Abstract
The volume of solid organ transplant in the United States is increasing, providing improved quality of life and survival for patients with organ failure. The growth of transplantation requires a systematized management of transplant outcomes assessment, especially with the movement towards value-based care. However, there are several challenges to analyzing outcomes in the current registry-based, transplant reporting system: (1) longitudinal data points are difficult to capture in outcomes models; (2) data elements are restricted to those that already exist in the registry data; (3) there is a delay in the release of outcomes report. In this article, we propose an informatics approach to solve these problems by utilizing a ‘common data model’ to integrate disparate data sources, data elements, and temporal data points. Adopting such a framework can enable multi-center outcomes analyses among transplant centers, nationally and internationally.
Introduction
Transplantation has made dramatic strides over the last few decades with significant improvements in post-transplant patient outcomes1,2. The introduction of center-specific survival rates and subsequently program-specific reports (PSR), is part of the effort to measure the risk adjusted performance of transplant centers3. While the risk adjustment models have evolved over time, the basic approach has not. These multivariable models use data that transplant centers self-report to the United Network for Organ Sharing (UNOS). This data is currently collected using a web-based data system (Transplant Information Electronic Data Interchange (TIEDI)) at pre-specified time points: wait-listing, transplantation, and subsequently pre-specified post-transplant time points. Similarly, donor data is also obtained either from organ procurement organizations (OPO) (for deceased donors) or transplant centers (living donors).
Despite ongoing efforts to analyze and manage transplant outcomes, challenges remain with the current data collection and outcomes reporting system. First, although the data collected by UNOS at wait-listing and transplantation are reasonably complete, subsequent longitudinal patient data is limited in scope, frequently incomplete as evidenced by the challenges of even meeting new UNOS reporting thresholds for living donor follow-up, and limited largely to hard endpoints such as allograft failure and patient death4. However, given that the results of the program evaluation are patient-facing, the choice of outcomes of interest needs to incorporate issues that patients value, including measures of patient safety and quality of life5,6. Additional relevant information such as genetic data, measures of socioeconomic status (beyond insurance status and education level), and environmental/ecological factors cannot be considered for inclusion when assessing transplant center performance7,8.
Given the challenges and volume of data requested by UNOS, transplant centers are reluctant to add any additional data reporting mandates – either in the form of additional data elements, time points, or patient-centered outcomes. The addition of data elements or change to data collection forms is a multistep process requiring a public comment period and approval from several groups within UNOS in addition to external federal agency approval resulting in a relatively slow process in revising the data elements being captured.
Lastly, program specific reports include transplants over a 2.5-year period ending a year prior to reporting. As a result, the delayed identification of poor performance precludes the ability of programs to rely on these reports for early identification of issues that adversely impact outcomes. For example, programs may be cited as having poor outcomes for events that may have occurred 3–4 years prior to the publication of the PSR. Continuous quality improvement efforts require centers to be able to evaluate their outcomes more immediately. The development of robust quality programs that utilize complex statistical and data management skills is probably beyond the abilities of many, perhaps even a majority of, transplant centers today. Existing efforts among a few transplant centers to perform real-time quality monitoring are limited in that they typically require a membership or are proprietary systems, often at great expense, that are of limited value to the larger transplant community4,9,10. In this article, we propose informatics solutions that can be leveraged to overcome the limitations of the current practice of transplant outcomes management.
The Proposed Solution: Utilizing the Big Data Approach
In the age of big data, the idea of integrating multiple data sources is commonplace. We propose that UNOS data can similarly be augmented by integrating multiple data sources. Presently, the Scientific Registry for Transplant Recipients (SRTR) not only receives UNOS data, but also integrates it with secondary data sources such as the National Death Index (NDI) from the National Center for Health Statistics (NCHS); the Surveillance, Epidemiology, and End Results (SEER); Social Security Death Master File (SSDMF); hospital-specific data; and Centers for Medicare and Medicaid Services – End Stage Renal Disease (CMS-ESRD)11. Additional data sources that could potentially enrich the outcomes model are however needed. Opportunities for further enrichment using other administrative and research datasets exist. For example, the United States Renal Data System (USRDS) contains data on all patients with end-stage renal disease (ESRD) in the United States and includes cross-sectional data from the 2728 Medicare form, subsequent claims data, and medication data from Medicare part D12. However, a richer and untapped source of data is the electronic health record (EHR) itself. This data source has the potential of introducing high volumes of data (i.e. vitals from devices, labs, clinical notes, etc.). Srinivas et al. recently demonstrated the value of augmenting UNOS data with EHR data, their institutional transplant database (structured data), and unstructured clinical text4. This approach, if widely accepted, could also benefit data already in registry datasets by decreasing the volume of missing data by using tools to automatically abstract some information, such as clinical notes.
More specifically, in the field of transplantation, there are research questions that can only be answered by integrating multiple data sources with the data available in the UNOS registry. For example, identifying the optimal strategy to assess cardiovascular disease in the prospective transplant patient could be feasible through this approach. Currently, neither the results of cardiovascular testing nor what testing a patient underwent prior to wait-listing or transplantation is captured in the UNOS registry. As a result, there is significant center-level practice variation – as reflected in the American College of Cardiology (ACC) and the American Heart Association (AHA) statement which was significantly hampered by the absence of reasonable underlying data.13,14 Linkage of outcomes to treatment modalities and cardiovascular disease measures would inform the identification of approaches to testing and risk stratification associated with better outcomes. But while this approach enables studying novel research questions, the implementation of this proposed approach requires augmenting disparate data sources, for which a ‘common data model’ is needed.
Common Data Model
A “data model” is the conceptual layout and organization of data within a database. It defines the required data elements and the relationships between different data elements15. Common data models (CDM) allow multiple databases to deploy the same layout of data and therefore support the integration of disparate data sources by ensuring consistency in data structure and unambiguous interpretation of data across different information systems.
The CDM would be necessary to permit the aggregation of data from disparate sources, easily allowing the introduction of new data elements and more data values with variable time points for transplant outcomes research16. Multiple studies have utilized the CDM in fields outside transplantation17,18. For example, Hripcsak, et al., identified the ordered sequence of medications prescribed for type 2 diabetes mellitus, hypertension, and depression using 250 million patients from 11 data sources and 4 countries17.
This example highlights the opportunity a CDM presents in answering transplant-specific research questions that require more granular and higher volume of data. As mentioned above, identifying the optimal strategy to assess cardiovascular disease in prospective transplant patients can be studied once we integrate data on the type of cardiovascular testing that a patient received prior to wait-listing or transplantation and the results of this testing. The CDM makes such a study feasible by integrating UNOS registry data with EHR data that captures the data elements not found in the registry. It also allows transplant centers that adopt the same CDM to easily conduct multicenter studies. By having identically formatted datasets, the same analysis methods and programming codes can be run across centers. Individual transplant centers need to share only the aggregate results which precludes the need to share individual patients’ protected health information between centers. This approach facilitates the aggregation of results across centers by a central coordinating center in addition to facilitating, if needed, a direct comparison of results between participating transplant centers. Currently, the study of the efficacy of treatment regimens for acute rejection that occur years after transplantation is limited almost exclusively to single-center studies. Similarly, our understanding of the impact of various infections on outcomes, or changes in immunosuppression in response to various clinical events remains restricted to observational analyses from single-center cohort studies that often have limited generalizability.
In summary, the benefits of taking the CDM approach include: (1) the ability to answer diverse research questions that are infeasible using registry data alone; (2) facilitation of large-scale multicenter studies to provide more generalizable clinical evidence; (3) no requirement to exchange patient data between transplant centers to conduct multicenter studies, thereby safeguarding patient privacy; (4) timely improvement in the practice of evidence-based clinical care. Nevertheless, there are limitations of taking the CDM-based approach, namely the potential loss of data while transforming source data to the CDM format if source data elements or values do not have the corresponding values in the target CDM19. However, previous studies have shown that the transformation can be done with minimal data loss20. Table 1 presents a comparison between a registry-only approach and a CDM-based approach.
Table 1.
Comparison of Registry-only Approach vs. CDM-based Approach
| Registry-only Approach | CDM-based Approach | |
|---|---|---|
| Data Time Points | Difficulty in capturing longitudinal data points | Can capture more frequent data points from various sources |
| Data Elements | Data elements restricted to existing registry elements | Can capture various data elements from various data sources (e.g. EHR, claims, etc.) |
| Outcome Status | Dependent upon data compilation: delayed release of outcomes reports | Rapid institutional access to outcomes status report |
The first consideration when choosing the optimal CDM for the transplant domain is whether to build a novel data model (“ground-up” approach) or modify an existing data model (“modification” approach). Using a “ground-up” approach allows us to design an ideal data model that fits the needs of the transplant community. However, this approach is time-consuming, limits the ability to integrate unplanned data elements from outside of transplant, and potentially leads to a data model that is too complex or cannot be easily integrated or applied to other data sources beyond those in transplant21. Although a ‘modification’ approach requires the customization of an existing data model, it is more time-efficient and has the potential benefit of a larger active user community that might have already addressed data integration issues faced within the clinical domain while also ensuring that data elements not unique to transplant have already been considered and included21. As a result, using a widely-accepted CDM and adding missing elements that would be necessary to study transplant-specific factors is the preferred approach.
The Observational Medical Outcomes Partnership (OMOP) CDM
Many existing CDMs are currently being used to conduct observational health research, including the Informatics for Integrating Biology and the Bedside (i2b2), Mini-Sentinel CDM, PCORnet CDM, and the Observational Medical Outcomes Partnership (OMOP) CDM. In the informatics domain, prior studies have compared existing CDMs to identify the optimal CDM for comparative effectiveness research (CER)15,21. Quality dimensions including whether the data model has the ability to accommodate the required data elements and domain; is extensible, scalable, and adaptable; is easily understandable; supports standard vocabularies; has an active community using and supporting the data model were used to evaluate the different CDMs15,21. These studies concluded that the OMOP CDM optimally satisfied the defined criteria and thus is the most appropriate model for CER studies15,21. The OMOP CDM’s data dictionary can be found in the Observational Health Data Sciences and Informatics (OHDSI) public wiki page22.
Using OHDSI for Transplant Outcomes Research
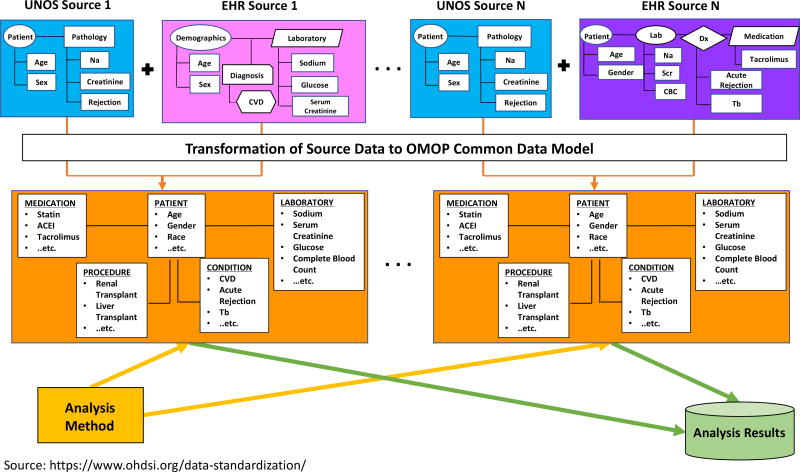
The OMOP CDM is now the data model adopted by the OHDSI collaborative23. OHDSI is a multi-stakeholder, multi-disciplinary collaboration whose goal is to generate evidence from large-scale, multi-site observational health data to promote better healthcare23. Over 140 researchers from 16 countries collaborate in the OHDSI community24. To achieve OHDSI’s goal, the international network of investigators that own observational health data have converted their clinical data to the OMOP CDM format23. Currently, there are over 650 million patient records within the OHDSI network, and several academic medical centers within the United States have converted their local EHR data into the OMOP CDM25–27. One advantage of joining the OHDSI network is that the analytic methods developed on top of the OMOP CDM by a single institution can be shared with other institutions within the network since they all have adopted the OMOP CDM. Another advantage of participating in OHDSI is that every resource is “open source,” meaning that all the related resources of OHDSI, including programming code and analysis results, are publicly available at no cost to any organization or individual23. Tools ranging from cohort creation to data characterization to novel statistical methods are just some of the examples of these publicly available products23,28,29. Figure 1 shows how data standardization makes these large scale multi-institutional studies possible.
Figure 1. Data Standardization using OMOP CDM.
Data extracted from sources with differing methods of data organization are combined with UNOS data and transformed to a common model that allows easy compilation and comparison of data from different centers for outcomes analyses. As we can see in the figure, the layout and shape that contains data are different to show the distinct structure between databases, and wordings for clinical concepts are different to show that disparate information systems represent the same clinical concepts differently. Once the disparate data sources are transformed to the OMOP CDM, the structure and clinical concept representation are standardized regardless of which data source you are dealing with. This allows institutions to apply the same analysis methods and aggregate results.
The aforementioned study by Hripcsak et al. was in fact produced by leveraging the OHDSI research network to create clinical evidence17,18. Similarly, collaboration between several national and international transplant centers will make large-scale studies possible in terms of both the number of subjects and the number of data elements23,17. For example, studies that address the racial variation in transplant outcomes can be conducted internationally to delineate optimal subgroup-specific treatment pathways or outcomes disparities.
Leveraging the OMOP CDM, OHDSI technology tools, and statistical methods would quickly facilitate the creation of a wide transplant-centered research network. As previously mentioned, the Healthcare Insurance Portability and Accountability Act (HIPAA) and patient privacy concerns can be major stumbling blocks to data aggregation across centers but are overcome in OHDSI by the use of a distributed model where data owners can store their data locally and apply the same query authored at one institution to all databases that follow the OMOP CDM format23,17. The results can then be aggregated centrally by the coordinating center, encompassing the results from all participating centers with no risk of breach of patient privacy23,17. This also facilitates compliance with the myriad of rules that govern sharing patient data across center, state, and national boundaries.
The OHDSI community, which includes national experts in clinical natural language processing (NLP), has enhanced the OMOP CDM to support NLP data from clinical text, which is important given that adding clinical unstructured data has been shown to improve the performance of transplant outcomes analyses4,30. Moreover, the OMOP CDM is now being used in many different national NIH-funded initiatives, such as the Electronic Medical Records and Genomics (eMERGE) Network and the All of Us research program (formerly known as the Precision Medicine Initiative)26,27. Both eMERGE, which aims to combine genomic and EHR data and facilitate the sharing of phenotypic algorithms, and the All of Us research program, which aims to accelerate precision medicine research, use the OMOP CDM to store participants’ clinical data26,27.
By leveraging tools and infrastructure from OHDSI, the transplant community can analyze large amounts of observational data and varied outcomes of interest. The transparency of the OHDSI tools, methodologies, and active international community would distinguish our transplant research framework from other existing efforts. We envision that an OHDSI-based, transplant research framework will be able to generate evidence to improve clinical care.
Implementation Cost, Feasibility, and Challenges
CDM implementation requires a team composed of the following roles: a CDM expert, a local data expert, a data engineer, clinician, and a business stakeholder. One or two technical personnel can take the role as a CDM expert, local data expert, and data engineer. Most importantly, a clinical steward and transplant center leadership support would be needed. The actual costs of implementing the CDM approach would therefore be the salary of these technical and clinical personnel; however, this can be shared across departments and may even be mitigated if other departments have already established an OMOP CDM. The software for OHDSI is open-source, so no additional software costs are incurred.
Implementing the CDM is technically feasible, but the biggest challenge would be transplant center motivation. Strong leadership by the clinical steward would therefore be required for the success of this transformation. Transplant centers need to keep in mind that taking the CDM approach will build an ecosystem that enables innovative research. Transforming the data sources into the OMOP CDM, as already said, would provide more data elements and data points to enrich the type of studies that can be conducted, enable multicenter trials which can produce generalizable results, and reduce the time spent to conduct large-scale studies. Additionally, the centers can derive benefits such as quality control, improved patient outcomes, and decreased healthcare cost by integrating different data sources.
Discussion
We propose the implementation of the OMOP CDM by transplant centers in order to provide analytic opportunities that extend well beyond what is feasible with current registry data. There are many strengths to our proposed approach. First, better risk adjustment models for assessing transplant center performance and patient outcomes would be achievable. For example, data elements such as cardiovascular risk burden, as measured by prior cardiovascular events or hypercoagulable conditions are not currently captured in the UNOS registry. Inclusion of such parameters would not only add to the model performance, but also perhaps remove the adverse incentive of transplant centers having to avoid transplanting patients with these unmeasured risk factors.
Second, augmenting UNOS data with the EHR will afford novel research opportunities. Questions focused on work flow or process measures such as referral rates, time to wait-listing, center level patient and organ selectivity, and quality of life measures among others would all potentially be available to study across centers. The study of variation in clinical practice is an essential step to identifying strategies and clinical approaches that are associated with the best outcomes.
Third, large scale studies to measure the impact of clinical variables on outcomes that are not in the current UNOS dataset would no longer be prohibitively expensive. With several academic medical centers already using OMOP for other research purposes, the effort to include transplant-specific data elements, which might already exist in some centers, would be minimal, allowing for such evaluations. For transplant centers without an OMOP CDM, converting EHR data may not be challenging. Current OHDSI members have shared their knowledge of converting their EHR data into OMOP. Most major EHR vendors are represented within the OHDSI community, making it relatively easy for transplant centers to adopt the OMOP CDM25. In fact, OHDSI working groups are currently developing guidelines on populating a CDM, dealing with conflicting data, and measuring data quality.
Finally, CDM-based tools either for quality or compliance monitoring internal to an institution could also be easily shared, allowing for quality assessment/monitoring. Additional tools could be disseminated easily across the transplant community, since each tool would have been developed leveraging the same CDM. In fact, the OHDSI community has designed tools to allow such dissemination in order to encourage reproducibility of research results.
Though there are these advantages of adopting the OMOP CDM and the OHDSI tools, there are competing factors worth considering. Adopting the OMOP CDM for a new site cannot be done without concerted effort. The transplant center will need institutional support in the terms of information technology resources and infrastructure. For example, an institution’s IT department would need to provide a server to host a database, access to EHR data, and analysts. Also, the integration of UNOS data into the OMOP CDM requires not only the effort of those who implement the integration but also the support from the community. We would need support from the transplant community and encourage more transplant centers to participate in this journey. Without the participation of multiple transplant centers, the goal of conducting a large-scale study with national and global transplant patients cannot be achieved. Currently, we are working on mapping key transplant-specific data found in UNOS into the OMOP CDM and will be releasing it to the public. We expect that this will be the first step in building a large research network among transplant centers and enable the identification of transplant cohorts aggregated from multiple institutions, thereby making it feasible for robust, large analyses that have been impossible until now.
Acknowledgments
This work was supported by the Laura and John Arnold Foundation (S.M.), American Society of Transplant Surgeons, American Society of Transplantation and Immunology Research Network (TIRN) (to S.M.), National Institute of Diabetes and Digestive and Kidney Diseases (R01DK11489301 and grant U01DK11606601) (to S.M.), and Health Resources and Services Administration (contract 234-2005-37011C). This work was also supported by the U.S. National Library of Medicine under Training Grant 2T15LM007079-26 (to S.C.).
Abbreviations
- ACC
American College of Cardiology
- AHA
American Heart Association
- CER
comparative effectiveness research
- CDM
common data model
- CMS-ESRD
Centers for Medicare and Medicaid Services – End Stage Renal Disease
- EHR
electronic health record
- eMERGE
Electronic Medical Records and Genomics
- ESRD
end-stage renal disease
- HIPAA
Healthcare Insurance Portability and Accountability Act
- i2b2
Informatics for Integrating Biology and the Bedside
- NCHS
National Center for Health Statistics
- NDI
National Death Index
- NIH
National Institutes of Health
- NLP
natural language processing
- OHDSI
Observational Health Data Sciences and Informatics
- OMOP
Observational Medical Outcomes Partnership
- OPO
organ procurement organization
- PSR
program-specific reports
- SEER
Surveillance, Epidemiology, and End Results
- SRTR
Scientific Registry for Transplant Recipients
- SSDMF
Social Security Death Master File
- TIEDI
Transplant Information Electronic Data Interchange
- UNOS
United Network for Organ Sharing
- USRDS
United States Renal Data System
Footnotes
Disclaimer
The content is the responsibility of the authors alone and does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.Grinyó JM. Why is organ transplantation clinically important? Cold Spring Harb Perspect Med. 2013;3(6):a014985. doi: 10.1101/cshperspect.a014985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.United Network for Organ Sharing. [Accessed February 17, 2017];US organ transplants, deceased donors set record in 2016. https://www.unos.org/us-organ-transplants-set-record-in-2016/. Published 2017.
- 3.Lin H-M, Kauffman HM, McBride MA, et al. Center-specific graft and patient survival rates: 1997 United Network for Organ Sharing (UNOS) report. JAMA. 1998;280(13):1153–1160. doi: 10.1001/jama.280.13.1153. [DOI] [PubMed] [Google Scholar]
- 4.Srinivas TR, Taber DJ, Su Z, et al. Big data, predictive analytics and quality improvement in kidney transplantation- a proof of concept. Am J Transplant. 2016 Nov; doi: 10.1111/ajt.14099. [DOI] [PubMed] [Google Scholar]
- 5.Krumholz HM. Outcomes research. Circulation. 2008;118(3):309–318. doi: 10.1161/CIRCULATIONAHA.107.690917. [DOI] [PubMed] [Google Scholar]
- 6.Ladner DP, Alonso EM, Butt Z, et al. NUTORC—a transdisciplinary health services and outcomes research team in transplantation. Transl Behav Med. 2012;2(4):446–458. doi: 10.1007/s13142-012-0176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard RJ, Cornell DL, Schold JD. CMS oversight, OPOs and transplant centers and the law of unintended consequences. Clin Transplant. 2009;23(6):778–783. doi: 10.1111/j.1399-0012.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- 8.Schold JD, Phelan M, Buccini LD. Utility of Ecological Risk Factors for Evaluation of Transplant Center Performance. Am J Transplant. 2016 doi: 10.1111/ajt.14074. [DOI] [PubMed] [Google Scholar]
- 9.American Society of Transplant Surgeons. [Accessed November 30, 2017];RAPID: Real-time Analytics and Process Improvement Dashboard. http://asts.org/resources/knowledge-base/rapid-real-time-analytics-and-process-improvement-dashboard#.WiAdk7Q-fUo.
- 10.American Society of Nephrology. [Accessed November 30, 2017];Data Harmonization in Kidney Transplant. https://www.asn-online.org/khi/project.aspx?ID=52.
- 11.Dickinson DM, Ellison MD, Webb RL. Data sources and structure. Am J Transplant. 2003;3(s4):13–28. doi: 10.1034/j.1600-6143.3.s4.3.x. [DOI] [PubMed] [Google Scholar]
- 12.Massie AB, Kuricka LM, Segev DL. Big data in organ transplantation: registries and administrative claims. Am J Transplant. 2014;14(8):1723–1730. doi: 10.1111/ajt.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatti NK, Karimi Galougahi K, Paz Y, et al. Diagnosis and Management of Cardiovascular Disease in Advanced and End-Stage Renal Disease. J Am Heart Assoc. 2016;5(8) doi: 10.1161/JAHA.116.003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lentine KL, Costa SP, Weir MR, et al. Cardiac disease evaluation and management among kidney and liver transplantation candidates: a scientific statement from the American Heart Association and the American College of Cardiology Foundation: endorsed by the American Society of Transplant Surgeons, American Society of Transplantation, and National Kidney Foundation. Circulation. 2012;126(5):617–663. doi: 10.1161/CIR.0b013e31823eb07a. [DOI] [PubMed] [Google Scholar]
- 15.Ogunyemi OI, Meeker D, Kim H-E, Ashish N, Farzaneh S, Boxwala A. Identifying appropriate reference data models for comparative effectiveness research (CER) studies based on data from clinical information systems. Med Care. 2013;51:S45–S52. doi: 10.1097/MLR.0b013e31829b1e0b. [DOI] [PubMed] [Google Scholar]
- 16.Embi PJ, Kaufman SE, Payne PRO. Biomedical informatics and outcomes research. Circulation. 2009;120(23):2393–2399. doi: 10.1161/CIRCULATIONAHA.108.795526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hripcsak G, Ryan PB, Duke JD, et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci. 2016;113(27):7329–7336. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duke JD, Ryan PB, Suchard MA, et al. Risk of angioedema associated with levetiracetam compared with phenytoin: Findings of the observational health data sciences and informatics research network. Epilepsia. 2017;58(8) doi: 10.1111/epi.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rijnbeek PR. Converting to a Common Data Model: What is Lost in Translation? Drug Saf. 2014;37(11):893–896. doi: 10.1007/s40264-014-0221-4. [DOI] [PubMed] [Google Scholar]
- 20.Matcho A, Ryan P, Fife D, Reich C. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug Saf. 2014;37(11):945–959. doi: 10.1007/s40264-014-0214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahn MG, Batson D, Schilling LM. Data model considerations for clinical effectiveness researchers. Med Care. 2012;50 doi: 10.1097/MLR.0b013e318259bff4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reich C, Ryan P, Belenkaya R, Natarajan K, Blacketer C. [Accessed January 30, 2018];OMOP Common Data Model v5.3 Specifications. https://github.com/OHDSI/CommonDataModel/wiki.
- 23.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574. [PMC free article] [PubMed] [Google Scholar]
- 24.Banda JM, Halpern Y, Sontag D, Shah NH. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Summits Transl Sci Proc. 2017;2017:48. [PMC free article] [PubMed] [Google Scholar]
- 25.Observational Health Data Sciences and Informatics. [Accessed December 9, 2017];OHDSI Collaborators. https://www.ohdsi.org/who-we-are/collaborators/
- 26. [Accessed November 6, 2017];About the All of Us Research Program. https://allofus.nih.gov/about/about-all-us-research-program.
- 27. [Accessed November 6, 2017];Electronic Medical Records and Genomics (eMERGE) Network. https://emerge.mc.vanderbilt.edu/
- 28.Observational Health Data Sciences and Informatics (OHDSI) [Accessed December 7, 2017];Propensity Score Evaluation. https://github.com/OHDSI/PropensityScoreEvaluation.
- 29.Observational Health Data Sciences and Informatics (OHDSI) [Accessed December 7, 2017];Patient Level Prediction. https://github.com/OHDSI/PatientLevelPrediction.
- 30.Observational Health Data Sciences and Informatics (OHDSI) [Accessed December 9, 2017];OHDSI Natural Language Processing Working Group. http://www.ohdsi.org/web/wiki/doku.php?id=projects:workgroups:nlp-wg.