SUMMARY
Maintenance of skeletal muscle mass requires a dynamic balance between protein synthesis and tightly controlled protein degradation by the calpain, auto-phagy-lysosome, and ubiquitin-proteasome systems (proteostasis). Several sensing and gene-regulatory mechanisms act together to maintain this balance in response to changing conditions. Here, we show that deletion of the highly conserved Rbfox1 and Rbfox2 alternative splicing regulators in adult mouse skeletal muscle causes rapid, severe loss of muscle mass. Rbfox deletion did not cause a reduction in global protein synthesis, but it led to altered splicing of hundreds of gene transcripts, including capn3, which produced an active form of calpain3 protease. Rbfox knockout also led to a reduction in autophagy flux, likely producing a compensatory increase in general protein degradation by the proteasome. Our results indicate that the Rbfox-splicing factors are essential for the maintenance of skeletal muscle mass and proteostasis.
Graphical Abstract
In Brief
Rbfox-splicing factors are highly conserved and expressed in brain, heart, and skeletal muscle. Singh et al. show that Rbfox proteins are essential for the maintenance of muscle mass and proteostasis, as Rbfox double knockout causes increased calpain and proteasome activity and reduced autophagy flux.
INTRODUCTION
Although much research in the muscle field has been devoted to understanding myogenesis, muscle regeneration, and myofiber function (Bassel-Duby and Olson, 2006; Comai and Tajbakhsh, 2014; Potthoff and Olson, 2007; Schiaffino et al., 2013; Yin et al., 2013), there has been a growing recognition of the importance of maintenance of muscle mass in adulthood. Skeletal muscle plays a crucial role in metabolism (James et al., 2017; Rai and Demontis, 2016) and directly influences quality of life in aging and in chronic disease (McLeod et al., 2016). Muscle tissue also faces special challenges: it must generate considerable force on demand and is continually exposed to mechanical, temfigperature, and oxidative stresses. Maintaining homeostasis under such conditions requires tight regulation of protein synthesis and turnover (Bell et al., 2016); failure of proteostasis occurs in many muscle diseases (Lecker et al., 2006; Nishino et al., 2000; Richard et al., 1995; Sandri et al., 2013), as well as the more general conditions of sarcopenia and cachexia (Bowen et al., 2015). Despite the common occurrence of these latter conditions, we do not fully understand the mechanisms that are critical for the maintenance of adult muscle mass.
We hypothesized that alternative splicing substantially contributes to muscle homeostasis in adults. Alternative splicing generates multiple protein isoforms from a single gene, thereby expanding the functional repertoire of proteins far beyond what could be expected from the relatively modest size of the genome (Kalsotra and Cooper, 2011; Wang et al., 2008; Yang et al., 2016). The resulting protein diversity is thought to be particularly important in tissues that must respond to highly variable conditions, such as the brain, heart, and skeletal muscle (Kalsotra and Cooper, 2011; Raj and Blencowe, 2015). Indeed, skeletal muscle has particularly high levels of alternative splicing. We and others have shown that the splicing factors Rbfox1 and Rbfox2, which are highly conserved from C. elegans to humans (Gallagher et al., 2011; Jin et al., 2003; Kuroyanagi et al., 2007; Venables et al., 2012), are required for muscle differentiation and function (Pe-drotti et al., 2015; Runfola et al., 2015; Singh et al., 2014); but, Rbfox factors have never been studied specifically in adult skeletal muscle.
In this study, we induced skeletal muscle-specific knockout of both Rbfox1 and Rbfox2 in adult mice (7 weeks of age or older). Our double-knockout mice suffered a rapid, severe loss of skeletal muscle mass (30%−50% within 4 weeks). RNA sequencing of control and double-knockout (DKO) skeletal muscles, 2 weeks after knockout induction, revealed that hundreds of transcripts were altered in expression and splicing. This loss of mass was attributable to an increase in proteolysis rather than a reduction in protein synthesis.
RESULTS
Deletion of Rbfox1 and Rbfox2 in Adult Mice Causes Rapid, Severe Muscle Loss
To determine the role of Rbfox-splicing factors in mature adult myofibers, we induced knockout of Rbfox1, Rbfox2, or both in 7-week-old mice using a tetracycline-inducible and skeletal muscle-specific Cre line (Rao and Monks, 2009). To quantify the knockout efficiency of Rbfox1 and/or Rbfox2, we performed western blot analysis using tibialis anterior (TA) muscle protein extracts from control (Rbfox1 f/f and Rbfox2 f/f), Rbfox1-knockout (Rbfox1f/f: ACTA1-rtTAcre/+), Rbfox2-knockout (Rbfox2 f/f: ACTA1-rtTAcre/+), and DKO (Rbfox1 f/f: Rbfox2 f/f ACTA1-rtTAcre/+) animals (Figure 1A). Rbfox1-only knockout led to a 2-fold upregulation of Rbfox2 protein in muscle, whereas Rbfox1 protein level did not increase significantly in Rbfox2- only-knockout muscle. We also tested the effects of Rbfox1 and/or Rbfox2 deletions on alternative splicing of Bin1 exon 10, which we identified previously as an Rbfox target in a myoblast culture cell line (Singh et al., 2014). Single knockout of either Rbfox1 or Rbfox2 diminished the inclusion of Bin1 exon 10 by <5%, but Rbfox DKO reduced inclusion of this exon by >65% (Figure 1A, bottom panel). These observations indicate that Rbfox1 and Rbfox2 paralogs compensate for one another’s loss of function and regulate overlapping splicing events, so the experiments described here were conducted primarily with the DKO animals.
Figure 1. Rbfox DKO in Adult Skeletal Muscle Causes the Rapid Loss of Muscle Mass by a Reduction in Fiber Size.
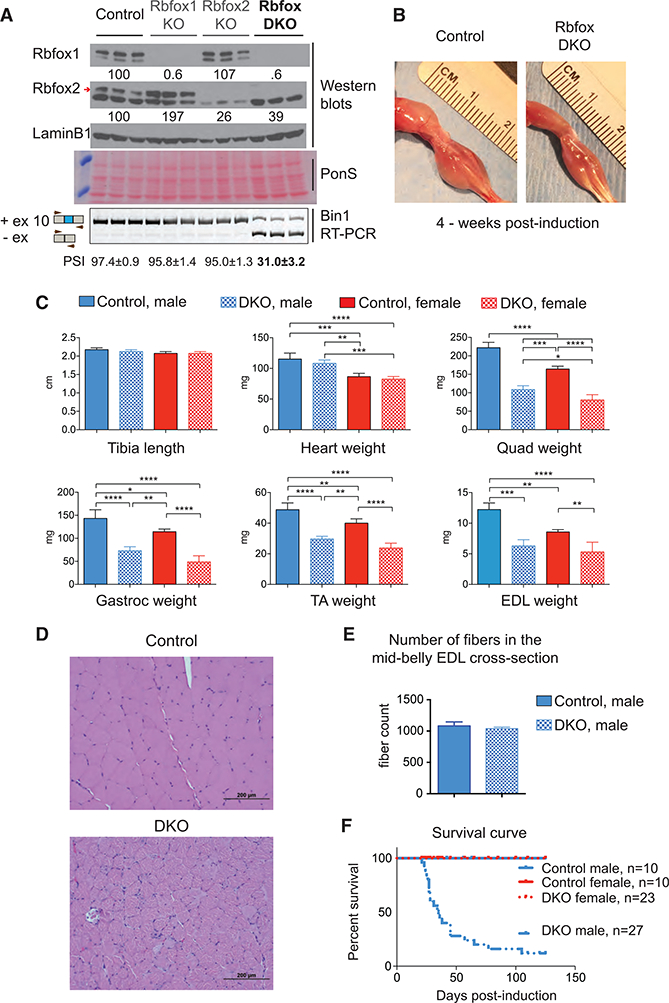
(A) Western blots for Rbfox1, Rbfox2, and LaminB1 from control (Rbfox1f/f and Rbfox2f/f), Rbfox1 (Rbfox1 f/f: ACTA1-rtTA,tetO-cre), Rbfox2 (Rbfox2 f/f: ACTA1-rtTA,tetO-cre), and double-Rbfox knockout (Rbfox1f/f, Rbfox2f/f,ACTA1-rtTA,tetO-cre) tibialis anterior (TA) muscles. Red arrow indicates confirmed Rbfox2 band. Ponceau S staining of representative blots was used as an additional loading control. RT-PCR using primers flanking Bin1 exon 10 (bottom panel) from the same TA muscle that was used for westerns is shown. Percent spliced in (PSI) values are indicated below (mean ± SD from n = 3).
(B)Representative image of the hind limbs from control and Rbfox-DKO mice 4 weeks after starting the dox diet.
(C) Average measurements of tibia length and weights of isolated heart and different skeletal muscles: quadriceps (Quad), gastrocnemius (Gastroc), TA, and extensor digitorum longus (EDL) from control and DKO male and female mice (n ≥ 3). The columns and error bars in graphs indicate mean ± SD; p values as indicated by asterisk were calculated by ANOVA, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
(D) H&E staining of quadriceps from control (top) and DKO (bottom) mice 4 weeks after the induction of Rbfox DKO.
(E) Total fiber number count from the mid-belly cross-section of EDL muscle from control and DKO mice 4 weeks after the induction of knockout (n = 3).
(F) Survival curves for control and DKO, male and female, animals.
Rbfox-DKO animals displayed a marked loss of muscle mass and total body weight, compared to age- and sex-matched control animals, within 4 weeks of knockout induction (Figures 1B and S1A). Tibia length was the same between aged-matched control and DKO animals (Figure 1C), but there were significant reductions in the weight of five predominantly fast-twitch skeletal muscles-quadriceps, gastrocnemius, TA, extensor digitorum lon-gus (EDL), and triceps-in both male and female mice (Figures 1C and S1A). The weight of the soleus, which contains approximately 40% type I or slow-twitch fibers, did not decrease in mice of either sex (Figure S1A), most likely because the knockout of Rbfox2 in the soleus was not as efficient as in fast-twitch muscle (data not shown). Consistent with the muscle specificity of the Cre recombinase (Rao and Monks, 2009), we did not observe a change in heart or liver weights (Figures 1C and S1A). Single knockout of Rbfox1 or Rbfox2 did not reduce skeletal muscle weight (Figure S1B).
Because ACTA1-rtTA Cre recombinase expression is slightly leaky in the absence of doxycycline (dox) (Rao and Monks, 2009), we measured the weight of the gastrocnemius muscle several times between starting the dox-containing diet at the age of 7 weeks until 21 days later. The DKO gastrocnemius muscle initially weighed 15% less in DKO mice than in uninduced animals, but by 21 days the DKO gastrocnemius weight was 41% less than that of control animals (Figure S1C). Based on these results, we conclude that Rbfox-splicing factors are required to maintain skeletal muscle mass in adult animals.
Histological analysis of Rbfox-DKO quadricep cross-sections showed non-uniform and dramatically reduced fiber size in DKO mice compared to controls (Figure 1D). We observed that few myonuclei were centrally located, which suggested that minimal degeneration-regeneration had occurred. We also did not observe infiltration of inflammatory cells in DKO muscles. The total number of muscle fibers in the mid-belly cross-sections of EDL muscles did not differ between DKO and control mice (Figure 1E). We conclude that a reduction in fiber size is the main reason for loss of muscle mass after Rbfox DKO.
Despite the overall similarity of muscle loss in the male and female mice, there was one notable difference between the sexes: 80% of the male Rbfox-DKO animals died as early as 3 weeks after knockout induction, but none of the control or DKO female animals died during the course of our experiments (Figure 1F).
Deletion of Rbfox1 and Rbfox2 Causes Loss of Muscle Strength, but Not Endurance
To evaluate muscle function, we measured the forelimb and alllimb grip strength in DKO male and female animals 2–8 weeks after starting the dox-containing diet. (The premature death of the males prevented us from studying them at a later time point.) Both sexes showed a significant reduction in forelimb and alllimb grip strength compared to age- and sex-matched control animals (Figure 2A). We also found a significant reduction in upside-down hanging time in both male and female DKO animals. Performance on a treadmill assay revealed no significant difference in endurance among control, DKO male, and DKO female mice (Figure 2A). We also measured the total activity of male and female mice in the vertical (rearing) and horizontal (ambulation and fidgeting) planes. Male DKO mice displayed significantly less rearing than control mice (Figures S2A and S2B), whereas female DKO mice displayed significantly greater horizontal plane activity and a trend for increased rearing compared to control females (Figures S2A and S2B). The activity differences between control and DKO animals in both sexes were predominantly due to movement in the active period (12 hr, night) (Figures S2A and S2B).
Figure 2. Rbfox1/2 Knockout in Adult Muscle Causes Reduced Muscle Strength and Altered Glucose Homeostasis.
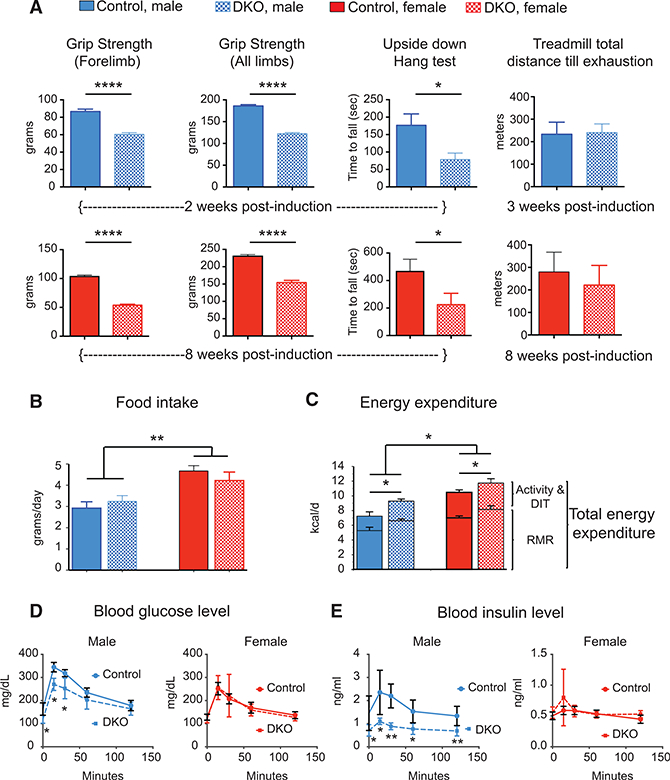
(A) Forelimb (left panels) and all limb (left middle panels) grip strength in control (Rbfox1f/f and Rbfox2f/f) and Rbfox-DKO (Rbfox1f/f, Rbfox2f/f, ACTA1-rtTA,tetO-cre) male (top panels) and female (bottom panels) mice, 2 weeks and 8 weeks after knockout induction, respectively. The right middle panels show the average time animals hang upside down on wire mesh. Treadmill performance (right panels) of control and DKO mice with increasing speed until exhaustion (at 10% incline). n≥9 for all panels. Error bars, mean ± SEM.
(B and C) Singly housed age-matched male and female animals were monitored for food intake (B), and indirect calorimetry was utilized to estimate total and resting energy expenditure (C) (n = 5 for male and n ≥ 3 for female). Values shown are least square means ± SE adjusted using analysis of covariance (ANCOVA) for differences in body weight (for food intake) and lean mass (for energy expenditure; fat mass does not contribute to variance). RMR, resting metabolic rate; DIT, diet-induced thermogenesis. Activity and DIT are not significantly different between genotype or gender.
(D and E) Glucose tolerance test in male (left panels) and female (right panels) mice 2 weeks after the induction of Rbfox knockout. (D) shows blood glucose level, and (E) shows insulin levels in control and DKO male (left) and female (right) mice (n = 5). The columns and error bars indicate mean ± SD; p values were calculated by multiple Student’s t tests and are indicated as follows: *p ≤ 0.05, **p ≤ 0.01, and ****p ≤ 0.0001.
DKO Adult Mice Expend More Energy
To understand how the loss in muscle mass affects energy metabolism, we performed indirect calorimetry in age-matched control and DKO male and female animals starting 10 days after knockout induction. We found no significant differences in 24-hr, weight-adjusted food intake between control and DKO animals, although females ate more than males (Figure 2B). Both male and female DKO animals, however, expended more energy than age-matched control animals (Figure 2C); this difference was attributable to differences in resting metabolic rate, and therefore, it was evident both in periods of rest (12 hr, day) and activity (12 hr, night) (Figure S2C).
Since skeletal muscle is a primary organ for maintaining glucose homeostasis, we performed glucose tolerance tests to determine the effect of Rbfox DKO on glucose metabolism in age-matched control and DKO animals 2 weeks after knockout induction. Before glucose injection, male, but not female, DKO mice had lower levels of serum glucose relative to control animals. Even after glucose injection, glucose levels remained lower in DKO male animals, but not in female animals (Figure 2D). Serum insulin levels were nearly half that of controls in the DKO male animals, while DKO females did not show a difference (Figure 2E). These results indicate that glucose homeostasis is altered in male Rbfox-DKO animals and that male DKO animals show a trend toward insulin hypersensitivity.
Deletion of Rbfox1 and Rbfox2 Causes Widespread Transcriptome Changes
To investigate the molecular mechanism by which Rbfox1/2 deletion produced these changes, we performed 100-bp paired-end RNA sequencing (RNA-seq) using polyadenylated RNA from the EDL and soleus muscles from two male DKO mice, 2 weeks after knockout induction, and two age-matched male controls. For each sample, we obtained over 170 million reads, 87% of which mapped to the genome (Table S1). Mapped reads were used to quantify transcriptome changes, including gene expression and alternative use of first, last, and internal exons. Gene expression changes for biological replicates correlated very highly (R2 = 0.99), demonstrating the reproducibility of RNA-seq and computational analysis of the data (Figure S3A) in the DKO EDL, which showed a significant loss of muscle mass, 832 genes were differentially expressed (false discovery rate [FDR] < 0.05), with 495 genes (~60%) showing upregulation and 337 genes (~40%) showing downregulation when compared to control animals (Figure 3A; Table S2). In the DKO soleus, which did not show significant changes in mass in males or females, 206 genes were differently expressed (FDR < 0.05), with 132 genes showing upregulation and 74 genes showing downregulation relative to their levels in control animals (Figure 3A; Table S2).
Figure 3. Rbfox Knockout in Skeletal Muscle Causes Widespread Transcriptome Changes.
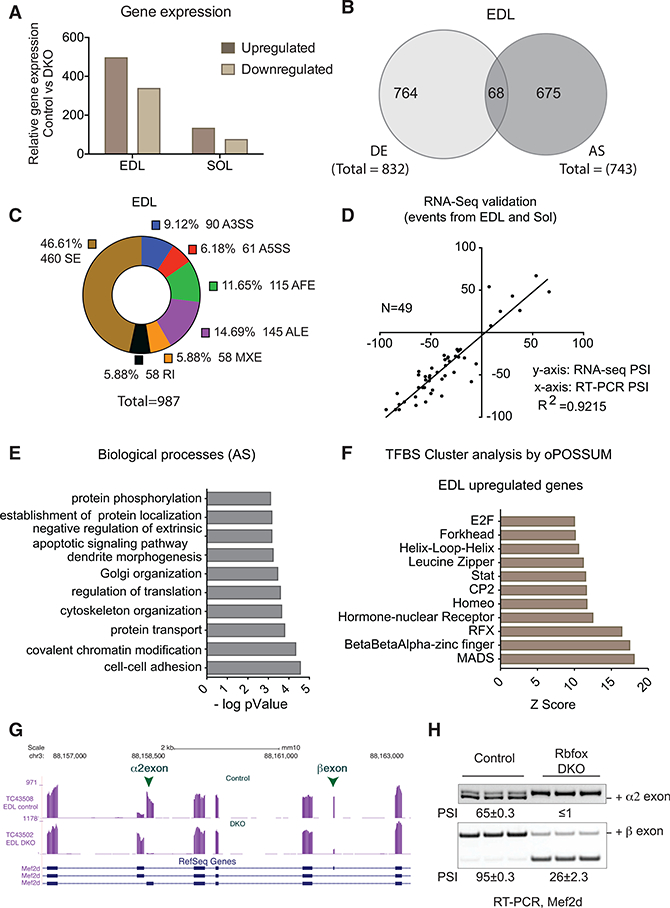
(A) Bar graphs showing gene expression changes (FDR < 0.05) in EDL and soleus muscles 2 weeks after the induction of Rbfox DKO compared to control muscles.
(B) Venn diagram showing differentially expressed (DE) and alternatively spliced (AS) gene transcripts in DKO compared to control EDL muscle (FDR < 0.05 for DE and PSI ≥ 0.2 for AS).
(C) Donut chart showing a description of tran- scriptome changes in EDL muscle 2 weeks after Rbfox knockout compared to control muscle. SE, cassette exons; A3SS, alternative 3ʼ splice site; A5SS, alternative 5ʼ splice site; AFE, alternative first exon; ALE, alternative last exon; MXE, mutually exclusive exons; RI, retained intron.
(D) 49 randomly selected alternative splicing events in EDL and soleus muscles were validated by RT-PCR, and ∆PSI was plotted against ∆PSI derived from RNA-seq data.
(E) DAVID gene ontology (GO) analyses showing biological processes enriched in genes that are alternatively spliced in Rbfox-knockout muscles.
(F) oPOSSUM analysis to determine the enrichment ofclustered transcription factor-binding sites (TFBSs) in genes that are upregulated after Rbfox knockout.
(G) RNA-seq tracks for Mef2d in control and Rbfox-DKO muscles showing that muscle-specific inclusion of α2 and β exons requires Rbfox proteins.
(H) RT-PCR showing reduced PSI for Mef2d α2 and β exons in DKO muscle when compared to control muscle (n = 3).
When we examined alternative splicing, we found that 743 genes showed different usage of exons between control and Rbfox-DKO EDL muscle, with a percent spliced in (PSI) cutoff of ≥20% (Figure 3B; Table S3) (Wang et al., 2008). There was little overlap, of only 68 genes, between genes that changed in overall mRNA levels (832 genes) and those that showed differential usage of alternative exons (743 genes) (Figure 3B). Similarly, of the 389 genes that showed alternative use of exons, only 9 differed in overall expression (FDR < 0.05) between DKO and control soleus muscles (Figure S3B; Table S3). The majority of alternative exons altered by Rbfox DKO was cassette-type in both EDL (460 exons) and soleus (146 exons) (Figures 3C and S3C). RT-PCR analysis of splicing for 49 alternative exons found a strong correlation (R2 = 0.92) between ΔPSl from RT-PCR and computed from the RNA-seq data (Figure 3D).
We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation tool to identify biological processes that are enriched among these groups of genes (Huang da et al., 2009). Functions that are enriched among those genes that were downregulated include oxidation-reduction, calcium handling, and cell signaling (Figure S3D). Enriched functions among upregulated genes include muscle differentiation and development, muscle contraction, cell adhesion, and transcription regulation (Figure S3E). Enriched categories among genes showing altered splicing include cell-cell adhesion; cytoskeleton organization; chromatin modification; and regulation of protein metabolism, including translation, transport, localization, and phosphorylation (Figure 3E).
We used a web-based tool, oPOSSUM-3, to identify over-represented clustering of transcription factor-binding sites (TFBSs) within 5,000 bases upstream and downstream of genes that showed altered expression in Rbfox-DKO muscle (Kwon et al., 2012). In upregulated genes, TFBS clustering analysis showed enrichment for the MADS family of transcription factors (Figure 3F). TFBS analysis identified 441 MADS-binding sites clustered in the promoter regions of 206 genes that were upregulated but only 19 binding sites in 17 genes that were downregulated in Rbfox-DKO muscle; this suggests that alterations in the MADS transcriptional program lead to the upregulation of its targets (Table S4).
It is worth noting that the MADS family includes the Mef2 transcription factors. We and others have shown that Mef2a and Mef2d splicing is coordinately and directly regulated by Rbfox proteins during myogenesis in culture (Runfola et al., 2015; Singh et al., 2014); we have also previously shown that the Mef2d splice variant expressed in adult skeletal muscle is required for late stages of muscle differentiation (Singh et al., 2014). Here we found that Rbfox-DKO muscle displayed significantly lower use of the β exon of Mef2a and both the α2 and β exons of Mef2d (Figures 3G, 3H, S3G, and S3H), representing a loss of muscle-specific isoforms. We conclude that Rbfox coordinately regulates splicing of Mef2a and Mef2d to maintain the expression and proper activity of muscle-specific isoforms (Sebastian et al., 2013).
Altered Proteostasis in Rbfox-DKO Muscle
Skeletal muscle mass is determined by the balance between rates of protein synthesis and protein degradation. The rate of protein synthesis in skeletal muscle is regulated by growth factor, amino acid, and insulin-signaling pathways (Figure 4A). We first quantified the phosphorylation status of Akt (S473) and S6 Kinase (S389), the key downstream phosphorylation events of insulin signaling, and we found no significant changes in the ratio of phospho/total protein levels in Rbfox-DKO mice when compared to control mice (3 weeks post-induction; Figures S4A and S4B). We also quantified the inhibitory phosphorylation of AKT on Gskβ (S9) and FoxO1 (S256), and we found no changes in phospho/total protein ratios of these proteins in Rbfox-DKO mice when compared to control mice (Figures S4A and S4B). Overall, the downstream insulin-signaling pathways that are important for muscle mass maintenance were not signif- icantly altered in Rbfox-DKO mice. We then measured the rate of protein synthesis in control and Rbfox-DKO TA muscles 2 weeks after knockout induction using puromycin in the surface sensing of translation (SUnSET) assay (Goodman and Hornberger, 2013; Schmidt et al., 2009). There was no reduction of puromycin- labeled peptides in DKO muscle compared to controls (Figure 4B), indicating that the loss of muscle in Rbfox-DKO skeletal muscle was not due to a reduction in protein synthesis. We therefore turned our attention to proteolytic mechanisms of muscle loss.
Figure 4. Altered Regulation of Calpain in Rbfox-DKO Muscles.
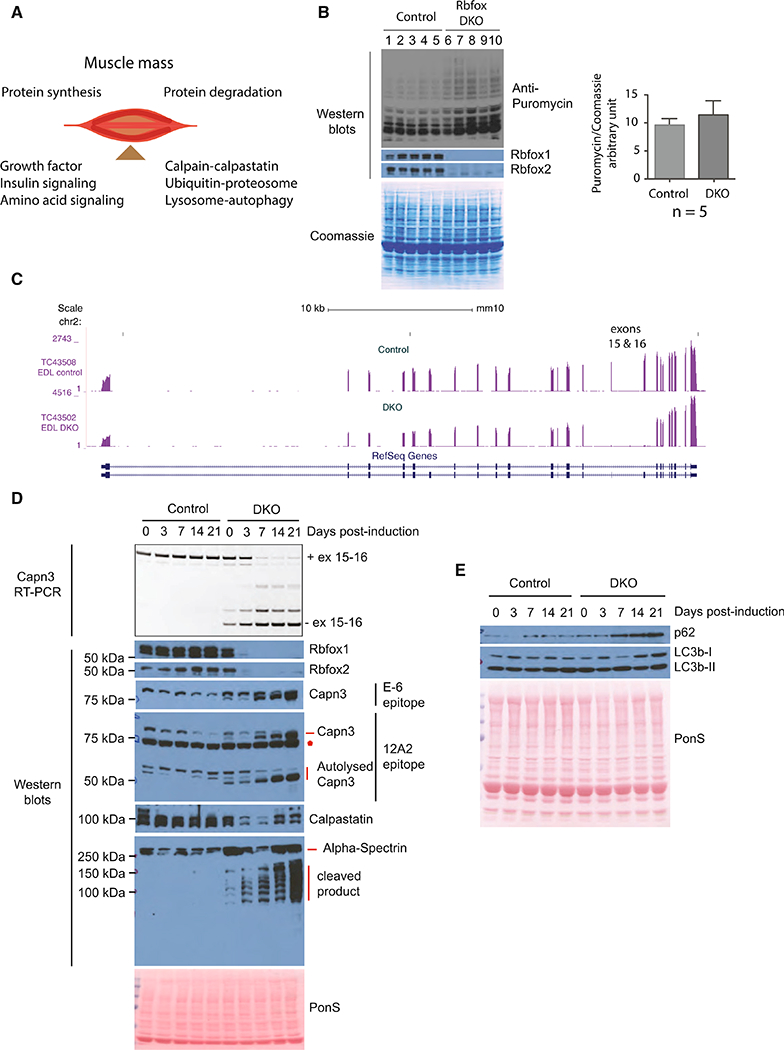
(A) Maintenance of skeletal muscle mass is a balance between protein synthesis and protein degradation. Major determinants of protein synthesis and protein degradation are indicated.
(B) Puromycin incorporation into nascent proteins in TA muscles was quantified in five control (lanes 1–5) and Rbfox-DKO animals (lanes 6–10) 2 weeks after the induction of Rbfox DKO. Western blots using antibody against puromycin, Rbfox1, or Rbfox2 and Coomassie staining of the blot are shown. Puromycin intensity/total Coomassiesignal was calculated asarelative measure oftotal protein synthesisand plotted in a bargraph (right panel). Thecolumnsand errorbars in graphs indicate mean ± SD.
(C) RNA-seq tracks showing Capn3 in control and Rbfox-DKO muscles. Exons 15 and 16 are nearly completely skipped in Rbfox-DKO muscles.
(D) RT-PCR for calpain 3 using primer pairs annealing in exons 14 and 17, and western blots using antibodies against indicated proteins in uninduced (0 day) and 3-, 7-, 14-, and 21-day-induced TAmusclesfrom control and DKO animals. ThesameTAmuscleswere used to prepare RNAand protein extractsfor RT-PCR and western blots.
(E) Western blots for the indicated proteins to assess the effect of Rbfox DKO on autophagy.
Protein degradation is tightly regulated by three major proteolytic systems: (1) Calpain-Calpastatin, (2) lysosome-auto- phagy, and (3) ubiquitin-proteasome (Figure 4A). We focused our analysis on alternatively spliced genes involved in the three major protein degradation systems. We prioritized targets based on robustness of splicing changes in DKO based on RNA-seq data, the presence of Rbfox motifs (TGCATG or GCATG) near the regulated exons and their correlation with exon inclusion (downstream motifs) or exclusion (motifs upstream) (Yeo et al., 2009), and direct Rbfox binding to targets in muscle culture based on our previous individual-nucelotide resolution cross-linking and immunoprecipitation (iCLIP) data (Singh et al., 2014). We found that two consecutive alternative exons (exons 15 and 16) in Capn 3 transcripts were almost completely skipped in Rbfox- DKO muscle and are likely direct Rbfox targets (Figure 4C). Auto- lytic cleavage within the IS1 region enables proteolytic Capn3 activity, and cleavage within the IS2 region of Capn3 is required for complete degradation and inactivation of Capn3 (Figure S4C) (Ono et al., 2014). Skipping of exons 15 and 16 in Rbfox-DKO muscle likely produces a Capn3 isoform that lacks the cleavage site within the IS2 region, thereby stabilizing active Capn3 and causing it to accumulate. We hypothesized that this contributes to severe muscle loss by tilting the protein balance toward protein degradation.
To determine whether the timing of Rbfox loss correlated with altered splicing of Capn3 and the initiation of muscle loss, we performed western blots for Rbfox1 and Rbfox2 and RT-PCR for splicing of Capn3 exons 15 and 16 using primers complementary to exons 13 and 17 in uninduced and DKO muscles at 3, 7, 14, and 21 days after starting the dox diet. Rbfox1 and Rbfox2 knockout was efficient 3 days after starting the dox diet, which correlates with increasing expression of Capn3 transcripts lacking exons 15 and 16 (Figure 4D). By 7 days post-induction, the majority of the mRNA expressed from the Capn3 gene lacked both exons. We performed western blots for Capn3 using two antibodies, one recognizing an epitope in the first 80 amino acids (E-6, Santa Cruz Biotechnology) and the other (12A2, Leica) recognizing an epitope within residues 355–370 (Figure S4C). Both antibodies recognize the full-length protein and 12A2 also detects the enzymatically active (auto- lysed) C-terminal fragment of Capn3 (Charton et al., 2016; Fanin et al., 2003; Richard et al., 1995). Both antibodies recognized a band corresponding to full-length protein at 94 kDa in control animals and a slightly smaller band predominating following Rbfox DKO, likely encoded by the mRNA lacking exons 15 and 16 (Figure 4D). Importantly, both the full-length isoform and the active autolysed C-terminal fragments (55 and 60 kDa) detected by 12A2 accumulated after induction of the DKO, correlating with the timing of rapid muscle loss. These results indicate that the loss of Rbfox proteins caused altered splicing and production of smaller isoforms of Capn3, which is autolytically cleaved, pro-teolytically active, and stably expressed in DKO muscles.
We next determined whether Capn3 substrates are degraded in Rbfox-DKO muscle. Several studies have used calpastatin as a Capn3 substrate (Ono et al., 2004, 2014). Calpastatin is also an endogenous inhibitor of Capn1 and Capn2; we predicted that reduced calpastatin levels in Rbfox DKO would lead to increased Capn1 and Capn2 activity. By western blotting, we found that calpastatin was stably expressed in control muscles, but it was reduced in DKO muscles within 3 days after starting the dox diet (Figure 4D). Alpha-spectrin is another substrate for Capn3 (and possibly Capn1 or Capn2) (Huang and Forsberg, 1998; Na- kajima et al., 2006; Takamure et al., 2005; Zhang and Bhavnani, 2006), and western blotting showed a prominent band at the expected size for alpha-spectrin in control muscles, but an increasing proportion of several smaller fragments appeared in DKO muscles after starting mice on the dox diet (Figure 4D). These results support the hypothesis that increased calpain activity in Rbfox-DKO muscle causes degradation of calpastatin and alpha-spectrin, and likely other calpain substrates, which in turn contributes to the loss of muscle mass.
To assess the contribution of autophagy to muscle loss in Rbfox-DKO muscle, we performed western blotting for auto-phagy markers in control and DKO animals at different time points after DKO induction. In lysosome-autophagy proteolytic pathways, the autophagosome must fuse with the lysosome for substrate degradation to occur. Autophagosome formation requires the activity of the microtubule-associated protein 1A/ 1B-light-chain 3 beta (LC3B), which is first proteolytically cleaved to form LC3B-I. LC3B-I is then converted to LC3B-II by lipidation, which is present in the autophagosome membrane. Thus, different LC3B isoforms are used as markers of autophagic activity (Mizushima et al., 2010). We also quantified levels of Sequestosome1 (Sqstm1 or p62), a ubiquitin-binding protein and adaptor molecule that binds and sequesters substrates into autophagosomes (Mizushima et al., 2010). Compared to control muscle, we found a trend for the accumulation of p62 and LC3B in DKO muscle over time (Figure 4E).
To characterize the lysosome-autophagy flux in vivo in Rbfox- DKO muscles, we injected mice with colchicine, a drug that de- polymerizes microtubules and impairs autophagy by inhibiting autophagosome-lysosome fusion (Ju et al., 2010). Mice were injected with colchicine or vehicle (water) for 2 consecutive days at day 4–5 (early) or day 12–13 (late) after DKO induction. The TA was harvested 48 hr after the first injection (day 6 after DKO for the early time point and day 14 for the late time point). At the early time point, there was no difference in the levels of p62 or LC3B-II between water-injected control and Rbfox-DKO mice. Compared to water-injected mice, colchicine-injected mice without Rbfox KO showed the predicted accumulation of LC3B-II and p62 at the early time point (Figures 5A and 5B). In contrast, colchicine-injected Rbfox-DKO mice failed to produce an increase in p62 or LC3B-II (Figures 5A and 5B), indicating an early Rbfox-dependent reduction in autophagy flux.
Figure 5. Autophagy Flux Is Reduced in Rbfox-DKO Muscle.
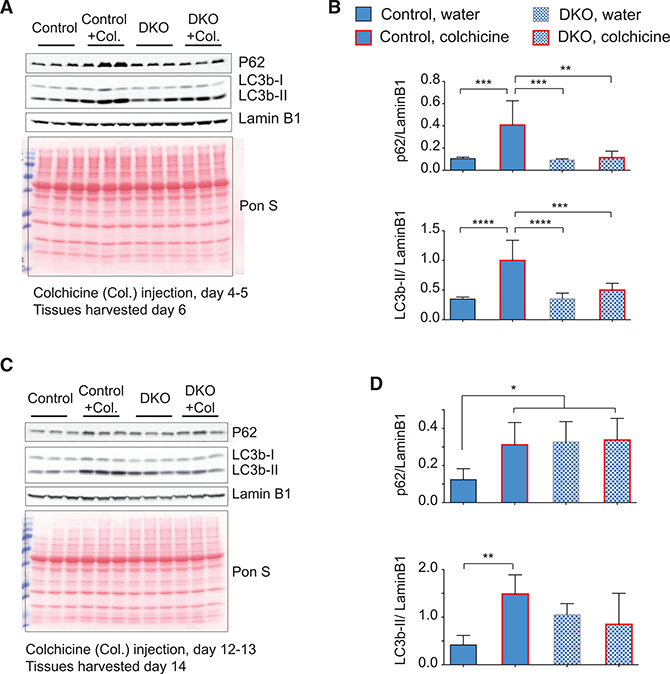
(A-D) Western blots using antibodies against the indicated proteins from TA muscles from control and DKO muscles. Control (Con) or Rbfox- knockout (DKO) mice were injected with colchicine (Col) or water at day 4–5 (A) or day 12–13 (C) following Cre induction, and muscles were harvested the next day for western blotting. Levels of p62 and LC3B-II were normalized to Lamin B1 and plotted in (B) and (D) for western blots in (A) and (C), respectively (n = 4 for the early time point and n ≥ 3 for the late time point). The columns and error bars indicate normalized mean intensity of p62 or LC3B-II ± SD. Red lines in the graph columns correspond to values from colchicine-injected mice. p values as indicated by asterisk were calculated by ANOVA, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
This early reduction in autophagy flux is expected to increase the accumulation of p62 and LC3B-II over time, and, consistently, we observed substantially increased p62 and LC3B-II levels in water-injected DKO mice in comparison to control mice, 2 weeks after the induction of KO. Compared to water-injected mice, colchicine-injected mice without Rbfox KO showed the predicted accumulation of LC3B-II and p62 at the late time point (Figures 5C and 5D). In DKO mice, already high levels of LC3B-II and p62 did not increase further after colchicine injection (Figures 5C and 5D), confirming that an early Rbfox-dependent reduction in autophagy flux is maintained at later time points. These results indicate that there is a marked reduction in autophagy flux in Rbfox DKO as early as day 4–6 after KO induction, which persists at later times.
The cellular proteins targeted for destruction by the protea-some are ubiquitinated by the activity of E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin-ligating) enzymes (Lecker et al., 2006). Several cases of muscle atrophy have been associated with the upregulation of E3 ubiquitin ligases, such Fbxo32 (atrogin-1) and Trim63 (MuRFI), which target sar- comeric components for degradation (Bodine and Baehr, 2014; Bodine et al., 2001). We performed western blotting for Murf-1, Trim 55 (MuRF-2), Trim 54 (MuRF-3), and Atrogin-1, and we found that Murf-1, Murf-3, and Atrogin-1 are expressed at similar levels in control and DKO muscles after the induction (Figure S4D). Murf-2 levels decreased slightly in the DKO, but overall we did not observe any increase in E3 ubiquitin ligases that are known to be upregulated in several muscle atrophy conditions. To determine the relative amount of proteins targeted for destruction by the ubiquitin-proteasome system in Rbfox-DKO muscle, we acutely inhibited the proteasome in mice using MG262, a potent inhibitor of proteasome (Frase et al., 2006; Lindsten et al., 2003). We quantified the relative levels of ubiqui- tinated proteins in control and Rbfox-DKO muscles in mice treated with vehicle (DMSO) or MG262 (5 μmol/kg). As expected, the treatment of MG262 caused a >2.5-fold increase in ubiquitinated proteins in control mice (Figures S5A and S5B). However, the DMSO-treated Rbfox-DKO mice displayed 50% more ubiquitinated protein compared to control mice after MG262 treatment. Compared to control muscles, there were more ubiq- uitinated proteins in Rbfox-DKO muscles in both DMSO- and MG262-treated groups, indicating that increased proteasome activity in Rbfox2-DKO muscles contributes to the loss of muscle mass in Rbfox-DKO mice.
From these experiments, we conclude that knockout of Rbfox proteins in skeletal muscle leads to increased calpain and proteasome activity along with impaired autophagic flux, thereby altering the main proteolytic systems in skeletal muscle. Such a dramatic imbalance of protein synthesis and degradation results in the net loss of protein and promotes atrophy.
DISCUSSION
We show that concurrent deletion of Rbfox1 and Rbfox2 in adult mouse skeletal muscle causes the dramatic and rapid loss of muscle mass. This response directly correlated with impaired autophagy flux, increased levels of ubiquitinated proteins that are targeted for destruction by the proteasome, and the formation of alternative isoforms of calpain 3 that are not degraded rapidly, leading to the accumulation of active calpain 3. Within 4 weeks of inducing the DKO, both male and female mice lost nearly half the muscle mass in the five fast-twitch muscles we measured. We also observed a significant decrease in whole- body lean mass (data not shown) after DKO induction, indicating ubiquitous loss of muscle mass in both male and female Rbfox- DKO mice. Despite the comparable muscle loss, however, only male mice had a shortened lifespan. The cause of this premature lethality is unclear, but we found alterations in glucose metabolism, trending toward insulin hypersensitivity, only in male animals. Interestingly, deletion of the estrogen receptor alpha (ERα) specifically in skeletal muscle of female mice causes an alteration in glucose homeostasis, increased fat accumulation, and impaired mitochondrial function (Ribas et al., 2016), so it may be that estrogen protects the female mice from death despite the dramatic loss of muscle mass after the induction of Rbfox knockout.
The inhibition of lysosome-autophagy causes loss of muscle mass in humans and mice, emphasizing the importance of auto- phagy in muscle growth, remodeling, and maintenance. Loss-of-function mutations in lysosome-associated membrane protein 2 (LAMP-2), a protein important for the fusion of autophagosome with lysosome, underlie Danon disease, in which patients present with skeletal muscle weakness, intellectual disability, and an accumulation of vacuoles with autophagic material in the heart and skeletal muscle (Bell et al., 2016; Masiero and Sandri, 2010; Raben et al., 2008; Tanaka et al., 2000). Deletion of Auto-phagy-related protein 7 (ATG7), a protein required for autophagosome formation, in skeletal muscle causes a loss of muscle mass in mice (Masiero et al., 2009). These mice show increased expression of atrogin-1 and Murf1 and a loss of muscle mass, which is caused, in part, by the compensatory upregulation of ubiquitin-proteasome proteolytic activity. We found that, in Rbfox, DKO caused a rapid reduction in autophagy flux and led to a greater accumulation of p62 and LC3B-II in DKO muscles over time (Figures 5A-5D). The reduction in autophagy flux in combination with increased calpain activity is expected to increase the demand for proteolytic activity. We propose this compensatory increase in proteolytic activity also contributes to the loss of muscle mass in Rbfox-DKO animals.
An isoform of Capn3 lacking IS1 or IS2 is proposed to be dominantly active with catastrophic effects (Beckmann and Spencer, 2008; Spencer et al., 2002). Consistent with this notion, muscle-specific overexpression of full-length Capn3 at high (over 25x wild-type) levels yields no observable phenotype, but overexpression of Capn3 lacking exon 6, which encodes most of the IS1 region, produces a severe phenotype that includes reduced muscle mass, smaller muscle fibers, kyphosis, and early death. The authors concluded that the phenotype is due to altered activity of Capn3 during the postnatal development (Kramerova et al., 2004; Spencer et al., 2002). Overexpression of a Capn3 cDNA lacking exon 15, which only lacks 6 amino acids in the IS2 region, leads to a subtle phenotype in the soleus, apparently without affecting other muscles, and Capn3 protease activity toward casein was much higher for muscle expressing the isoform lacking exon 15 compared to wild-type Capn3 or a Capn3 isoform lacking the full IS1 domain due to missing exon 6 (Spencer et al., 2002).
In our study, we induced Rbfox DKO in animals older than 7 weeks so that we would not affect the normal postnatal function of Capn3. Rbfox knockout switched the expression of full-length Capn3 to an isoform lacking both exons 15 and 16 (114 nt) that encode the majority (44 of 71 amino acids) of the IS2 region, which is required for the complete degradation of Capn3 (Figure S4). The smaller Capn3 isoform is autolytically processed to generate a catalytically active form of Capn3, and we propose that stable expression of the shorter, active form of Capn3 contributes to muscle loss in Rbfox-DKO mice. To determine whether knockdown of Capn3 would rescue muscle loss in the Rbfox DKO, we injected 6-week-old Rbfox-DKO mice with AAV-expressing control or Capn3 small hairpin RNAs (shRNAs) (targeting all isoforms) in contralateral hind limb gastrocnemius muscles. Rbfox knockout was induced 10 days after AAV injection, and muscles were harvested 4 weeks after knockout induction. We did not observe any significant changes in control muscles in response to Capn3 knockdown, but the muscle mass was reduced further (~13%) in Rbfox DKO after Capn3 knockdown in contrast to our expectations that the loss of Calpain3 activity should at least partially rescue muscle loss (data not shown). While these experiments confirm that Capn3 is playing a role in muscle maintenance, we believe it is hard to interpret these results in the Rbfox-DKO background given results from several labs indicating that either increased or decreased Calpain3 activity can cause muscle loss. Additional experiments would be required to identify the relevance and function of different Capn3 isoforms in adult skeletal muscle.
Calpains are nonprocessive proteases that cleave substrates at specific sites (Ono and Sorimachi, 2012). It has been predicted that over 1,000 proteins are calpain targets, yet fewer than 100 substrates have mapped calpain cleavage sites because cal-pain-cleaved fragments are short-lived (Ono and Sorimachi, 2012; Piatkov et al., 2014). Despite the critical role Capn3 plays in skeletal muscle biology, it is not clear which proteins are in vivo substrates for Capn3; the majority of its substrates have been identified through biochemical or cell culture studies (Huang and Forsberg, 1998; Nakajima et al., 2006; Ono et al., 2004, 2014; Takamure et al., 2005; Taveau et al., 2003; Zhang and Bhavnani, 2006). We found that levels of two of the previously identified substrates, calpastatin and alpha-spectrin, are reduced within 7 days after the induction of Rbfox knockout. Reduced calpastatin protein level would imply less inhibition of Capn1 and 2, strongly supporting a hypothesis that Capn3 is the master regulator of the calpain-calpastatin system in skeletal muscle (Ono et al., 2004). It is interesting that increased levels of Calpn3 lacking exons 15 and 16 are as detrimental to myofibers as loss-of-function mutations in Capn3, which are the most common cause of limb girdle muscular dystrophy type 2A (LGMD2A), a childhood-onset, progressive disease. The dramatic skeletal muscle phenotype in the Rbfox-DKO mouse provides clear evidence not only of the importance of alternative splicing for the maintenance of adult muscle mass but also of the sensitivity of muscle to alterations in proteostasis, as has been observed for the brain (Navone et al., 2015; Smith et al., 2015; Sulistio and Heese, 2016).
EXPERIMENTAL PROCEDURES
Mice
The Institutional Animal Care and Use Committee at Baylor College of Medicine approved all animal procedures. All mouse lines in this study were obtained from Jackson Laboratory (Bar Harbor, ME) and in the C57BL/6J genetic background. Previously described floxed Rbfox lines were mated with a muscle-specific, dox-inducible Cre line to generate single-Rbfox1 (Rbfox1 f/f: ACTA1-rtTAcre/+) or Rbfox2 (Rbfox2f/f: ACTA1-rtTAcre/+) and double-Rbfox (Rbfox1f/f: Rbfox2f/f: ACTA1-rtTAcre/+) knockout (Gehman et al., 2011, 2012; Pedrotti et al., 2015; Rao and Monks, 2009). For inducing the knockout, 7-week or older control (floxed mice) and littermate single- and/or DKO mice werefedadox-containing diet (2 g/kg, Bio-Serv, NJ) for 1 weekand thentrans-ferred to normal chow (5V5R, LabDiet, MO). Mice were anesthetized with iso-flurane, and tissueswere isolated, weighed, and snap frozen in liquid nitrogen for further analysis.
RNA Isolation, RNA-Seq, and Analysis of RNA-Seq Data
RNA was isolated from mouse muscles using TRIzol reagent (Invitrogen) or RNeasy fibrous tissue mini-kit (QIAGEN, Germantown, MD). We used RNA from two biological replicates from control and Rbfox-DKO EDL and soleus muscles at 14 days after knockout induction. The poly(A) RNA was isolated for paired-end 100-bp sequencing as previously described (Singh et al., 2014). The analysis of the RNA-seq data was performed by AccuraScience (Johnston, IA). RNA-seq reads were aligned to the genome using TopHat, Edge R was used for quantification of gene expression changes, MISO was used for quantification of alternative splicing changes, and PSI was calculated, as previouslydescribed (Katz et al., 2010; Robinson et al., 2010;Trapnell et al., 2010; Wang et al., 2008). To validate RNA-seq alternative splicing data, we performed standard RT-PCR using primers that flank specific alternative exons, and PCR products were separated on a 5% acrylamide gel. The gel was imaged and PSI was calculated as previously described (Singh et al., 2014).
Detection of Puromycin-Labeled Peptides
Rbfox knockout was induced in 7-week-old control and Rbfox-DKO male animals, and 2 weeks latermicewere injected intraperitoneally (i.p.) with puromycin (0.04 μmol/g body weight) or water (Goodman and Hornberger, 2013; Schmidt et al., 2009). TA muscle was harvested 45 min after the injection for western blotting with anti-Puromycin (3RH11) antibody (Kerafast, MA). The same western blots were stained with the Coomassie for the normalization of puromycin signal by total protein signal by densitometry.
Autophagy Flux Experiment
Autophagy flux was determined as described (Ju et al., 2010). Mice were injected i.p. with 0.4 mg/kg ofcolchicine(Sigma, St. Louis, MO)for 2 consecutive days at a 24-hr interval, and theywere sacrificed approximately 48 hrafterthe first injection. The relative levels of LC3B-II and p62 were quantified on western blots and normalized to Lamin B1.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq datafrom control and Rbfox DKO EDL and soleus muscle reported in this paper is Sequence Read Archive (SRA): SRP151185.
Supplementary Material
Highlights.
Proteostasis in adult skeletal muscle requires alternative splicing via Rbfox1 and Rbfox2
Rbfox1 and Rbfox2 regulate hundreds of splicing targets in skeletal muscle
Rbfox1 and Rbfox2 loss produces an active and stable isoform of calpain 3 protease
Autophagy flux is markedly decreased in muscle lacking Rbfox1 and Rbfox2
ACKNOWLEDGMENTS
This project was supported by the Mouse Phenotyping Core at Baylor College of Medicine with funding from the NIH (UM1HG006348). Measurements of body composition, energy balance, and food intake were performed in the Mouse Metabolic Research Unit at the USDA/ARS Children’s Nutrition Research Center, Baylor College of Medicine (https://www.bcm.edu/research/advanced-technology-core-labs/lab-listing/mouse-metabolic-research-unit) and supported by funds from the USDA ARS (USDA CRIS 6250–51000054). R.K.S. is supported by a post-doctoral fellowship and Scientist Development Grant from the American Heart Association (12POST11770017 and 15SDG25610021). This project is funded by grants from the Muscular Dystrophy Association (MDA276796) and NIH to T.A.C. (R01HL045565, R01AR060733, and R01AR045653). We thank A. Vainshtein and V. Brandt for helpful suggestions and careful reading of the manuscript. We also thank members of the Cooper lab for discussion and help throughout the project.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.06.017.
AUTHOR CONTRIBUTIONS
R.K.S. and T.A.C. conceived the study, designed the experiments, interpreted data, and wrote the manuscript. R.K.S. also performed the experiments. A.M.K. helped with mouse muscle function tests and validation of RNA-seq data, and M.L.F. contributed to the design, data analysis, and interpretation of energy balance studies.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Bassel-Duby R, and Olson EN (2006). Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem 75, 19–37. [DOI] [PubMed] [Google Scholar]
- Beckmann JS, and Spencer M (2008). Calpain 3, the ‘‘gatekeeper’’ of proper sarcomere assembly, turnover and maintenance. Neuromuscul. Disord 18, 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RA, Al-Khalaf M, and Megeney LA (2016). The beneficial role of proteolysis in skeletal muscle growth and stress adaptation. Skelet. Muscle 6,16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, and Baehr LM (2014). Skeletal muscle atrophy and the E3 ubiq- uitin ligases MuRF1 and MAFbx/atrogin-1.Am. J. Physiol. Endocrinol. Metab 307, E469–E484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708. [DOI] [PubMed] [Google Scholar]
- Bowen TS, Schuler G, and Adams V (2015). Skeletal muscle wasting in cachexia and sarcopenia: molecular pathophysiology and impact of exercise training. J. Cachexia Sarcopenia Muscle 6, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charton K, Suel L, Henriques SF, Moussu JP, Bovolenta M, Taille- pierre M, Becker C, Lipson K, and Richard I (2016). Exploiting the CRISPR/Cas9 system to study alternative splicing in vivo: application to titin. Hum. Mol. Genet 25, 4518–532. [DOI] [PubMed] [Google Scholar]
- Comai G, and Tajbakhsh S (2014). Molecular and cellular regulation of skeletal myogenesis. Curr. Top. Dev. Biol 110, 1–73. [DOI] [PubMed] [Google Scholar]
- Fanin M, Nascimbeni AC, Fulizio L, Trevisan CP, Meznaric-Petrusa M, and Angelini C (2003). Loss of calpain-3 autocatalytic activity in LGMD2A patients with normal protein expression. Am. J. Pathol 163, 1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frase H, Hudak J, and Lee I (2006). Identification of the proteasome inhibitor MG262 as a potent ATP-dependent inhibitor of the Salmonella enter- ica serovar Typhimurium Lon protease. Biochemistry 45, 8264–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, and Conboy JG (2011). Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol 359, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M Jr., Mody I, and Black DL (2011). The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet 43,706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M Jr., Otis TS, and Black DL (2012). The splicing regulator Rbfox2 is required for both cerebellardevelopment and mature motorfunction. Genes Dev. 26, 445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, and Hornberger TA (2013). Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc. Sport Sci. Rev 41, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, and Forsberg NE (1998). Role of calpain in skeletal-muscle protein degradation. Proc. Natl. Acad. Sci. USA 95, 12100–12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- James HA, O’Neill BT, and Nair KS (2017). Insulin Regulation of Proteo-stasis and Clinical Implications. Cell Metab. 26, 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Ya-suda K, and Inoue K (2003). A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22, 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Varadhachary AS, Miller SE, and Weihl CC (2010). Quantitation of ‘‘autophagic flux” in mature skeletal muscle. Autophagy 6, 929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, and Cooper TA (2011). Functional consequences of developmentaliy regulated alternative splicing. Nat. Rev. Genet 12, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, and Burge CB (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerova I, Kudryashova E, Tidball JG, and Spencer MJ (2004). Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet 13, 1373–1388. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Mitani S, and Hagiwara M (2007). The Fox-1 fam- ilyand SUP-12coordinately regulatetissue-specificalternative splicing invivo. Mol. Cell. Biol 27, 8612–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon AT, Arenillas DJ, Worsley Hunt R, and Wasserman WW (2012). oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2, 987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, and Mitch WE (2006). Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol 17, 1807–1819. [DOI] [PubMed] [Google Scholar]
- Lindsten K, Menendez-Benito V, Masucci MG, and Dantuma NP (2003). A transgenic mouse model of the ubiquitin/proteasome system. Nat. Bio- technol 21, 897–902. [DOI] [PubMed] [Google Scholar]
- Masiero E, and Sandri M (2010). Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6, 307–309. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, and Sandri M (2009). Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515. [DOI] [PubMed] [Google Scholar]
- McLeod M, Breen L, Hamilton DL, and Philp A (2016). Live strong and prosper: the importance of skeletal muscle strength for healthy ageing. Biogerontology 17, 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, and Levine B (2010). Methods in mammalian autophagy research. Cell 140, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima E, David LL, Bystrom C, Shearer TR, and Azuma M (2006). Calpain-specific proteolysis in primate retina: Contribution of calpains in cell death. Invest. Ophthalmol. Vis. Sci 47, 5469–5475. [DOI] [PubMed] [Google Scholar]
- Navone F, Genevini P, and Borgese N (2015). Autophagyand Neurodegeneration: Insights from a Cultured Cell Model of ALS. Cells 4, 354–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, et al. (2000). Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906–910. [DOI] [PubMed] [Google Scholar]
- Ono Y, and Sorimachi H (2012). Calpains: an elaborate proteolytic system. Biochim. Biophys. Acta 1824, 224–236. [DOI] [PubMed] [Google Scholar]
- Ono Y, Kakinuma K, Torii F, Irie A, Nakagawa K, Labeit S, Abe K, Suzuki K, and Sorimachi H (2004). Possible regulation of the conventional calpain system by skeletal muscle-specific calpain, p94/calpain 3. J. Biol. Chem 279, 2761–2771. [DOI] [PubMed] [Google Scholar]
- Ono Y, Shindo M, Doi N, Kitamura F, Gregorio CC, and Sorimachi H (2014). The N- and C-terminal autolytic fragments of CAPN3/p94/calpain-3 restore proteolytic activity by intermolecular complementation. Proc. Natl. Acad. Sci. USA 111, E5527–E5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti S, Giudice J, Dagnino-Acosta A, Knoblauch M, Singh RK, Hanna A, Mo Q, Hicks J, Hamilton S, and Cooper TA (2015). The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function. Hum. Mol. Genet 24, 2360–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkov KI, Oh JH, Liu Y, and Varshavsky A (2014). Calpain-generated natural protein fragments as short-lived substrates ofthe N-end rule pathway. Proc. Natl. Acad. Sci. USA 111, E817–E826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, and Olson EN (2007). MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140. [DOI] [PubMed] [Google Scholar]
- Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, and Plotz P (2008). Suppression ofautophagy in skeletal muscle uncoversthe accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet 17, 3897–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M, and Demontis F (2016). Systemic Nutrient and Stress Signaling via Myokines and Myometabolites. Annu. Rev. Physiol 78, 85–107. [DOI] [PubMed] [Google Scholar]
- Raj B, and Blencowe BJ (2015). Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 87, 14–27. [DOI] [PubMed] [Google Scholar]
- Rao P, and Monks DA (2009). A tetracycline-inducible and skeletal muscle-specific Cre recombinase transgenic mouse. Dev. Neurobiol 69, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas V, Drew BG, Zhou Z, Phun J, Kalajian NY, Soleymani T, Daraei P, Widjaja K, Wanagat J, de AguiarVallim TQ, et al. (2016). Skeletal muscle action of estrogen receptor a is critical for the maintenance of mitochondrial function and metabolic homeostasis in females. Sci. Transl. Med 8, 334ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. (1995). Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81, 27–40. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfola V, Sebastian S, Dilworth FJ, and Gabellini D (2015). Rbfox proteins regulate tissue-specific alternative splicing of Mef2D required for muscle differentiation. J. Cell Sci 128, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Coletto L, Grumati P, and Bonaldo P (2013). Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci 126, 5325–5333. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, and Sandri M (2013). Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 280, 4294–4314. [DOI] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Sebastian S, Faralli H, Yao Z, Rakopoulos P, Palii C, Cao Y, Singh K, Liu QC, Chu A, Aziz A, et al. (2013). Tissue-specific splicing of a ubiqui-tously expressed transcription factor is essential for muscle differentiation. Genes Dev. 27, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Xia Z, Bland CS, Kalsotra A, Scavuzzo MA, Curk T, Ule J, Li W, and Cooper TA (2014). Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol. Cell 55, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HL, Li W, and Cheetham ME (2015). Molecular chaperones and neuronal proteostasis. Semin. Cell Dev. Biol 40, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MJ, Guyon JR, Sorimachi H, Potts A, Richard I, Herasse M, Chamberlain J, Dalkilic I, Kunkel LM, and Beckmann JS (2002). Stable expression of calpain 3 from a muscle transgene in vivo: immature muscle in transgenic mice suggests a rolefor calpain 3 in muscle maturation. Proc. Natl. Acad. Sci. USA 99, 8874–8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulistio YA, and Heese K (2016). The Ubiquitin-Proteasome System and Molecular Chaperone Deregulation in Alzheimer’s Disease. Mol. Neurobiol 53, 905–931. [DOI] [PubMed] [Google Scholar]
- Takamure M, Murata KY, Tamada Y, Azuma M, and Ueno S (2005). Cal- pain-dependent alpha-fodrin cleavage atthe sarcolemma in muscle diseases. Muscle Nerve 32, 303–309. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, LÜllmann- Rauch R, Janssen PM, Blanz J, von Figura K, and Saftig P (2000). Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906. [DOI] [PubMed] [Google Scholar]
- Taveau M, Bourg N, Sillon G, Roudaut C, Bartoli M, and Richard I (2003). Calpain 3 is activated through autolysis within the active site and lyses sarcomeric and sarcolemmal components. Mol. Cell. Biol 23, 9127–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Vignal E, Baghdiguian S, Fort P, and Tazi J (2012). Tissue- specific alternative splicing of Tak1 is conserved in deuterostomes. Mol. Biol. Evol 29, 261–269. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, King- smore SF, Schroth GP, and Burge CB (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, et al. (2016). Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 164, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, and Gage FH (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol 16, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, and Rudnicki MA (2013). Satellite cells and the muscle stem cell niche. Physiol. Rev 93, 23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Bhavnani BR (2006). Glutamate-induced apoptosis in neuronal cells is mediated via caspase-dependent and independent mechanisms involving calpain and caspase-3 proteases as well as apoptosis inducing factor (AIF) and this process is inhibited by equine estrogens. BMC Neurosci. 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


