Abstract
An ongoing technological revolution is continually improving our ability to carry out very high- resolution studies of gene expression patterns. Current technology enables the global gene expression profiles of single cells to be defined, facilitating dissection of heterogeneity in cell populations that was previously hidden. In contrast to gene expression studies that use bulk RNA samples and provide only a virtual average of the diverse constituent cells, single-cell studies allow the molecular distinction of all cell types within a complex population mix, such as a tumour or developing organ. For instance, single-cell gene expression profiling has contributed to improved understanding of how histologically identical, adjacent cells make different differentiation decisions during development. Beyond development, single-cell gene expression studies have enabled the characteristics of previously known cell types to be more fully defined and facilitated the identification of novel categories of cells, contributing to improvements in our understanding of both normal and disease-related physiological processes and leading to the identification of new treatment approaches. Although limitations remain to be overcome, technology for the analysis of single-cell gene expression patterns is improving rapidly and beginning to provide a detailed atlas of the gene expression patterns of all cell types in the human body.
The gene expression pattern of a cell defines its protein components. Essentially, all cells of the human body contain the same set of ~20,000 genes, but different cells express different sets of these genes, leading to between-cell differences in the expression of membrane components, ion transporters, cytoskeletal elements, growth factors, receptors and transcription factors. The gene expression profile therefore describes in exquisite detail the phenotype of a cell, which underlies its molecular functions.
Historically, gene expression studies have been lim-ited to the analysis of pooled populations of cells, which was necessary to obtain sufficient RNA for analysis. For example, the combined expression pattern of all cells of a tumour would be examined in aggregate to identify perturbed molecular pathways. However, a tumour contains a heterogeneous population of cells, including vascular cells, fibroblasts, invading immune cells and rapidly dividing cancer cells as well as more quiescent cancer stem cells, and the resulting gene expression profile of a pooled population of tumour cells therefore provides only an ensemble average of the cell types present. Analysis of pooled cell populations does not enable identification of the cell types that express certain genes but instead provides a virtual average of the multiple cellular components, which may well say very little about any specific cell type present. Similar problems are encountered when pooled populations are used to assess gene expression associated with other disease conditions.
Cell heterogeneity is also a feature of organ development, wherein progenitor cells that are often histologically indistinguishable undergo diverse differentiation decisions to become specific cell types. Analysis of the gene expression of pooled populations of progenitor cells does not enable distinction of the signals that drive a progenitor down a particular differentiation pathway; for instance, the signals that determine whether a nephron progenitor cell becomes a podocyte or a proximal tubule cell. To better understand the signals that drive cell differentiation and cell fate decisions, developmental biologists need to define the early gene expression events associated with the lineage choice of individual cells.
The past decade has witnessed powerful technological advances, enabling gene expression analysis to be carried out at much higher resolution than previously possible. Indeed, the expression level of every gene, even in a single cell, can now be defined. This technology, known as single- cell RNA sequencing (scRNA- seq), enables rapid determination of the precise gene expression patterns of tens of thousands of individual cells. Such analysis of the constituent parts — the single cells — provides much more meaningful insights into cell behaviour than analysis of aggregated blocks. For example, scRNA- seq of tumour cells can enable separation of tumour fibroblasts from the endothelial and cancer cells on the basis of their gene expression signatures. Moreover, each cell type can be further divided into subtypes; for example, tumour fibroblasts might be separated into fibroblast subtypes1. Such single- cell studies have also enabled identification of previously unknown cell types2–4 and have provided insights into the heterogeneity of non- cancer cell populations within tumours5, highlighting the power of this research tool.
This technological revolution is providing stunning new insights into the underlying mechanisms of organ development and disease. Just as the Human Genome Project previously defined our full complement of genes, several Human Cell Atlas Projects, including the Human Cell Atlas project6, the Human BioMolecular Atlas Program and the Chan Zuckerberg Initiative Atlas Project, are currently underway, each devoted to defining how each cell type in the body makes differential use of this set of genes. Application of these new tools to the analysis of normal, developing and diseased tissue will enable a much deeper understanding of the human body by providing insights into how a single cell, the zygote, develops into a complete person and how perturbed molecular pathways and processes can lead to disease. In this Review, I discuss the fundamental concepts of single- cell RNA analysis, including discussion of the experimental strategies, and the strengths and weaknesses of different technologies. I also describe specific applications of the technology for the study of development, cancer and normal and diseased kidneys.
Methodological overview
Principle of RNA sequencing
RNA sequencing — whether of single cells or of pooled populations of cells — is a powerful method for analysing gene expression patterns and involves the reverse transcription of RNA into cDNA, which then undergoes high- throughput DNA sequencing. Genes that are highly expressed within a sample produce more RNA, more cDNA and more DNA sequence reads than genes that are more weakly expressed. Thus, RNA sequencing provides a digital readout of gene expression, with the number of DNA sequence reads aligning with the expression level of a particular gene in a sample.
The concept of scRNA- seq is the same, except that single cells must first be isolated, and owing to their minute RNA contents, a powerful amplification process is required to generate sufficient cDNA for sequencing (Fig. 1). As with traditional RNA sequencing, a higher number of DNA sequence reads for a specific gene corresponds to higher expression of that gene within the cell. Remarkably, current scRNA-seq approaches enable expression levels of all genes to be defined.
Fig. 1 |. General strategy for scRNA- seq.

First the organ or tissue of interest is dissociated to make a single-cell suspension. Single cells are then captured for single-cell RNA sequencing (scRNA- seq) analysis. The single cells are then lysed, and the RNA is reverse transcribed to synthesize cDNA , which must then be amplified, often by PCR , to make sufficient material to generate cDNA libraries for sequencing. The resulting sequence reads are assigned to cells via cell- specific barcodes incorporated into the cDNA through the primers used for reverse transcription and are aligned to specific genes.
High- throughput single-cell sequencing
A number of scRNA-seq methods allow high-throughput analysis of large numbers of cells. The introduction of the Fluidigm C1 microfluidics system in 2012 revolutionized scRNA-seq, providing gene expression data for up to 96 cells in a single run, over a time frame of ~1 day. High-throughput7 Fluidigm IFC chips, introduced more recently in ~2015, can examine up to 800 cells at once. Following cell lysis, reverse transcription and amplification in microchambers, cDNA libraries are produced that are tagged with a cell- specific barcode, which enables the resulting DNA sequence reads to be assigned to specific cells. This approach provides high- quality gene expression readouts but is relatively expensive per cell compared with other available methods.
Microdroplet approaches.
The most popular current scRNA- seq methods use microdroplets in place of microchambers6–8 (Fig. 2). Use of microfluidics technology enables hundreds of thousands of microdrops to be inexpensively generated. These aqueous microdrops, surrounded by oil, have a volume of ~2 nanolitres and contain a bead with a uniquely barcoded set of oligonucleotides and a single cell. Following cell lysis, the bar-coded oligonucleotides hybridize to the polyA tails of the released mRNA. For Drop-seq, the beads, along with attached oligonucleotides and annealed mRNAs, are all released from the drops and combined into a single tube, and reverse transcription is then carried out. For the Chromium and InDrop technologies, the hydrogel beads dissolve in the microdrops, releasing the oligo-nucleotides to hybridize to the mRNAs, and the reverse transcription reactions are carried out within the drops. In either case, the bead- specific barcode is incorporated into the cDNA, thereby enabling the subsequent DNA sequence reads to be aligned with a specific cell. All the microdroplet-based systems allow the beads8 or cDNAs9,10 to be pooled and processed through subsequent reactions together, minimizing labour and reagent costs. In fact, using these methods, RNA sequencing (RNA- seq) data, including DNA sequencing costs, can be generated for approximately US$1 per cell. A cross-platform comparative study of Drop- seq, Fluidigm 800 cell IFC and the Chromium system found that they performed comparably11, with each technology dividing mouse embryonic kidney cells into similar clusters with closely overlapping sets of markers. Nevertheless, both microdroplet technologies are considerably less expensive per cell than the Fluidigm method.
Fig. 2 |. Microdroplet- based scRNA- seq.

A microfluidics system is used to make microdroplets, which contain cells mixed with beads that are encapsulated in oil. Each bead has oligonucleotides that are uniquely barcoded for that bead and are in a solution that contains a mild detergent, which lyses the cells after mixing. The RNAs from the lysed cell anneal to the bead oligonucleotides, and subsequent reverse transcription incorporates the bead- specific barcode into the cDNA , thereby allowing the sequences of those cDNAs to be assigned to a specific cell. scRNA-seq, single- cell RNA sequencing.
The Chromium microdrop system, mentioned above, offers some advantages over the Drop-seq method. First, the data obtained by the Chromium system are somewhat higher quality, as it detects more genes per cell than the Drop-seq method. This greater sensitivity, with reduced technical noise, is particularly important when trying to separate very closely related cell types because noisy data can conceal very subtle differences in gene expression patterns (see below). Second, the Chromium system provides gene expression profile data for a much higher percentage of input cells. With the Chromium system, almost every cell finds itself in a microdrop that includes a bead, but with Drop-seq, the vast majority of cells end up in drops without a bead and therefore go unseen. This is because for the Drop- seq method, both the barcoded beads and the cells become incorporated into drops in a random fashion. The beads and cells must therefore be sufficiently diluted to prevent drops from receiving two beads or two cells. Two cells together with a bead in a single drop would result in the transcripts from both cells having the same bead barcode and hence assigned to a single cell. A somewhat lesser problem, two beads in one drop with one cell, would end up giving the transcripts from one cell to two distinct barcodes, thereby counting that cell twice. Therefore, Drop- seq protocols typically aim to incorporate about one cell in every 20 drops, so that only one drop in 400 on average receives two cells. Moreover, the bead concentration of the majority of Drop- seq protocols gives at most one bead per ten drops. Thus, this system suffers from double Poisson distribution; the net result, combined with normal bead loss during handling, is that only ~5% of Drop- seq input cells give RNA- seq data. By contrast, almost all drops generated by the Chromium system have a single bead, as the large hydrogel beads used by this system can be loaded at very high, back- to-back concentrations. By carefully adjusting the bead input flow rate, it is possible to place one bead in each drop. The system still suffers from a single Poisson distribution with the potential for two cells to be incorporated in a single drop; however, almost every cell finds itself in a drop with a bead. The net result is that over 50% of input cells into the Chromium system give rise to RNA- seq data, about a tenfold higher success rate than that of Drop- seq. This difference is particularly important when the number of starting cells is limited. Another advantage of the commercial Chromium system over the Drop-seq method is the ease of its set-up and operation. The main disadvantage of the Chromium system compared with Drop- seq is the considerably higher cost of reagents.
Non-microfluidics approaches.
In addition to the high-throughput microfluidics methodologies described above, scRNA- seq can also be carried out in microwell plates by performing RNA- seq on cells isolated by simply picking them or via fluorescence- activated cell sorting (FACS). A variety of amplification chemistries can be used with the microtitre plate format, including SMART- seq2 chemistry12, Cel- seq13, MARS- seq14 and STRT- seq15. SMART- seq2 and STRT- seq amplify cDNA products of reverse transcription with PCR, whereas Cel- seq and MARS-seq incorporate a bacterial virus promoter into the cDNA, which is then amplified by in vitro transcription. These microtitre0 plate methods are not restricted by cell- size constraints of the Fluidigm system, which requires different micro-fluidic devices for cells of different sizes. Although microdroplet methods are less sensitive to cell size than the Fluidigm method, as the droplets are very large compared to cells, they are prone to clogging during the generation of microdroplets with certain cell types, like neurons and muscle cells. In addition, the equipment requirements and set-up costs are minimal for microtitre plate methods. Approaches that enable full coverage of cDNA sequencing, for example, SMART-seq2, can also facilitate analysis of alternative splicing patterns, enabling the identification of distinct transcripts with potentially very different functions. By contrast, other scRNA- seq methods, such as Drop-seq, InDrop, Chromium and 800 cell IFC Fluidigm, provide sequence information only for the 3′ or 5′ ends of the cDNAs and not the entire transcript, and therefore cannot be used for the analysis of alternative splicing patterns. Of interest, a 2017 modification of the STRT- seq system, termed STRT- seq-2i, uses a 9,600 microwell array platform, allows microscopic confirmation of single cells and generates high-quality scRNA- seq data at a competitive cost of ~$1 per cell, including DNA sequencing16.
CytoSeq is another powerful scRNA-seq technology17, whereby single cells are allowed to settle by gravity into microwells, upon which a bead suspension is then placed at saturation concentration so that each well receives a bead. The beads, similar to Chromium and Drop-seq technologies, carry uniquely barcoded oligonucleotides to prime reverse transcription and are sized so that only one can fit in a microwell. Excess beads are removed, the cells are lysed and the mRNA from each cell then hybridizes to the bead oligonucleotides, similar again to the Chromium and Drop- seq technologies. The advantages of this system are that it can be used without the complicated microfluidics systems required for Fluidigm, Drop-seq and the Chromium systems. It is also easily scalable by using plates with more microwells. A variant of this method, Microwell-seq, was used to carry out scRNA-seq on over 400,000 mouse cells to create a single-cell atlas for most major organs, including the kidney18.
Yet another scRNA- seq technology, SPLiT- seq, further reduces equipment requirements and can cut the cost of library construction for sequencing to $0.01 or less for each cell19. With this method, the cells themselves, following gentle fixation and permeabilization, serve as microdrops. The single cells are randomly divided into the wells of a 96-well plate, and the reverse transcription reaction is carried out within the cells with well- specific primers. The barcodes are ligated onto the cDNAs in a sequential and combinatorial fashion. All the cells in a single well of the 96-well microtitre plate have a short well- specific oligonucleotide sequence ligated to all their cDNAs. The cells of the entire microtitre plate are then pooled into a single tube, mixed thoroughly and randomly distributed into the wells of yet another 96-well plate. Once again, short well-specific oligonucleotides are ligated to the cDNAs to further contribute to the barcodes. Three rounds of this pooling and splitting process, followed by a fourth round with 24 independent PCRs, gives a total of 21,233,664 possible barcode combinations. All the cDNAs within one cell travelled the same path during the split–pool process of barcoding; therefore, they all have the same cell-specific barcode. The resulting scRNA- seq data quality seems similar to that of the Chromium and Drop- seq methods. The major advantage of SPLiT-seq is therefore its low cost for generating cDNA libraries; at present, however, the major cost component of scRNA-seq is the DNA sequencing. As DNA sequencing costs continue to drop, the inexpensive library construction costs of technologies such as SPLiT-seq will become increasingly important and could facilitate the scRNA-seq analysis of very large numbers of cells, perhaps even billions.
Experimental design
scRNA- seq experiments must be designed carefully, with consideration of numbers of cells, reads per cell and biological replicates. Each of these variables contributes to the final statistical power of the analysis. Higher numbers of each component (that is, cells, readouts and biological replicates) are always better than lower numbers, but return must be optimized for money spent. Different technologies will have different thresholds for the maximum number of sequence reads per cell, beyond which additional sequencing provides little additional information. The choice of technologies should depend on the degree of similarity of cell types being analysed. In attempting to resolve extremely similar cell types, it might be necessary to use a more expensive system that gives a higher resolution of gene expression patterns. The degree of biological variability, and hence the number of biological replicates required, will be experiment- dependent. Consultation with a statistician is recommended as there is no one size fits all solution to these questions.
Challenges of single-cell studies
Working with limited material
The primary challenge for single- cell studies is the very small amount of material available to work with. Each cell contains only ~10 picograms of total RNA on average, of which only ~0.1 picograms is mRNA20. For many genes, there are only tens of transcripts per cell. All the scRNA- seq technologies described above therefore require an amplification step to generate sufficient cDNA for sequencing from the picogram amounts of RNA in a single cell. Unfortunately, cDNA amplification is never perfectly linear, leading to disproportional representation of all cDNAs present in a cell. To help overcome this problem, almost all current scRNA- seq technologies incorporate a unique molecular identifier (UMI) into the primer oligonucleotide used for reverse transcription. For example, the oligonucleotides attached to the beads used for Drop- seq each have an identical 12-base bead- specific barcode that allows the eventual DNA sequence reads to be aligned to the single cell in the drop with that bead. In addition, however, every oligonucleotide on each bead has an 8-base UMI barcode that is different for every oligonucleotide. The presence of these UMI barcodes means that all the DNA sequence reads with the same UMI can be compressed to a single hybridization event between an mRNA and a specific oligonucleotide. The ability to count the initial cDNA product of reverse transcription rather than all the amplicons avoids bias in our interpretation of gene expression levels if, for example, cDNA from one hybridized mRNA amplifies poorly owing to high GC content, whereas cDNA from another hybridized mRNA amplifies particularly well. This compression step during data deconvolution can remove most amplification- based distortions of the gene expression data.
Capturing single cells
Another challenge of scRNA-seq is the process of breaking down an organ or tissue into single cells for analysis. One approach is simple micromanipulation21, for example, using forceps or micromanipulators, a micropipette and a microscope, to isolate single cells from histological sections or an early embryo. This approach can be effective and ensures that each sample is indeed a single cell, but it is labour intensive and low throughput22,23. Laser capture microdissection (LCM) can also be used to isolate single cells by using a laser beam to excise cells from cryostat sections24,25. An advantage of this approach is that it can also provide some spatial information for the cell of interest, but like micromanipulation, it is low throughput and labour intensive. In addition, it is technically challenging to use LCM to capture the contents of a single cell cleanly, without any contamination from flanking cells and without damaging the cell RNA. If the cell type of interest is quite rare, then FACS can be used for enrichment. FACS is high throughput and can also be used to place single cells in 96-well plates, which is an important first step for some scRNA-seq methods.
Another approach takes advantage of the very high-throughput nature of currently available scRNA- seq technologies by simultaneously examining massive numbers — up to millions — of single cells dissociated from the tissue of interest. In this manner, every cell type present in the starting tissue is examined, unless it is exceptionally rare. This approach is also marker free and does not require cell- specific tags, for example, green fluorescent protein (GFP) for FACS- based cell selection, and can be applied to almost any starting tissue. Despite the many advantages of this approach, the fact that all spatial information is lost during the cell dissociation process presents a challenge. Reconstruction of a three-dimensional single cell-resolution gene expression atlas with this approach therefore requires a second step whereby cell types are bioinformatically assigned spatial positioning on the basis of their known gene expression signatures. In the developing mouse or human kidney, for example, cap mesenchyme progenitor cells can be identified by their expression of Cited1, Six2, Eya1 and Osr1; stromal progenitor cells can be identified by the expression of Foxd1; podocytes by Mafb; and proximal tubule cells by Hnf4a. Spatial positioning of newly identified cell types requires in situ hybridization or immunofluorescence studies. Such analyses can produce a remarkable virtual organ, with all transcription factors, growth factors, other secreted proteins and receptors fully defined for every cell type (Fig. 3). This strategy has been used, for example, to create a virtual embryo of the genes expressed by cells within developing Drosophila larvae, providing a powerfully useful tool for the research community26.
Fig. 3 |. Creation of a single-cell-resolution virtual organ.

The organ of interest, which could be normal, mutant or diseased, is first subjected to dissociation to generate a single- cell suspension. Many thousands of the single cells are then used for single- cell RNA sequencing (scRNA- seq) gene expression profiling and the resulting data analysed to define cell types, which are then spatially assembled to reconstruct a virtual organ. The virtual organ includes all cell types and provides a complete gene expression pattern for each cell, thereby defining the expression of transcription factors, growth factors, receptors and potential pathogenic pathways that contribute to disease. Plot of dissociated cells adapted from Development, 144, Adam, M. et al. Psychrophilic proteases dramatically reduce single-cell RNA- seq artefacts: a molecular atlas of kidney development (2017), with permission from Elsevier.
A difficulty with the above- described approach is that the enzymes commonly used for tissue dissociation, including trypsin, collagenase, TrypLE, dispase, liberase and pronase, all require incubation at 37° C. This is also the temperature at which the mammalian enzymes within the cells of interest, including the transcriptional machinery, are maximally active. We therefore expect that the cells will change their gene expression patterns in response to the foreign environment during the dissociation process. Indeed, early response genes, including multiple members of the FOS and JUN families, show dramatic elevations in gene expression after only a few minutes of dissociation at 37°C (ReF.27). Therefore, gene expression artefacts of the cell separation process almost certainly taint, in some measure, most scRNA- seq data sets generated to date.
One way to minimize these dissociation artefacts is to carry out RNA- seq on nuclei instead of whole cells. Nuclei can be prepared using mechanical homogenization, such as with a Dounce homogenizer, on ice. Sequencing of nuclear RNA has been used, for example, to analyse gene expression in autopsy samples, where dead and broken cells have leaked out cytoplasmic RNA, and for muscle cells or neurons that are too large to use with the Fluidigm C1 system28–32. The major disadvantage of nuclear RNA sequencing is that only a small fraction of cellular RNA (generally 10–20%) is in the nucleus, so technical noise (see below) is increased. Nuclear RNA analyses will also include a higher proportion of unprocessed transcripts that still contain introns, which can complicate analysis. In addition, some concern exists regarding the degree to which nuclear RNA content reflects that of cytoplasmic RNA. Despite these remaining challenges, this approach has enormous potential as a powerful tool for the analysis of gene expression of cells for which the isolation and analysis of cytoplasmic RNA is problematic.
In addition to nuclear RNA analysis, preservation of in vivo gene expression patterns during cell dissociation can be achieved through the use of transcription inhibitors. In particular, addition of α-amanitin — which inhibits transcription by RNA polymerase II — can reduce gene expression artefacts during cell dissociation33. One disadvantage of this approach is the slow cellular uptake of α- amanitin, which can take hours and requires the presence of specific transporters34,35. In addition, transcriptional inhibitors do not block RNA turnover, which also influences RNA content.
Another approach to minimize dissociation-induced artefacts in gene expression is to carry out the single-cell dissociation using cold- adapted proteolytic enzymes. In contrast to thermophilic organisms that can survive extreme heat, for example, deep-sea hydrothermal vents and other geothermal heated regions, psychrophilic, or cryophilic, organisms flourish in extreme cold. Just as we use DNA polymerase from thermophiles for PCR because it can survive the high- temperature step used to denature DNA, we can use proteases from psychrophilic microorganisms to carry out cell dissociation at near ice temperatures, thereby better preserving in vivo gene expression patterns27.
Of note, some cells in a tissue are more easily dissociated than others. Immune cells, for example, tend to be only loosely attached to their neighbours and are easily freed. On the other hand, podocytes are firmly attached to glomerular capillaries through their many tentacle-like extensions. Likewise, mesangial cells are securely embedded in extracellular matrix and are also notoriously difficult to dissociate. Equally important, some cells are more fragile than others and can be easily killed by rigorous enzymatic and/or mechanical disruption methods that are required to release difficult- to-dissociate cells. These points exemplify some of the challenges of organ dissociation, as the resulting single- cell suspension will rarely provide proportional representation of the many starting cell types.
Noise
Important limitations exist in the amount of information that can be obtained by scRNA- seq. One important limitation is a consequence of the biological noise that results from the pulsatile, bursting nature of gene expression36–38. Genes are not expressed in a steady-state manner but rather are actively transcribed in a sporadic fashion in short bursts, meaning that for a given cell, the levels of various gene transcripts are in a constant state of flux. This gene expression chatter is a consequence of the fundamental nature of gene expression and is unavoidable. A second limitation resides in the methodological measurement of transcript levels when starting with the limited amount of RNA present in single cells. Current estimates suggest that most scRNA- seq technologies detect only about 10–20% of the mRNA molecules that are actually present15,39. This poor sensitivity is therefore an area in need of improvement. Low levels of gene expression, in particular, are difficult to detect, which can be problematic given that the average gene expresses surprisingly few transcripts — generally only 10–30 — per cell. Inefficiencies in the reverse transcription and amplification steps therefore add a considerable layer of technical noise to single- cell gene expression studies. Twenty per cent of noise from scRNA- seq studies is estimated to be of biological origin, whereas the remaining 80% is thought to be the result of technical limitations40.
Given the noise associated with scRNA- seq, it is perhaps surprising that the gene expression profiles obtained using this technology are usually sufficient to allow unsupervised clustering of the cells. Unsupervised clustering allows the software analysis programme to search for patterns and similarities in groups of cells without any input from the investigator with regard to known marker genes expressed by the different cell types. Once similar cells are clustered, or grouped together, then all the gene expression data from all the cells of a single cell type are combined to obtain a robust measure of the gene expression profile. This cluster and combine strategy provides important data complementation (Fig. 4). Genes with low expression levels might be detected in only a small fraction of the cells of a cluster but would be seen in the aggregate. For example, a rare transcript present at only one copy per cell might be detected in only 10% of cells, but in a cluster of 100 cells, 10 cells would show expression of this gene. In the combined view of the cells in a cluster, this gene would be identified as being expressed. Enormous statistical power therefore exists in analysing large numbers of cells, and increasing the number of cells in each cluster improves the effectiveness of this complementation approach and the output of the analysis. Thus, perhaps unexpectedly, for single- cell studies, we examine single cells to avoid the ensemble average effect of pooling multiple distinct types of cells, but once the single cells are divided into clusters, the data from each group are pooled to give a more sensitive and complete representation of the gene expression pattern of that cell type.
Fig. 4 |. Use of cluster and combine methodology to define cell types.

Gene expression profiles for individual cells are noisy and incomplete, with the holes in each shape representing the missing information. In this example, there are three different cell types, as indicated by different colours and shapes. Although the data sets for each individual cell are incomplete, they are sufficient to allow clustering of similar cells into groups. The data for all cells within a group are then combined to give a robust view of that cell type. The gene expression information missing in one cell can be provided by other cells in that group. Thus, through complementation, each cell contributes something to the total picture, resulting in a combined profile of the cell type that is very complete, enabling detection of very low gene expression levels.
Data analysis
Identification of expressed genes.
The data sets obtained using scRNA-seq technologies are extremely challenging, partly owing to the combination of biological and technical noise. Owing to technical limitations, microdroplet- based technologies, such as Drop- seq and InDrop, typically detect only 1,000–3,000 genes expressed per cell, which is only a fraction of the number of genes actually expressed. Genes that are expressed but the transcripts for which are not detected for technical reasons are termed dropouts. Further, a gene is considered expressed even if only a single cDNA sequence read is obtained that aligns to that gene. Indeed, for many genes that are considered expressed, only a single read is observed, providing a very imperfect measure of gene expression level. The fact that scRNA- seq data are so remarkably useful despite these shortcomings is a tribute to the data analysis programmes that have been developed.
Analysis of gene expression patterns first requires the scRNA- seq data to be deconvoluted, with one of the paired DNA sequence reads assigned to a cell- specific barcode and the other aligned to a gene sequence, to pair the expression of particular genes to individual cells. As described earlier, many of the scRNA-seq techno logies, including Drop-seq and the Chromium system, use UMI barcodes that allow compression of sequence reads to individual barcoded oligonucleotides, enabling mRNA hybridization events to be counted and eliminating nonlinearities in subsequent amplification steps. The net result is a digital gene expression readout for each cell, including the total number of DNA sequence reads identified for every gene.
Quality control.
Quality control steps during data analysis include the removal of cells that have a low number of genes that are considered to be expressed in order to eliminate the weakest data sets. A typical threshold is a minimum of 500 genes identified as being expressed per cell8. In addition, cells with very high proportional readouts for mitochondrial DNA are removed, based on the assumption that these cells are likely to be dead as cells with a leaky cytoplasmic membrane lose cytoplasmic RNA, but not mitochondrial transcripts, which remain contained in the mitochondria. Red blood cells are also often removed based on their high haemoglobin expression.
Analysis strategies.
An important step in the analysis of scRNA- seq data is dimensionality reduction. The number of dimensions in a set of gene expression data is represented by the expression levels of all genes. If 2,000 genes are considered to be expressed in a cell, then there are 2,000 dimensions. Dimensionality reduction involves the use of various predictive model algorithms that transform high dimensional space to a space with fewer dimensions. Principal components analysis, for example, carries out a linear transformation, but many nonlinear dimensionality reduction methods exist8,41–43.
Reiterative clustering is a common strategy used in the analysis of scRNA- seq data. As previously noted, the data obtained through scRNA- seq are sufficiently informative to enable cells to be grouped according to their gene expression pattern. This clustering is carried out on the basis of complex gene expression signatures, involving many genes, and not the restricted expression of only a few cell type- specific marker genes. This approach overcomes the problem of gene dropout and allows cells to be grouped even if the expression of a key marker gene goes undetected. An initial round of clustering typically divides cells into the most distinct groups. For example, an analysis of cells in the developing kidney might initially divide the cells into stromal cells and various types of nephron epithelial cells. A second round of analysis can then be carried out, perhaps, for example, using only the collecting duct cells, to divide them into subtypes, which would include β-intercalated cells, principal cells and other cell types (Fig. 5). Further such rounds of analysis can be performed in an attempt to define additional cell sub-types, but at a certain threshold, this process begins to ‘cluster on noise’, with the supposed cell heterogeneities actually based on data noise. One approach to determine when this threshold has been reached is to visually examine a heat map illustrating the differences in gene expression between the different clusters to look at the level of reproducibility of gene expression between clusters. Real cell subtypes will show consistency in their differential gene expression, whereas noise- based clustering will show little reproducibility. In addition, real differences in gene expression patterns between cell subtypes will substantiate with the use of orthogonal technologies, such as immunofluorescence or fluorescence in situ hybridization (FISH), whereas noise-based differences will fail to validate.
Fig. 5 |. Use of cluster and subcluster methodology to define cell subtypes.

A common strategy for analysis of scRNA- seq data is to carry out an initial unsupervised clustering, which divides cells into the most distinct groupings. In this example, the cells of the developing kidney are separated into separate categories, including collecting duct cells, stromal cells, endothelial cells, podocytes, cells from the loop of Henle, and so on. A group of cells of particular interest, for example, collecting duct cells, can then be separated out and subjected to another round of clustering to define subtypes of cells. The collecting duct subtypes in this example include principal cells, which express Scnn1b, β- intercalated cells, which express Slc26a4, and other unknown cell subtypes. Plot of dissociated cells adapted from Development, 144, Adam, M. et al. Psychrophilic proteases dramatically reduce single-cell RNA- seq artefacts: a molecular atlas of kidney development (2017), with permission from Elsevier. UB, ureteric bud.
Data analysis software.
Software for the analysis of scRNA- seq data is an area of rapid development and has been reviewed in detail else where44,45. The programmes in use include Seurat8, Sincera46, AltAnalyze47, Monocle48, Backspin49, Pagoda50, scLVM51, NMF52, SCDE53, RaceID2 (ReF.54), HiLoadG- HC55, RC1, SCell56 Wishbone57, Wanderlust58, DPT59, p- Creode60 and others40,61, all of which have been applied to scRNA-seq data with success. My laboratory has used Seurat, Sincera and AltAnalyze and found all three to give excellent and comparable results.
Applications
Single-cell gene expression profiling is rapidly becoming a standard analytical tool for researchers in many disciplines, including neurobiology, immunology and cardiology, to name just a few. In this Review, we focus on the use of this technology for the study of development, cancer and the kidney.
Development
Our understanding of development and the gene expression programmes that drive organ formation has important practical consequences. For example, using the principles that drive normal embryonic kidney development, stem cells can be induced to form kidney organoids, which are quite small yet include the many differentiated cell types found in a developing kidney62,63. Furthermore, in Nobel prize- winning work, adult fibroblasts were induced to de- differentiate and convert into stem cells through the forced expression of just four transcription factors64. These advances raise the exciting possibility that at some point in the future, it might be possible to take skin cells from a patient and use them to make immuno- compatible replacement organs.
scRNA- seq is a very powerful tool for the study of development. Early in organ development, collections of cells are often present that look histologically identical yet will differentiate in diverse directions to form distinct cell types. Better understanding of the precise gene expression programmes that drive these distinct differentiation pathways would improve understanding of the process of organ development. Gene expression analyses that examine pools of cells are inappropriate in this context, as the analyses would pool cells that ultimately differentiate down different pathways. Single- cell studies, however, can identify differential gene expression patterns associated with different lineage decisions in adjacent cells.
Multilineage priming.
A pioneering paper published in 2003 examined the gene expression patterns of single cells in the developing pancreas65. Using the technology then available, the researchers monitored the expression levels of about 100 selected genes in 60 cells. This study was distinctly ahead of its time in concept, recognizing the potential of single-cell studies to define the underlying basis of differentiation decisions, despite the histological similarity of cells early in organogenesis. One of the most interesting outcomes of this early work was the discovery that during pancreas development individual cells expressed genes associated with multiple possible future differentiated cell types65. For example, single progenitor cells expressed genes associated with both exocrine and endocrine lineages, whereas some single cells expressed multiple endocrine hormones, even though such combinations of gene expression do not occur in the adult. Such multilineage priming, whereby progenitor cells co- express genes of multiple distinct lineages, has also been seen in the haematopoietic system, where the finding was described as gene expression promiscuity66. A similar process of stochastic promiscuous gene expression followed by more restricted lineage specific transcription patterns occurs during development of the early mammalian embryo, as the epiblast and primitive endoderm lineages are established67.
Multilineage priming also occurs in kidney development. The adult metanephric kidney initially forms as a result of cross- inductive interactions between the nephric duct and the metanephric mesenchyme68. The ureteric bud outgrowth from the nephric duct undergoes branching morphogenesis to give rise to the collecting ducts and induces the flanking cap mesenchyme progenitor cells to undergo nephrogenesis. These progenitor cells undergo mesenchyme- to-epithelial transition to form the renal vesicle, which gives rise to the S- shape body and eventually all the epithelial cells of the nephron.
scRNA- seq analysis of the renal vesicle shows that these cells are at an interesting intermediate stage of lineage decision. For example, cells at one end of the renal vesicle, the distal region that abuts the collecting duct, show almost no expression of genes associated with podocytes, indicating that they have already excluded this potential lineage69. At the opposite proximal end of the renal vesicle, however, many cells show stochastic yet robust expression of multiple podocyte genes, showing that these cells are apparently actively considering this direction of differentiation. Many of these same cells, however, also express multiple genes associated with other lineages, including, for example, proximal tubules (Fig. 6). It is clear that the lineage selection process requires both absolute repression of inappropriate genes and further activation of appropriate lineage specific genes, but exactly how these apparently poised gene states are established and subsequently turned off and on during differentiation remains an open question.
Fig. 6 |. Multilineage priming.
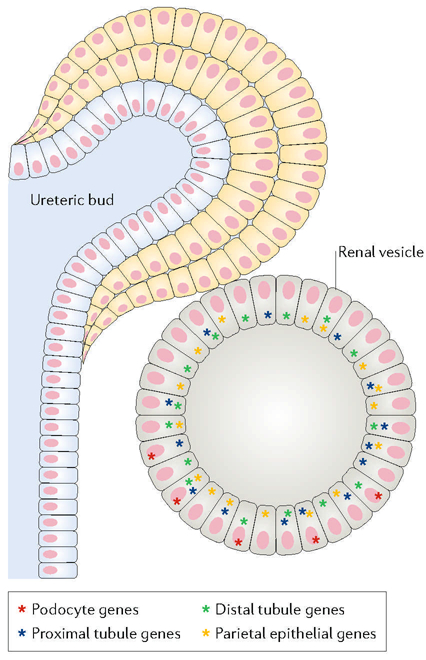
During development, cells show the stochastic expression of genes associated with potential future differentiation directions. For example, single- cell RNA sequencing (scRNA- seq) of cells of the renal vesicle, which will give rise to all the epithelial cells of the nephron, shows that some cells express multiple genes that are normally expressed only in differentiated podocytes. The expression levels are robust, but the expression patterns are seemingly stochastic, with some cells expressing some podocyte markers, other cells expressing other podocyte markers, and other cells expressing none. The situation is similar for genes associated with cells of the proximal tubule, with multiple cells in the renal vesicle showing stochastic expression of different subsets of proximal tubule-associated genes. Furthermore, some cells that express multiple podocyte genes also express multiple proximal tubule genes, suggesting that they retain the potential to differentiate in either direction. Parietal epithelial and distal tubule genes also show apparent stochastic expression patterns.
Atlases of organ development.
Single-cell transcriptional profiling can be used to create a high- resolution gene expression atlas of organ development. Such an atlas would define all the transcription factors, growth factors and receptors expressed in each cell type at different stages of development. It could globally characterize transcriptional transition states as cells differentiate and elucidate the crosstalk between flanking cell types. In sum, it could describe the gene expression programmes that drive development.
One general strategy for creation of such an atlas, as described earlier, is to dissociate the cells of a developing organ, perform high- throughput scRNA- seq on thousands of cells, for example, using a microdroplet-based technology, and use a bioinformatics-based approach to cluster cells into types and subtypes in an unsupervised manner. Spatial reconstruction of the single-cell atlas — that is, repositioning the analysed cells within a three-dimensional model of the developing organ — can be carried out manually based on prior knowledge of the gene expression signatures of different cell types or computationally by combining the newly obtained data with an established data set that distinguishes cell types70,71. New cell types would need to be positioned within the organ on the basis of immunofluorescent or in situ hybridization analysis of cell type specific markers.
Such scRNA-seq analysis of early kidney development revealed stromal cells as an important cell source of the glial cell line- derived neurotrophic factor (GDNF) — a growth factor that is essential for proper branching morphogenesis of the ureteric bud11,72,73. This finding was quite unexpected as the cap mesenchyme nephron progenitors had been thought to be the sole source of GDNF68,74. Nevertheless, use of three different scRNA-seq approaches — Drop- seq, Fluidigm and the Chromium system — to study gene expression in single cells of mouse kidney at embryonic day (E) 14.5 all defined stromal cells as a major source of GDNF. This finding was further confirmed using in situ hybridization and immunohistochemistry.
scRNA- seq studies of the developing lung have also characterized bipotential cells, identified markers of novel cell types, found transcriptional regulators and defined full gene expression programmes of the many cell types present at multiple stages of development46,55,75,76. The lungMAP consortium (see also the LGEA Web Portal) is using scRNA- seq to create a high- resolution gene expression atlas of the developing mouse and human lung75. Through the use of scRNA-seq, the adult mouse lung mesenchymal cell population, for example, was divided into five subtypes, including the alveolar niche, which supports alveolar growth and regeneration, and a myofibrogenic progenitor population, which contributes to a population of pathogenic myofibroblasts77. These studies provided the first comprehensive atlas of the gene expression patterns of all cell types in the developing lung, further defining the multiple transcription factor programmes that underlie lung development and the interactions between growth factors and their receptors that take place during development.
Implications for organ and organoid development.
In similar fashion to the above- mentioned studies, single-cell studies of the heart have defined the changing gene expression patterns of the endocardial, fibroblast and cardiomyocyte lineages during development7. Comparative analysis of mouse embryonic stem cell (ESC)-derived cardiomyocytes showed that they had a level of maturity comparative to that of fetal cardiomyocytes aged E14.5–E18.5. In general, gene expression analyses have shown that induced pluripotent stem cell (iPSC) and ESC- derived cell types and organoids are not as differentiated as originally believed and that further work is needed to understand how to drive their maturation. Of particular interest, the above- mentioned study of the developing heart also examined perturbed gene expression patterns present in the developing hearts of mice with heterozygous deletion of Nkx2.5, which encodes a transcription factor that is essential for normal heart development. Using scRNA- seq, the researchers identified a defect in the maturation of cardiomyocyte and endocardial cells, even though Nkx2.5 is only expressed in cardiomyocytes, demonstrating that cardiomyocyte crosstalk is required for proper endocardial maturation7.
The liver has impressive regenerative capacity, suggesting that it is a prime candidate for patient- derived organ replacement therapy. Indeed, human iPSCs can be used to generate liver bud- like tissue that, when transplanted, can extend life in a mouse model of liver failure78. Deeper understanding of the molecular drivers of liver development could advance liver organoids to the point where they could be used for human therapy. An elegant scRNA- seq study of the changing gene expression profiles of iPSCs as they formed liver buds79 defined a sequence of gene expression patterns that were present during the differentiation of iPSCs to hepatocytes, with the progressive downregulation of pluripotency genes and upregulation of genes controlling endoderm formation. However, similar to other studies that have assessed organoid maturity, the gene expression pattern of iPSC-derived hepatocytes more closely resembled that of human fetal hepatocytes than those from adult liver. The factors that influence vascularization of iPSC- derived liver buds were also investigated because vascularization is key to the survival and growth of organoids, which can only grow to millimetre size without vascularization. Human liver organoids vascularize following transplantation into mice, and scRNA- seq analysis of transplanted organoids suggested roles for genes involved in hypoxia, inflammation and matrix remodelling in this vascularization process80. Furthermore, analysis of complementary receptor and ligand expression in all cell types of the liver organoids defined potential crosstalk between the developing endothelial, mesenchymal and hepatocyte cells. Such insights into the processes underlying liver development have potential to define growth factor cocktails to facilitate the generation of improved liver organoids.
Thus, scRNA- seq is a very powerful tool for the study of developmental processes. It can define the transcription factor codes that define different cell types and drive differential gene expression. It can characterize the differentiation states of the multiple cell types present in organoids and provides a global analysis of potential ligand–receptor interactions. It can lend a much deeper understanding of normal developmental mechanisms and aid in the generation of improved organoids that could in time be used for the purposes of organ replacement therapy.
Cancer
As noted earlier, tumours contain a heterogeneous mix of cells that include cancer, vascular, immune and fibroblast cell types. Each of these cell types can be further divided into subtypes. Different types of infiltrating immune cells, for example, can stimulate both cancer cells and surrounding stromal cells through the production of epidermal growth factor (EGF), transforming growth factor- β (TGFβ), tumour necrosis factor (TNF), fibroblast growth factors (FGFs), interleukins and chemokines80. Multiple types of cancer- associated fibroblast cells also exist, some of which can secrete proliferation- inducing factors and pro- inflammatory agents that recruit further immune cells81. Multiple types of stromal cells are also present, contributing to the tumour microenvironment and driving cancer cell proliferation through paracrine and juxtacrine signalling. The study of gene expression patterns of stromal cells within the tumour microenvironment can provide prognostic value separate to that provided by the study of gene expression of intrinsic cancer cells82,83. scRNA-seq technology is clearly a very useful tool to dissect the properties of the multiple cell types within and surrounding tumour. Below, I briefly review a few key studies illustrating the power of this approach.
One study that used scRNA-seq to study gene expression in normal human colorectal tumours and matched normal mucosa1 identified ΤGFb1 to be the most upregulated differentially expressed regulatory gene in cancer-associated fibroblasts (CAFs). TGFβ secretion by CAFs contributed to activation of the TGFβ signalling pathway in tumour epithelial cells, suggesting crosstalk between these two cell types. Of interest, using analysis of gene expression patterns, the researchers could divide fibroblasts into three subtypes: normal (from control mucosa), those that expressed markers of myofibroblasts (that is, ACTA2, TAGLN and PDGFA) and those that expressed a distinct gene signature, including MMP2, DCN and COL1A2, with unknown function. Further, the scRNA- seq results could be used to reanalyse previous bulk RNA- seq data sets, thereby dividing the tumours into three distinct groups. One of the groups showed a strong epithelial cell type signature and weak fibroblast and myeloid signatures, and correlated with the best survival rates, demonstrating the translational potential of this technology1.
A separate study of human oligodendroglioma — a rare and incurable type of glioma — used scRNA-seq to identify a novel type of glioma-specific microglia, which expressed pro-inflammatory cytokines (IL1A, IL1B, IL8 and TNF), cytokines (CCL3, CCL4) and early response genes that distinguished them from the canonical M1 and M2 microglia84. These newly recognized micro-glia provide yet another example of cancer- associated cells that likely play a key part in the disease process. In addition, characterization of the cancer cells identified three types, including astrocytes, oligodendrocytes and a cluster of cells that expressed genes congruent with neural progenitor cells, suggesting that these cells have properties of stemness. Almost all the dividing cancer cells within the oligodendroglioma expressed these stemness markers, suggesting that these cells are responsible for fuelling tumour growth, giving rise to both astrocytes and oligodendrocytes, and supporting the cancer stem cell model.
scRNA- seq analysis of human breast cancer tissue has defined the variable expression of genes associated with epithelial- to-mesenchymal transition, stemness, angiogenesis, proliferation and tumour recurrence85. scRNA- seq has also been used to define genes that are differentially expressed by cell subtypes within a breast tumour. These studies revealed that the majority of non-cancer cells within these tumours were immune cells, including cells with the immune-suppressive characteristics of exhausted or regulatory T cells85. The exhausted T cell phenotype, indicating loss of T cell function, also emerged in a study of liver cancer that focused on T cells86. Patients with late-stage disease showed T cells with a higher level of exhaustion; scRNA- seq analyses enabled the complete gene expression patterns for these T cells to be defined and novel exhaustion marker genes to be described.
scRNA- seq has also been used to examine the multicellular composition of melanoma87. Non- malignant cells clustered according to cell type, for instance, T cells, B cells, macrophages, endothelial cells, natural killer cells and CAFs. Of interest, however, malignant cells from each patient clustered separately, indicating a high degree of intertumour heterogeneity. Malignant cells could be separated into cycling and noncycling cells, and the researchers characterized a subset of KDM5B- expressing, slow cycling, drug- resistant, stem- like melanoma cells. The researchers also studied the heterogeneity of malignant cells as a function of treatment with RAF proto-oncogene serine/threonine-protein kinase and MAP/ERK kinase (MEK) inhibitors and identified a change in the distribution of cell types reflecting differing sensitivities of the cells to treatment. Further relationships between non- malignant tumour cells, such as CAFs, and malignant gene expression patterns and drug sensitivity were also found, and CAF genes implicated in T cell infiltration were identified. Once again, this example shows how single-cell analysis can provide deeper insights into the biology of cancer than would be possible with bulk analysis of aggregate pools of multiple cell types.
One pioneering study used scRNA- seq to define cancer cell heterogeneity in metastatic clear cell renal carcinoma88 with the aim of defining active signalling pathways that could help guide therapeutic strategy. The researchers examined a limited number of cells from a single patient, including 34 from a metastasis, 36 from a patient metastasis- derived xenotransplant and 46 from a primary tumour- derived xenotransplant. Different drug target pathways were active in the primary and metastatic xenotransplants, and these reflected different drug sensitivities. The metastatic cells showed sensitivity to drugs that targeted epidermal growth factor receptor (EGFR) (gefitinib, erlotinib and afatinib), SRC family kinases (dasatinib) and BRAF–MEK (selumetinib), whereas the primary tumour cells were more sensitive to drugs that targeted MET (tivantinib, foretinib and crizotinib) and phosphoinositide 3-kinase (PI3K) (BKM120). Detailed analysis of cell heterogeneity within the primary and metastatic tumours suggested that treatment with both afatinib and dasatinib would be beneficial; indeed, this combination treatment had a synergistic antitumour effect on metastatic xenografts. The results from this study provide proof- of-principle that insights into cell heterogeneity achieved using scRNA-seq can be used to design therapeutic approaches that are optimal for individual patients.
Normal and diseased kidneys
Single- cell studies of adult kidneys, both normal and diseased, have lagged behind other areas, such as cancer and immunology, but new studies are emerging. Two studies have used the Fluidigm 96 cell IFC system to examine the gene expression profiles of very small numbers of mouse podocytes (20 cells)89 and mesangial cells (33 cells)90. Although these studies provide an interesting first look at the genes expressed by these cells, they are limited by the low numbers of cells examined.
One study used scRNA- seq to examine cells of the collecting duct, which have a major role in maintaining electrolyte and fluid balance. Given the relative rarity of collecting duct cells, FACS enrichment was first used to isolate them91, followed by scRNA- seq analysis to examine gene expression in 74 principal cells, 87 type A intercalated cells and 23 type B intercalated cells. Highlighting the power of scRNA- seq, the findings revealed possible crosstalk between the cell types. For example, Notch2 receptor was identified as being expressed in principal cells, whereas its ligand Jag1 was expressed in intercalated cells. Similarly, Kit was found to be expressed in intercalated cells and its ligand Kitl in principal cells. The researchers could also define all the channels and transporters expressed by the different cell types. In addition, this study delineated the receptors for humoral factors expressed by the different collecting duct cell types, including the extracellular calcium sensing receptor by type B intercalated cells and the V2 vasopressin and type 1 prostaglandin E2 receptors by principal cells. Overall, this work represents an important step forward in defining the characteristics of collecting duct cells and furthers our understanding of their physiologic regulation.
A more comprehensive scRNA- seq analysis used the Chromium microdroplet system to examine the gene expression patterns of 57,979 cells across the adult mouse kidney92. Clustering analysis divided the cells into 16 groups, and further analysis divided three of these clusters into a total of eight subclusters. Gene expression analysis identified 18 previously known cell types and 3 novel cell types. One of these novel cell types represented a transitional state between collecting duct principal and intercalated cells. This finding is of particular interest given that these cells have different functions: principal cells are responsible for the reabsorption of sodium and water and the secretion of potassium whereas α- intercalated and β- intercalated cells are responsible for acid and alkali secretion, respectively. The researchers documented the conversion potential of the principal and intercalated cells using a transgenic lineage tag system92 and observed cell plasticity, with a fraction of cells labelled with one cell type tag undergoing transition to the other lineage as measured by immunofluorescence. Further, the researchers demonstrated that transgene-mediated activation of the Notch pathway could drive the transition of intercalated cells to principal cells. This work therefore provides an exhaustive gene expression atlas of the many cell types of the normal adult kidney, connects disease- associated genes to specific cell types and demonstrates a surprising differentiation state flexibility in the adult kidney.
Another study examined single-cell gene expression in the zebrafish kidney2. In the adult zebrafish, the kidney, rather than bone marrow, is the site of haematopoiesis. Although this study mostly focused on haematopoiesis, the researchers also used the microdroplet- based InDrop system to examine the gene expression profiles of 1,699 non-haematopoietic kidney cells10. Among other achievements, the researchers were able to characterize mucin- producing cells, which make mucin barriers that reduce chances of infection. In contrast to the adult mammalian kidney, which cannot make new nephrons, the zebrafish adult kidney shows a remarkable ability for regeneration. Using scRNA- seq, the researchers were able to define the gene expression patterns of zebrafish adult nephron progenitors, showing notable similarity to mammalian embryonic cap mesenchyme nephron progenitor cells, with expression of Six2, Eya2 and Osr1 (ReF.10). Nevertheless, these adult zebrafish progenitor cells had some unique features, including the expression of a large number of genes encoding collagen matrix-associated proteins, as well as genes that are normally expressed in human CD31-negative stromal stem cells. This study therefore defines a unique resident nephron progenitor cell population present in the adult zebrafish kidney that helps to explain its regenerative capacity. Similar cells are not seen in the adult mammalian kidney.
scRNA- seq has also been used to investigate disease processes underlying renal manifestations of systemic lupus erythematosus74,93, an autoimmune disease that affects the skin, kidneys, joints and other tissues. Using 899 cells from 12 skin and 16 kidney biopsy samples from patients with lupus nephritis, the study detected only ~700 genes as being expressed per cell — a low number given that most cells express ~5,000–10,000 genes. Further, clustering analysis failed to identify podocytes or mesangial cells, suggesting that these cell types — which as mentioned earlier are difficult to dissociate — were not isolated by the dissociation process. Despite these limitations, the researchers identified a gene expression signature characterized by interferon response genes that correlated with disease severity. Of interest, the skin samples from patients with lupus nephritis showed a similar signature of interferon response genes, suggesting that this more accessible tissue provides useful biomarkers of disease state in patients with lupus nephritis.
Thus, scRNA- seq is beginning to provide important insights into normal kidney physiology, defining the complete receptor expression profiles for principal and intercalated collecting duct cells, and identifying cell– cell crosstalk. It is shedding light on the mechanisms that give the adult zebrafish kidney its surprising regenerative capacity. scRNA- seq is also providing insights into pathogenic pathways, identifying novel biomarkers and possible therapeutic targets for the diseased kidney. Nevertheless, application of this technology in the field of kidney research is still in the early stages, and further work is needed.
Conclusions
scRNA- seq is a powerful technology that can provide high- resolution analysis of biological systems. It provides deep scrutiny into the gene expression character of diverse cell types, lending insight into all the biological processes being carried out. A chief limitation at present is the noisy nature of the resulting data, primarily owing to the technical limitations of working with such small amounts of RNA. This noise makes it difficult to distinguish very similar cell types and is an area that is in need of technological improvement. Another limitation is the cost of scRNA-seq experiments: although currently available systems run at ~$1 per cell, which seems quite reasonable, the combined cost is considerable when many thousands of cells are analysed. The most expensive component of the analysis is the DNA sequencing; fortunately, we are in the midst of a remarkable revolution in DNA sequencing technology that is rapidly driving down this cost. Reduced sequencing costs will facilitate deeper scRNA- seq analysis of each cell, with more reads per cell, as well as the inclusion of ever greater numbers of cells in an scRNA- seq study, which will increase the statistical power in these analyses. This technology has already provided fascinating insights into the processes underlying various developmental, physiological and disease systems. Further studies in this area will enable even more profound understanding of these processes, leading to the development of atlases describing the expression of genes in cells throughout the body and contributing to the field of personalized medicine.
Key points
RNA sequencing of single cells (scRNA- seq) allows the global gene expression patterns of individual cells to be defined.
Almost all tissues and organs include a heterogeneous mix of cell types; the heterogeneity of these cell populations can be defined through the use of scRNA- seq.
scRNA- seq can fully define the expression of transcription factors, growth factors, receptors, solute transporters and other proteins for each cell type present, providing insights into cell function and cell–cell crosstalk.
scRNA- seq is an increasingly powerful tool for the analysis of development as well as normal and disease processes.
Splicing patterns
Sequences recognized by RNA processing enzymes of the spliceosome, which splice out introns. introns almost always begin with the bases gU and terminate with Ag, but additional sequences around splice sites are required to provide sufficient specificity.
Early response genes
Genes that are activated rapidly in response to a variety of stimuli, including stress and growth factors. About 40 immediate early response genes exist, including members of the FOS and JUN families.
Footnotes
Competing interests
The author declares no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Nephrology thanks L. Oxburgh, K. Kiryluk and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Related links
Chan Zuckerberg Initiative Atlas Project: https://www.chanzuckerberg.com/human-cell-atlas
Human BioMolecular Atlas Program: https://commonfund.nih.gov/hubmap
LGEA (Lung Gene Expression Analysis) Web Portal: https://research.cchmc.org/pbge/lunggens/mainportal.html
lungMAP consortium: https://www.lungmap.net/
References
- 1.Li H et al. Reference component analysis of single- cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet 49, 708–718 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Tang Q et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single- cell resolution using RNA sequencing. J. Exp. Med 214, 2875–2887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron M et al. A single- cell transcriptomic map of the human and mouse pancreas reveals inter- and intra- cell population structure. Cell Syst 3, 346–360. e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villani AC et al. Single- cell RNA- seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puram SV et al. Single- cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624. e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regev A et al. The Human Cell Atlas. eLife 6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLaughter DM et al. Single- cell resolution of temporal gene expression during heart development. Dev. Cell 39, 480–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macosko EZ et al. Highly parallel genome- wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein AM et al. Droplet barcoding for single- cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magella B et al. Cross- platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev. Biol 434, 36–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picelli S et al. Smart- seq2 for sensitive full- length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Hashimshony T, Wagner F, Sher N & Yanai I CEL- Seq: single- cell RNA- Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Jaitin DA et al. Massively parallel single- cell RNA- seq for marker- free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam S et al. Quantitative single- cell RNA- seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hochgerner H et al. STRT- seq-2i: dual- index 5’ single cell and nucleus RNA- seq on an addressable microwell array. Sci. Rep 7, 16327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan HC, Fu GK & Fodor SP Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science 347, 1258367 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Han X et al. Mapping the mouse cell atlas by Microwell- seq. Cell 172, 1091–1107. e17 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg AB et al. Single- cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y & Navin NE Advances and applications of single- cell sequencing technologies. Mol. Cell 58, 598–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuppers R, Zhao M, Hansmann ML & Rajewsky K Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 12, 4955–4967 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang F et al. mRNA- Seq whole- transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Xue Z et al. Genetic programs in human and mouse early embryos revealed by single- cell RNA sequencing. Nature 500, 593–597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamme F et al. Single- cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J. Neurosci 23, 3607–3615 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frumkin D et al. Amplification of multiple genomic loci from single cells isolated by laser micro- dissection of tissues. BMC Biotechnol. 8, 17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaiskos N et al. The Drosophila embryo at single- cell transcriptome resolution. Science 358, 194–199 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Adam M, Potter AS & Potter SS Psychrophilic proteases dramatically reduce single cell RNA- seq artifacts: a molecular atlas of kidney development. Development 144, 3625–3632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacar B et al. Nuclear RNA- seq of single neurons reveals molecular signatures of activation. Nat. Commun 7, 11022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.See K et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA- regulated de-differentiation and cell cycle stress- response in vivo. Nat. Commun 8, 225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao R et al. Nanogrid single- nucleus RNA sequencing reveals phenotypic diversity in breast cancer. Nat. Commun 8, 228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnaswami SR et al. Using single nuclei for RNA- seq to capture the transcriptome of postmortem neurons. Nat. Protoc 11, 499–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Habib N et al. Div- Seq: Single- nucleus RNA- Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Velthoven CTJ, de Morree A, Egner IM, Brett JO & Rando TA Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep 21, 1994–2004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bensaude O Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription 2, 103–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letschert K, Faulstich H, Keller D & Keppler D Molecular characterization and inhibition of amanitin uptake into human hepatocytes. Toxicol. Sci 91, 140–149 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Ross IL, Browne CM & Hume DA Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell Biol 72, 177–185 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Ozbudak EM, Thattai M, Kurtser I, Grossman AD & van Oudenaarden A Regulation of noise in the expression of a single gene. Nat. Genet 31, 69–73 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svensson V et al. Power analysis of single- cell RNA- sequencing experiments. Nat. Methods 14, 381–387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JK, Kolodziejczyk AA, Ilicic T, Teichmann SA & Marioni JC Characterizing noise structure in single- cell RNA- seq distinguishes genuine from technical stochastic allelic expression. Nat. Commun 6, 8687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usoskin D et al. Unbiased classification of sensory neuron types by large- scale single- cell RNA sequencing. Nat. Neurosci 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Hyvarinen A & Oja E Independent component analysis: algorithms and applications. Neural Netw 13, 411–430 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Pierson E & Yau C ZIFA: dimensionality reduction for zero- inflated single- cell gene expression analysis. Genome Biol. 16, 241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bacher R & Kendziorski C Design and computational analysis of single- cell RNA- sequencing experiments. Genome Biol. 17, 63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stegle O, Teichmann SA & Marioni JC Computational and analytical challenges in single- cell transcriptomics. Nat. Rev. Genet 16, 133–145 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Guo M, Wang H, Potter SS, Whitsett JA & Xu Y SINCERA: a pipeline for single- cell RNA- seq profiling analysis. PLoS Comput. Biol 11, e1004575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsson A et al. Single- cell analysis of mixed- lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeisel A et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single- cell RNA- seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Fan J et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buettner F et al. Computational analysis of cell- to-cell heterogeneity in single- cell RNA- sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol 33, 155–160 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Zhu X, Ching T, Pan X, Weissman SM & Garmire L Detecting heterogeneity in single- cell RNA- Seq data by non- negative matrix factorization. PeerJ 5, e2888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kharchenko PV, Silberstein L & Scadden DT Bayesian approach to single- cell differential expression analysis. Nat. Methods 11, 740–742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grun D et al. De novo prediction of stem cell identity using single- cell transcriptome data. Cell Stem Cell 19, 266–277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Treutlein B et al. Reconstructing lineage hierarchies of the distal lung epithelium using single- cell RNA- seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz A et al. SCell: integrated analysis of single- cell RNA- seq data. Bioinformatics 32, 2219–2220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Setty M et al. Wishbone identifies bifurcating developmental trajectories from single- cell data. Nat. Biotechnol 34, 637–645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bendall SC et al. Single- cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haghverdi L, Buttner M, Wolf FA, Buettner F & Theis FJ Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Herring CA et al. Unsupervised trajectory analysis of single- cell RNA- seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst 6, 37–51. e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shin J et al. Single- cell RNA- seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Taguchi A et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Takahashi K & Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Chiang MK & Melton DA Single- cell transcript analysis of pancreas development. Dev. Cell 4, 383–393 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Miyamoto T et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Ohnishi Y et al. Cell- to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol 16, 27–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McMahon AP Development of the mammalian kidney. Curr. Top. Dev. Biol 117, 31–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brunskill EW et al. Single cell dissection of early kidney development: multilineage priming. Development 141, 3093–3101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single- cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Achim K et al. High- throughput spatial mapping of single- cell RNA- seq data to tissue of origin. Nat. Biotechnol 33, 503–509 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Costantini F & Shakya R GDNF/Ret signaling and the development of the kidney. Bioessays 28, 117–127 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Skinner MA, Safford SD, Reeves JG, Jackson ME & Freemerman AJ Renal aplasia in humans is associated with RET mutations. Am. J. Hum. Genet 82, 344–351 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dressler GR Advances in early kidney specification, development and patterning. Development 136, 3863–3874 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ardini- Poleske ME et al. LungMAP: The molecular atlas of lung development program. Am. J. Physiol. Lung Cell. Mol. Physiol 313, L733–L740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Du Y et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72, 481–484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zepp JA et al. Distinct mesenchymal lineages and niches promote epithelial self- renewal and myofibrogenesis in the lung. Cell 170, 1134–1148. e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takebe T et al. Vascularized and functional human liver from an iPSC- derived organ bud transplant. Nature 499, 481–484 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Camp JG et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Balkwill F, Charles KA & Mantovani A Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell 7, 211–217 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Hanahan D & Coussens LM Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Finak G et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med 14, 518–527 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Galon J et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Tirosh I et al. Single- cell RNA- seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung W et al. Single- cell RNA- seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun 8, 15081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng C et al. Landscape of infiltrating T cells in liver cancer revealed by single- cell sequencing. Cell 169, 1342–1356. e16 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single- cell RNA- seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim KT et al. Application of single- cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol 17, 80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu Y et al. Genome- wide identification of genes essential for podocyte cytoskeletons based on single- cell RNA sequencing. Kidney Int 92, 1119–1129 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Lu Y, Ye Y, Yang Q & Shi S Single- cell RNA- sequence analysis of mouse glomerular mesangial cells uncovers mesangial cell essential genes. Kidney Int 92, 504–513 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Chen L et al. Transcriptomes of major renal collecting duct cell types in mouse identified by single- cell RNA- seq. Proc. Natl Acad. Sci. USA 114, E9989–E9998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park J et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease . Science. 10.1126/science.aar2131 (2018) [DOI] [PMC free article] [PubMed]
- 93.Der E et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2, 93009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


