNVs in coding regions of the genome. An automated variant filtering pipeline was used to narrow down the number of putative diagnostic variants, by ruling out common and non-functional variants, and then comparing variants against an in-house database of genes consistently implicated in specific developmental disorders, the Developmental Disorders Genotype-to-Phenotype database (DDG2P). This database includes more than 1000 genes that have been consistently implicated in specific developmental disorders and is updated regularly with newly implicated genes. (Wright et al, 2015)
By interrogating the DECIPHER database, where DDD results can be scrutinised, we identified eleven patients with an inactivating mutation of CTNNB1. We then contacted the responsible clinicians to obtain detailed phenotypic data on these patients, and received responses for ten patients. Consent was obtained for publication of photographs.
Clinical Features
The patients’ ages range from 3-27 years, with three males and seven females. In all patients symptoms occurred within the first year of life. Table 1 shows the pertinent clinical features for each patient.
Table 1.
Representation of the phenotypic data collected for each of the 10 patients with CTNNB1 mutations.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| DECIPHER ID | 267,844 | 278,755 | 259,002 | 259,408 | 259,162 | 275,999 | 264,405 | 272,034 | 267,130 | 259,248 |
| Mutation | c.1801C> T p.Gln601Ter |
c.1603C> T p.Arg535Ter |
c.1981C> T p.Arg661Ter |
c.1603C> T p.Arg535Ter |
c.1038_1044delGCTATCTinsGCT p.Val349AlafsTer9 |
c.999C> G p.Tyr333Ter |
c.1420C> T p.Arg474Ter |
c.799_809 delGAAGGAGCTAA insGAA p.Gly268TrpfsTer5 |
c.1612C> T p.Gln538Ter |
c.998dupA p.Tyr333Ter |
| Mutation type | Nonsense | Nonsense | Nonsense | Nonsense | Frameshift | Nonsense | Nonsense | Frameshift | Nonsense | Nonsense |
| Sex | Male | Male | Female | Male | Female | Female | Female | Female | Female | Female |
| Age of onset of symptoms | Birth | 3 months | Birth | Birth | Within 1st year | Birth | 7 months | 3 months | Within 1st year | 9 months |
| Age at diagnosis | 6 years 2 months | 3 years 3 months | 9 years 2 months | 14 years | 11 years | 27 years | 13 years | 7 years | 4 years 6 months | 9 years |
| Maternal Age | 27 | 39 | 27 | 28 | 32 | 21 | 29 | 29 | 32 | 32 |
| Paternal Age | 27 | 43 | 30 | 38 | 28 | 22 | 34 | 34 | 33 | 41 |
| Prenatal issues | Intrauterine growth retardation | None | Microcephaly and intrauterine growth retardation | None | Intrauterine growth retardation | None | None | None | Gestational diabetes: Exposure to anticonvulsant drugs | None |
| Gestation (weeks) | 39 | 40 | 38 | 41 | 40 | 39 | 41 | 42 | 41 | 40 |
| Birthweight (SD) | 2.098 kg (2.8SD) | 3.05 kg (1.05SD) | 2.53 kg (1.15SD) | 3.4 kg (0.32SD | 2.7 kg (1.6SD) | 2.92 kg (0.66SD) | 3.118 kg (0.63SD) | 3.1 kg (0.67SD) | 2.345 kg (2.05SD) | 2.98 kg (0.95SD) |
| Neonatal issues | Floppy | Low oxygen saturations | Poor feeding, reflux | Persistent Pulmonary Hypertension | Strabismus | Difficulty feeding, floppy | None | None | In SCBU for 21 days | Small anterior fontanelle, irritable |
| Growth Parameters (SD) | At 3 years: OFC 46.6 cm (3.6SD) Weight 11.4 kg (2.74SD) Height 87.2 cm (2.82SD) |
At 21 months: OFC 43 cm (5.25SD) Weight 10.45 kg (1.29SD) Height 83.5 cm (0.31SD) |
At 5 years: OFC 42 cm (8.18SD) Weight 15.2 kg (1.62SD) Height 99 cm (2.43SD) |
At 6 years: OFC 50.4 cm (1.93SD) Weight 26 kg (1.17SD) |
At 4 years: OFC 48.9 cm (2.18SD) At 11 years: OFC 51.1 cm (2.6SD) Height 0SD Weight 0SD |
At 26 years: OFC 52.7 cm (2.03SD) Weight 118 kg (3.25SD) Height 152 cm (1.96SD) |
At 2 years: OFC 45 cm (3.35SD) Weight 12.15 kg (0.14SD) Height 86.3 cm (0.11SD) At 9 years: OFC 47 cm (5.32SD) |
At 3 years: OFC 42.6 cm (6.67SD) Weight 13 kg (1.16SD) Height 92.1 cm (1.29SD) |
At 3 years: OFC (4.96SD) Weight 12.84 kg (1.46SD) Height 95.5 cm (0.68SD) |
At 4 years: OFC 46.9 cm (4.19SD) Weight 15.45 kg (1.03SD) Height 99 cm (1.71SD) |
| Developmental progress | Smiled e4 months; Head control - 9 months; Rolling back to front from 2 years; Bottom shuffles but unable to walk at age 6 years 2 months. |
Smiled e4 months; Sat supported - 8 months; Rolling - 9 months; not walking at 3 years 3 months, trying to crawl. |
Smiled e6 weeks; Rolling - 6 months; Sat - 11 months; Walked - 30 months |
Sat e15 months Walked with rollator - age 4; Can’t walk unaided at age 14 years |
Late sitting; Crawled - 13.5 months; Walked - 42 months, ataxic |
Sat independently - 30 months; Walked - 4e5 years, ataxic |
Sat independently 13 months; Walked - 42 months |
Sat - 18 months; Walked - 36 months, broad based gait |
Social smile e24 weeks; Sat independently - 23 months; Walked - 2.5e3 years, ataxic |
Social smile - 3 weeks; Sat independently - 14 months; Walked 50 months, still has walking difficulties |
| Speech and Language development | Can say Dad and Mum with understanding; has several Makaton signs and can point correctly to many parts of the body when asked. | Makes lots of noises but no words. | First words 3e4 years | Single words at age 14 years | Single words at age 5 years, talks in sentences at age 11 years | First words 4e5 years; can speak in sentences as adult but speech very unclear | First words 4e5 years | No speech | First words 3e4 years | Significant speech delay e first words age 4; more fluent speech age 6; said to be 3 years behind with verbal skills |
| Fair skin (ethnic origin) | Yes e but parents reported being as fair as children. (White British) | Yes (Scottish Caucasian) | Yes (Middle Eastern e Iran/Lebanon) | Yes (White British) | No (Egyptian) | Yes (White British) | Described as clear, has pigmented patch right knee (Caucasian) | Yes (Caucasian) | Normal for family (Caucasian) | No (Moroccan Sephardic Jewish) |
| Hair | Very blonde | Very blonde | Very slow growth and fine | Fair, not sparse, has cowlick | Fair and sparse, especially over temples | Fair as child, unusual hair whorl pattern, has fine hair as adult | Normal | Fine, blonde hair | Normal | Dark |
| Dysmorphic features | Slightly prominent nose in earlier childhood; thin upper lip. | Thin upper lip and prominent lower lip; long smooth philtrum; small ears; brachycephaly. |
Wide spaced teeth; sacral dimple; tented and thin upper lip; 5th finger clinodactyly; low set ears |
Low set ears; short philtrum; thin upper lip; high palate; prominent chin | Periorbital fullness; slightly low set ears with fleshy lobes; full tip to nose | Thin lower lip; downslanting palpebral fissures; prominent columella; large chin; wide spaced teeth | Deep set eyes with darker periorbital skin; epicanthic folds; short prominent nose with bulbous tip; smooth philtrum; thin upper lip; high palate; micrognathia with pointed chin; gum hypertrophy |
Thin upper lip; slightly low columella | Prominent eyes | Low set ears |
| Peripheral spasticity | Unsure | Yes | No | Yes | No | Yes | Yes | Yes | Yes | Yes |
| Truncal hypotonia | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes |
| Autistic features/behavioural problems | Occasional temper; frustration. Can bite others and himself; large appetite; constantly active e head rocking movements. |
No | Some autistic traits - obsessional behaviour, global delay | Aggressive outbursts; self harm; poor sleep | Stereotypies e.g. clapping repeatedly, temper tantrums, aggressive to family members | Aggressive, temper tantrums, self injurous (biting, picking) | Severe behavioural problems/ADHD; aggressive; poor sleep; teeth grinding; mouths objects |
Yes | Yes | No ASD but violent outbursts associated with difficulty expressing emotions |
| Seizures | No | No | No | No | No | No | No | No | No | ?possible absence seizures in early childhood; EEG normal |
| Additional clinical features | Single supernumerary upper incisor; bilateral orchidopexies |
Strabismus and hypermetropia, dystonic posturing | Poor balance; oromotor dyspraxia; slightly delayed bone age; hypermobile joints; glue ear |
Truncal obesity; absent left testis; brachydactyly; Achilles tendon contracture; high arched palate |
Strabismus and hypermetropia acquired teeth early, from 3 months | Asthma; left clubfoot; increased dermatoglyphic whorls |
Bilateral strabismus | Intermittent loss of skills | Strabismus; congenital ichthyosiform erythroderma (probably paternally inherited) | Dystonic posturing |
Craniofacial
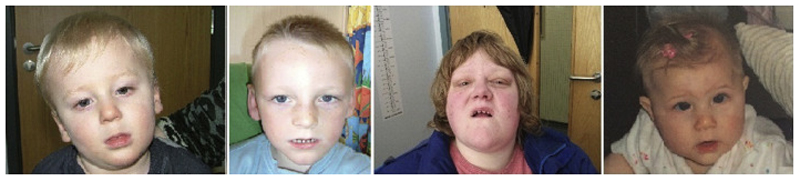
Mild dysmorphic features have been previously reported. Our patients displayed a range of features, with a thin upper lip being the most common (7/10 patients). Other features seen include low set ears, wide spaced teeth, prominent columella (previously noted in older patients) and a prominent nose (Fig 1). Dubruc et al’s case report noted that a patient with a whole gene deletion of CTNNB1 had fair, sparse hair and fair skin, and so we specifically asked about these features when collecting clinical details. 6/10 patients were reported to have fair skin, and 7/10 patients were reported to have fair and/or fine hair compared to other family members, with two of those having an unusual hair pattern such as a cowlick or unusual hair whorl.
Figure 1.
Photographs of 4 patients with CTNNB1 mutations; from left to right, patient 2 (age 20 months), patient 5 (age 8 years), patient 6 (age 27 years) and patient 8 (age 7 months). Note fair and fine hair, and thin upper lip.
Growth
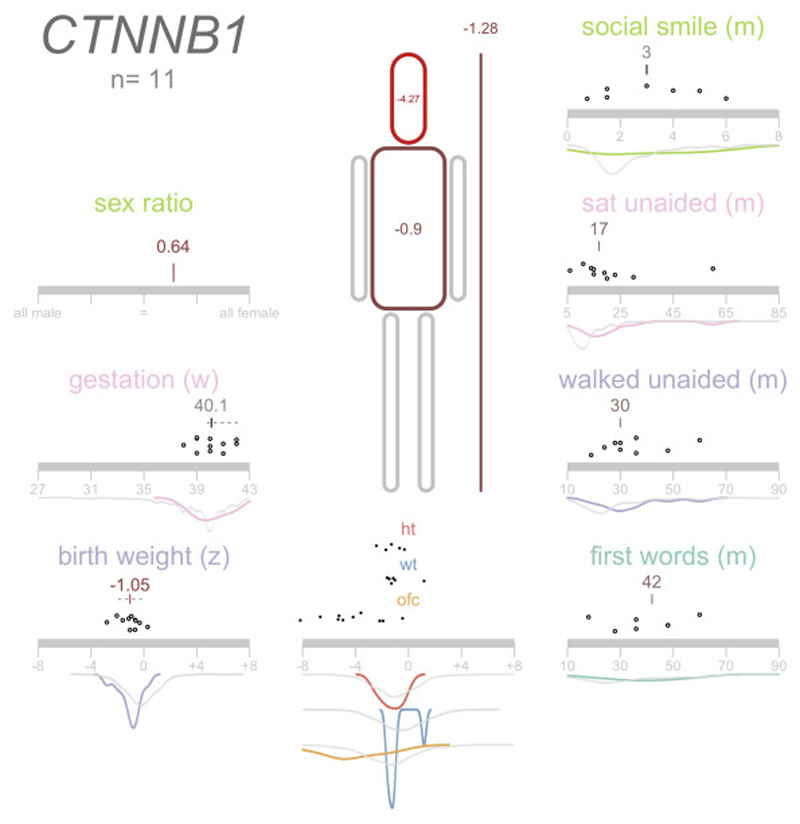
3/10 patients were noted to have intrauterine growth retardation on antenatal scanning. Postnatal microcephaly was a common finding, with 7/10 patients having a head circumference of -3 standard deviations or smaller. Figure 2 visualises the growth parameters and developmental progress of the 11 patients ascertained through the DDD study.
Figure 2.
Phenicon demonstrating data collected for DDD patients with CTNNB1 mutations.
Development
All of the patients have significant motor delay, with the earliest age of walking at 2.5 years. 3/10 patients are unable to walk independently, the oldest being 6 years 2 months of age. All of the patients have significant speech delay, as has previously been reported. Behavioural problems are a common feature, with 9/10 patients reported to have issues such as aggressive outbursts and poor sleep. Visual problems have previously been reported, and 4/10 patients have strabismus, with 2/10 also affected by hypermetropia.
Neurology
As noted in previous reports, 9/10 patients were noted to have truncal hypotonia, usually presenting within the first year of life. 2/10 patients had problems feeding in the neonatal period. Peripheral spasticity is a common feature which seems to present after initial hypotonia, with 7/10 patients developing this feature. 2/10 patients were noted to have broad based or ataxic gait, which became evident at 3 to 4 years of age. Dystonic posturing was also noted in 2/10 patients, presenting at 18 months to 2 years of age. None of our patients were reported to have seizures; there was a query regarding possible absence seizures in 1 patient, but EEG was normal. All 10 patients have had MRI brain scans, which were normal except for 1 patient, who was noted to have atrophy of the left temporal lobe.
Mutations
In our patients, 8 nonsense and 2 frameshift de novo mutations were detected in the CTNNB1 gene by whole exome sequencing of family trios (Table 1). These are all predicted to lead to premature truncation of the protein. This concurs with previously reported mutations and deletions, which were all de novo and caused loss of function of the gene.
Discussion
Our case series reinforces the phenotypic features previously reported in conjunction with inactivating CTNNB1 mutations, including intellectual disability, postnatal microcephaly, truncal hypotonia and peripheral spasticity, mild dysmorphic features and behavioural problems. An additional patient with CTNNB1 loss of function mutations identified by the DDD study also displays typical phenotypic features when looking at the DECIPHER database; we have not included this patient in our clinical report as we were not able to collect further updated phenotypic data for them. This patient (DECIPHER ID 264159), who has a de novo frameshift variant (c.1686_1690delGGGGGinsGGGG; p.Val564SerfsTer6), has clinical features listed on DECIPHER including delayed speech and language development, truncal hypotonia and abnormal hair pattern. Our case series appears to show an excess of female patients (7 females as opposed to 3 males). On looking at the previously described 22 cases, 12 were female and 10 male, so there doesn’t appear to be a significant gender bias in those affected by inactivating CTNNB1 mutations, although these are small numbers from which to infer such a finding and this may become more obvious as more cases are identified. In the DDD cohort, 11 cases were found from the 4,293 families where analysis has been completed, giving a frequency of 0.0026.
β-catenin has been shown to play a role in skin development and the formation of hair follicles in mice (Huelsken et al, 2001). Changes in hair and skin have only been, so far, described in detail by Dubruc et al (2014) in a patient who has a whole gene deletion of CTNNB1. Winczewska-Wiktor et al (2016) also mention that their patient has a right frontal hair upsweep, which would further support our findings. We have demonstrated in our patients that hair and skin anomalies are a common feature associated with CTNNB1 inactivating mutations, specifically fair skin, and fair, fine hair, with unusual hair patterning, which is out of keeping with the familial background.
In summary, our case series confirms findings previously described in association with CTNNB1 inactivating mutations, and further delineates specific skin and hair findings, which may be helpful in identifying likely affected patients.
Acknowledgements
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network
Footnotes
Web resources
This study makes use of data generated by the DECIPHER community. A full list of centres who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Accession numbers
Accession numbers on the DECIPHER database are as follows: 267844, 278755, 259002, 259408, 259162, 275999, 264405, 267130, 264159, 259248 and 272034.
References
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001 Apr;128(8):1253–64. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012 Nov 15;367(20):1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- Dong F, Jiang J, Mcsweeney C, Zou D, Liu L, Mao Y. Deletion of CTNNBl in inhibitory circuitry contributes to autism-associated behavioral defects. Hum Mol Genet. 2016 Apr 30; doi: 10.1093/hmg/ddw131. pii: ddw 131. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruc E, Putoux A, Labalme A, Rougeot C, Sanlaville D, Edery P. A new intellectual disability syndrome caused by CTNNB1 haploinsufficiency. Am J Med Genet A. 2014 Jun;164A(6):1571–5. doi: 10.1002/ajmg.a.36484. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. Beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001 May 18;105(4):533–45. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Kuechler A, Willemsen MH, Albrecht B, Bacino CA, Bartholomew DW, van Bokhoven H, van den Boogaard MJ, Bramswig N, Büttner C, Cremer K, Czeschik JC, et al. De novo mutations in beta-catenin (CTNNB1) appear to be a frequent cause of intellectual disability: expanding the mutational and clinical spectrum. Hum Genet. 2015 Jan;134(1):97–109. doi: 10.1007/s00439-014-1498-1. [DOI] [PubMed] [Google Scholar]
- Tucci V, Kleefstra T, Hardy A, Heise I, Maggi S, Willemsen MH, Hilton H, Esapa C, Simon M, Buenavista MT, McGuffin LJ, et al. Dominant β-catenin mutations cause intellectual disability with recognizable syndromic features. J Clin Invest. 2014 Apr;124(4):1468–82. doi: 10.1172/JCI70372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winczewska-Wiktor A, Badura-Stronka M, Monies-Nowicka A, Nowicki MM, Steinborn B, Latos-Bieleńska A, Monies D. A de novo CTNNB1 nonsense mutation associated with syndromic atypical hyperekplexia, microcephaly and intellectual disability: a case report. BMC Neurol. 2016 Mar 12;16:35. doi: 10.1186/s12883-016-0554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, Bevan AP, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385(9975):1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]