Abstract
Background: Gallbladder carcinoma (GBC) is a subtype of biliary tract malignancy with poor prognosis and high fatality rate. The present study was designed to uncover somatic and rare germline mutations in GBC to reveal the disease biology and understand the clinical importance of mutation profile in terms of prognostics and actionability. Materials and Methods: We performed ultra-deep sequencing across 409 cancer-related genes in 11 GBC patients of North-Indian descent. NGS data analysis was performed using Ion Reporter and several other publicly available resources and databases. Results: We identified 184 nonsynonymous somatic and 60 rare germline mutations in bona-fide cancer drivers such as SMAD family member 4 (SMAD4), lysine methyltransferase 2C (KMT2C), and tumor protein p53 (TP53). All the early-onset cases or hypermutated cases harbored mutation(s) in critical DNA-repair genes. Additionally, we detected 9 novel genes with high-impact somatic mutations in GBC. Conclusion: Our results indicated the significance of inherited rare germline mutations in DNA-repair pathway genes in addition to acquired somatic mutations in GB carcinogenesis.
Keywords: Gallbladder neoplasms, biliary tract cancer, targeted therapy, next generation sequencing, targeted therapy
Gallbladder carcinoma (GBC) is a type of biliary tract cancer, characterized by rapid progression and high mortality. Significant risk factors for GBC include presence of gallstones, chronic inflammation, anomalous pancreatobiliary ductal junctions, advancing age and female gender (1). Incidence and prevalence of GBC exhibit remarkable ethnic and geographical variability. This variability has been attributed to genetic predisposition and lifestyle habits or environmental exposure. The high incidence of GBC in the Indo-Gangetic belt and its poorly characterized molecular pathology have resulted in its reputation as an enigmatic Indian carcinoma (2). Until 2010, the genomic studies of GBC were limited to scanning a few common mutations in well-known cancer-associated genes such as tumor protein p53 (TP53) and KRAS proto-oncogene, GTPase (KRAS) (3-5). Advancements in massively parallel sequencing technology, or next-generation sequencing (NGS), have propelled the area of cancer genomics and have greatly improved the understanding of tumorigenesis and tumor evolution linked with disease progression (6,7). Application of NGS provides a means of high-throughput identification of cancer drivers and other genes that may be clinically relevant and or actionable for precision medicine (6). During the past decade, several large-scale cancer mutational landscape studies have contributed to the understanding of the existence of clonal variability within and across tumors in various common cancer types (8). However, GBC remains one of the understudied cancer types, with only a few studies on a limited number of samples (9-11). These studies and previous candidate gene-sequencing investigations indicate that GBC tumors from patients from different geographic regions may have different genetic architectures, suggesting the need for mutation exploration using a high-throughput NGS approach from varied populations (5,12).
The current treatment for patients with GBC is operative resection. However, resection is limited to cases that present at an early stage. GBCs which have progressed are largely incurable and the treatment mostly relies on focal radiation therapy and/or generic chemotherapy (12). The overall survival rate for patients with advanced GBC (stage 3 and 4) is very poor, and fewer than 10% survive for 5 years post diagnosis (13). Meanwhile, molecular and translational research studies of other extensively studied cancers such as breast, lung, melanoma, and leukemia, have shown that one of the most prominent reasons for generic treatment failure is the existence of molecular stratification across different tumors (8). However, due to limited somatic and germline mutational investigation in GBC, there is a gap in our understanding of molecular pathobiology of these tumors. Thus, GBC requires deep cancer genome excavation of signature mutations, especially cancer-related genomic regions, in order to understand genetic molecular heterogeneity (activation or inactivation of genes due to various mutations) and to define targetable molecular nodes of the tumor. Further sequencing of more and more tumors is of utmost importance in this field.
The present study was designed to uncover the spectrum of somatic as well as rare germline mutations and to understand disease biology and clinical importance of mutation profile in terms of prognostics and actionability of the detected mutation profile.
Materials and Methods
We recruited GBC cases only from the Indo-Gangetic region of northern-India (one of the hot-spots of GBC incidence) in order to study a cohort with similar lifestyle habits and exposures, and to reduce ethnicity-associated stratification across the samples. We then explored mutational profile of primary GB carcinomas through massively parallel sequencing with the AmpliSeq comprehensive cancer panel. This gene panel targets the exons of 409 oncogenes and tumor suppressor genes that are frequently mutated in various cancer types. It has been designed to simultaneously investigate the coding DNA sequences across multiple gene families that have been implicated in other cancer types.
Tissue selection and DNA extraction. The study was carried out in accordance with relevant guidelines and regulations, following approval by the ethical committee (code: 2012-169-EMP-66) of Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS) Lucknow, India. The informed consent was obtained from all the patients to be included in the study. Hematoxylin and eosin stained tissue sections of formalin-fixed paraffin-embedded (FFPE) or fresh tumor samples were reviewed by two pathologists. The tumor area was selected and marked. Only specimens with a minimum of 90% tumor content were selected for this study. Areas with a high degree of necrosis and intra-tumoral fibrosis were excluded from the study. Unstained tissue sections (5 µm-thick) were cut and deparaffinized and tumor tissue specimen was scarped for DNA extraction. Normal DNA was obtained from healthy tissue surrounding tumors or blood samples. Blood DNA was extracted by Qiagen blood extraction kit (Qiagen, Hilden, Germany). DNA was extracted using QIAamp DNA FFPE Tissue Kit (Qiagen). The Qubit DNA HS assay kit was used to quantify purified DNA.
Library preparation for Ion Proton sequencing. Library preparation for the comprehensive cancer panel was performed following the Ion Torrent Ampliseq Comprehensive Cancer Panel (Thermo Fisher Scientific, MA, USA) as per the manufacturer’s instructions. Sample barcoding was accomplished using Ion Xpress Barcode Adapter 1–96 kit (Thermo Fisher Scientific). The library prepared in each pool was quantified using a 2100 Bioanalyzer System (Agilent Technologies, South Clara, CA, USA). To clonally amplify the 409-gene panel DNA library on Ion Spheres (Thermo Fisher Scientific), emulsion polymerase chain reaction (PCR) was performed using Ion PI Template OT2 kit v2 and Ion One Touch 2 system (Thermo Fisher Scientific) as per the manufacturer’s instructions.
Sequencing data analysis using Ion Proton. Identification of sequence variants was performed using Ion Reporter Software Version 4.6 (Thermo Fisher Scientific). Human genome build 19 (hg19 or GRCh37) was used as the reference for alignment of the sequencing reads. After alignment, variant calling was performed using AmpliSeq CCP tumor-normal pair pipeline. Somatic single nucleotide variants (SNVs) having functional effect (frameshift insertion, stoploss, frameshift deletion, missense, frameshift block substitution, nonsense) with filtered coverage ≥50 reads and alternate allele reads count ≥10 were used as thresholds for filtering. Considering the risk of deamination-mediated genomic alterations, we removed the variants having alternative allelic frequency ≤0.05 and alternative allele supporting reads ≤50 in C>T and G>A mutations from the FFPE samples (presence of low frequency alternative allele in normal (germline) sample considered strongest red flag for removal as well).
For filtering germline variants, we utilized variant calls from blood and non-tumor control samples. We applied the above-mentioned filters in addition to population variant allelic frequency (<0.001%) from the ExAC database (http://exac.broadinstitute.org/) and SIFT (http://sift.jcvi.org/) and PolyPhen-2 (http://genetics.bwh. harvard.edu/pph2/) score (0.0≤SIFT≤0.05 OR 0.85≤PolyPhen≤1.0) (14-16). The read alignment and sequence variants were confirmed against the reference genome utilizing the Integrative Genomics Viewer (IGV) (17). Strand biases and sequencing errors were also considered to detect reliable sequence variants.
Deleteriousness assessment of variants. There are several tools to predict the deleterious impact of variant in different diseases. Many of them are enlisted by the American College of Medical Genetics and Genomics (ACMG) in its recommendations for pathogenicity detection documents (18). All the predictive tools have issues of low specificity and prediction of several false-positives. Even the larger consortiums such as the International Cancer Genome Consortium (ICGC) and the Cancer Genome Atlas (TCGA) follow deleterious calls following the outcome of predictors. The ACMG has stringent recommendations for calling a variant clinically “pathogenic” or “likely pathogenic”. In this study, deleteriousness of a mutation or variant was assessed based on sequential decision through several definitive and predictive analyses. Weight was given to the availability of references in deleterious mutation information bases such as HGMD (www.biobase-international.com/hgmd), and ClinVar annotations (19,20). General population frequency estimates of the mutations were referred from ExAC and 1000 genome database (21). We assessed the variants for their truncating amino acid product and thus loss or alteration of functional domains which makes them candidates for deleterious variant classification. Assessment of missense mutations was achieved through several stepwise predictive steps. Initial screens for relatively rare variants were carried out through PolyPhen-2 and SIFT recommended cut offs. Using Panther (http://www.pantherdb.org/pathway/) and MutPred (http://mutpred.mutdb.org/), we predicted the possible impact of a SNV on some of the 3D structural features of the protein and tried to predict potential candidates of deleterious mutations and classified them as “likely deleterious” (22,23).
Results
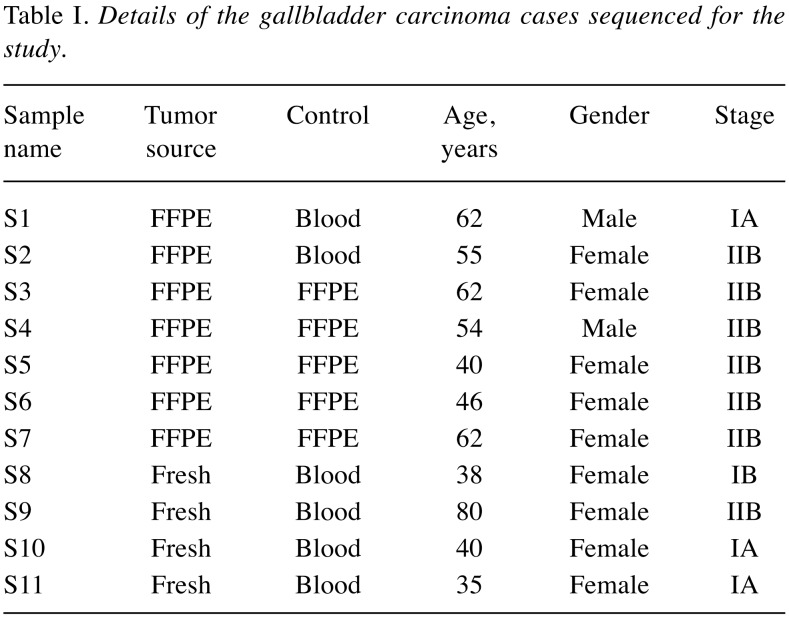
Characteristic profile of the study subjects. After obtaining written informed consent, blood, adjacent non-cancerous and tumor tissues were collected from 11 patients with GBC who underwent a comprehensive staging and curative resection at SGPGIMS, Lucknow, India. Data on demography, risk factors, clinical, radiological and histopathological features, and disease status at follow-up were collected. The characteristics of patients with GBC included in this study are listed in Table I.
Table I. Details of the gallbladder carcinoma cases sequenced for the study.
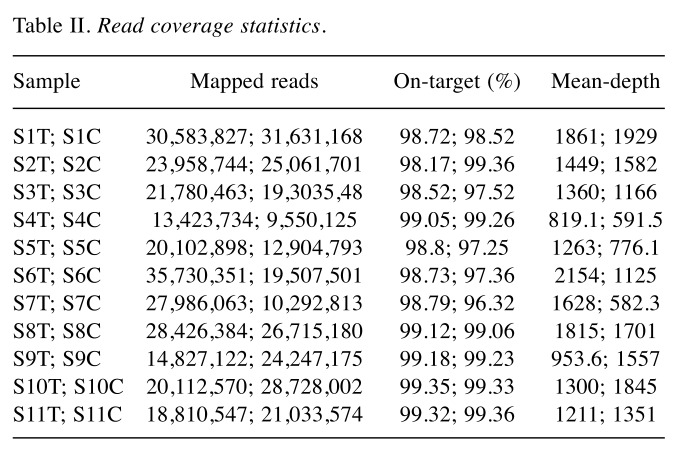
Targeted deep sequencing. We performed ultra-deep sequencing of 409 cancer related protein coding genes in 11 patients with GBC including tumor tissues (seven FFPE preserved and four fresh frozen) and the respective paired-normal or non-neoplastic tissues or blood (Table I). The extracted DNA from tumor tissues and normal samples were used to prepare an amplicon-based targeted library (Ion Torrent Ampliseq Comprehensive Cancer Panel). The Ion proton sequencing yielded sequencing depth ~1365X on average (range, 582X- 2154X) (Table II). Somatic variants were identified by comparing paired normal and tumor tissues; variants calls were performed on Ion Reporter by launching AmpliSeq CCP tumor-normal pair variant analysis pipeline. Confident variant calls were made using cascades of Ion Reporter filters.
Table II. Read coverage statistics.
We identified 184 somatic point mutations in cancer-related genes (172 unique mutation loci) after quality assessment. Among these 184 somatic SNVs, 180 were missense, three nonsense and one splice site/regulatory mutations. Functional annotations of genetic variants from public databases such as COSMIC, and dbSNP were provided by the Ion Reporter data analysis workflow (24).We removed two samples (samples S6 and S7) from downstream analysis for uneven coverage issues, finally reporting 182 confident high-quality nonsynonymous somatic mutations in 170 unique loci (Figure 1 and Figure 2); 126 transitions (C>T=48, T>C=23, G>A=38, A>G=17) and 56 transversions (A>C=6, C>A=12, A>T=6, T>A=3, G>T=7, T>G=7, G>C=6, C>G=9). C>T/G>A substitutions followed by T>C/ A>G represented the dominant somatic transition substitution for GBCs; we observed a higher proportion of transition (69.23%) compared to transversion (30.77%) events in the enriched targeted region of sequencing (after deamination filter).
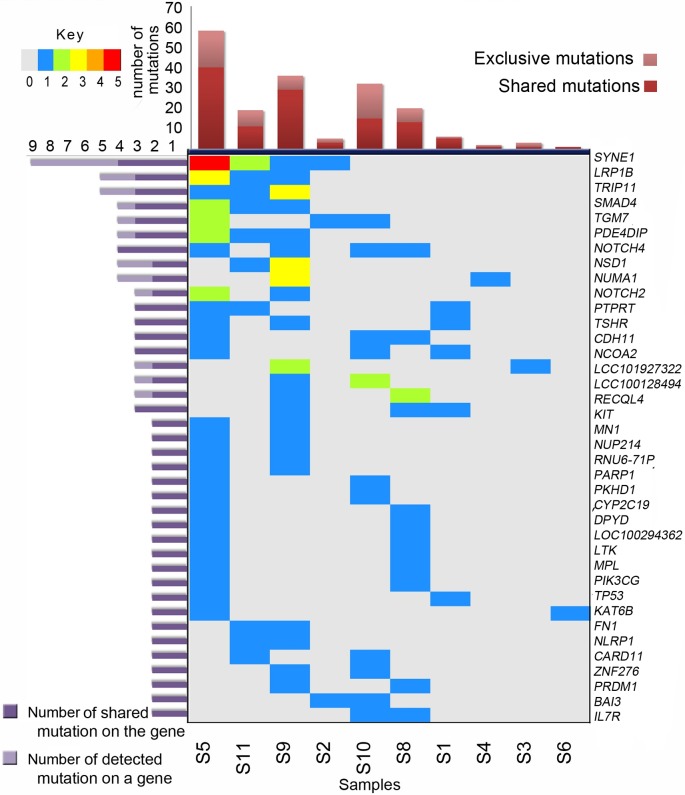
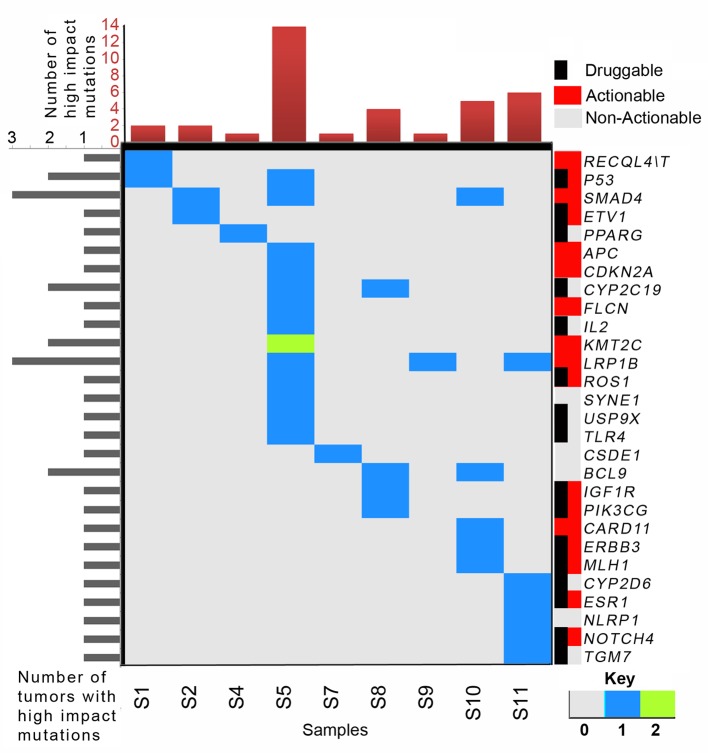
Figure 1. Frequently mutated genes and sample-wise mutation frequency in gallbladder cancer. Genes are arranged according to the total number of mutations per samples. Oncoplot shows the numbers of mutations per sample represented through different colors. Color keys are provided in top left panel. Samples are not arranged according to number of mutations. We did not detect good quality high-impact somatic mutation in S7. The bar plot at the top depicts the total number of detected somatic mutations.
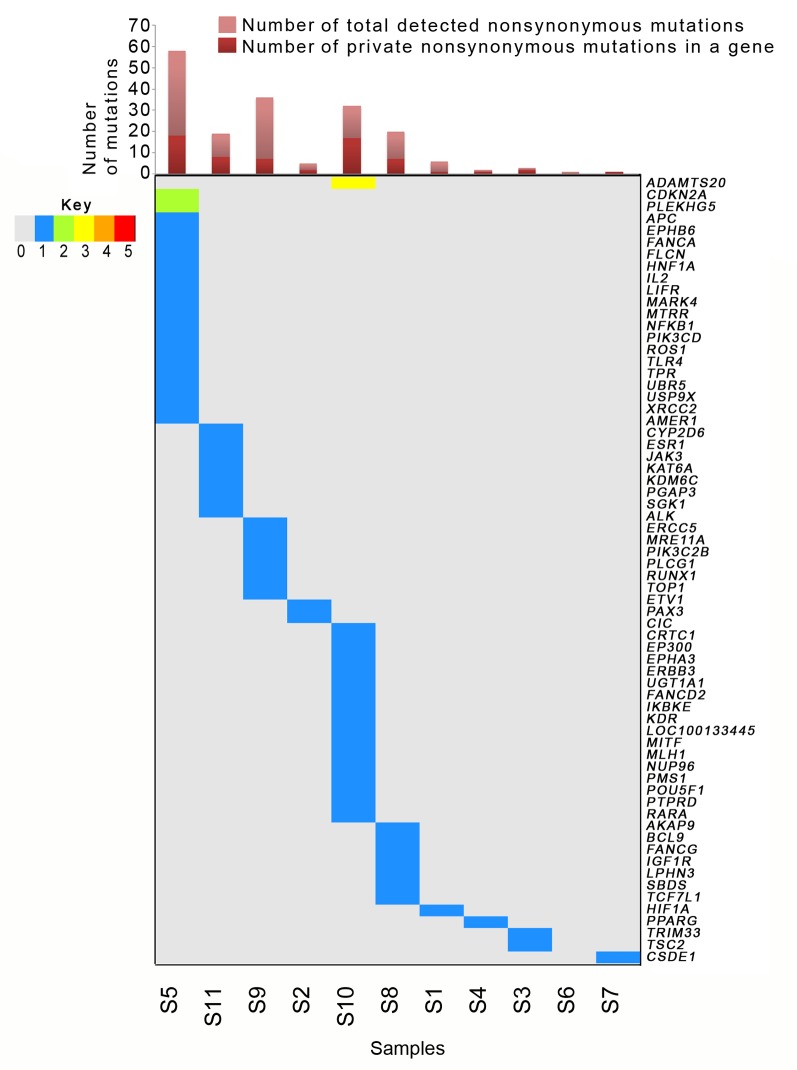
Figure 2. Distribution of private somatic mutations in different genes across gallbladder cancer tumors. Oncoplot presents genes which were mutated in only one sample. Numbers of mutations per sample are represented through different colors. Color keys are provided in the left panel. Samples are arranged according the frequency of mutations in each gene. Singleton mutations were most frequent in sample S5 followed by samples S10, S11, S8 and S9. The bar plot at the top depicts the total number of detected somatic mutations. S6 had no exclusive mutation.
Somatic mutation spectra across samples. Mutation load and spectrum of mutations across samples varied considerably, representing intertumoral heterogeneity in GBCs. Although most of the FFPE samples suffered overall low detectability of mutations (four per sample), sample S5 was an exception (Figure 1 and Figure 2). Fresh samples (S8-S11) had much better and even coverage across their normal and diseased tissues and had on average higher numbers of detected mutations (average 27 mutations per sample). Although sample 5 had DNA of FFPE origin, it was observed to have a higher number of identified mutations (n=58) and thus qualified as a representative- of hyper-mutator phenotype (Figures 1 and 2). In contrast, samples 6 and 7 were observed to carry only one detectable somatic mutation each [lysine acetyltransferase 6B (KAT6B) and cold-shock domain containing e1 (CSDE1) mutation, respectively]. Tumor S5 was found to harbor deleterious/ likely deleterious mutations in DNA-repair pathway genes such as poly ADP ribose protein 1 (PARP1), Fanconi anemia complementation group A (FANCA) and x-ray repair cross complementing 2 (XRCC2) (Figures 2 and 3). A mutation in folliculin (FLCN) in the same tumor may also be linked with the hyper-mutator phenotype. We were able to identify 31 and 30 somatic mutations in two other well-covered samples (samples S9 and S10 respectively). The number of mutations in S9 and S10 were clearly higher than mutations in two other well-covered samples, S8 and S11. The higher frequency of mutations in sample S10 can be linked to the mutations in several DNA repair-associated genes such as MutL homolog 1 (MLH1) and PMS1 homolog 1, mismatch repair system component (PMS1). Sample S10 also harbored mutations in poly(ADP-ribose) polymerase 1 (PARP1), FANCA (3’ UTR) and FANCD2 genes, whereas in sample S9, we detected a heterozygous but likely deleterious TP53 germline mutation (25).
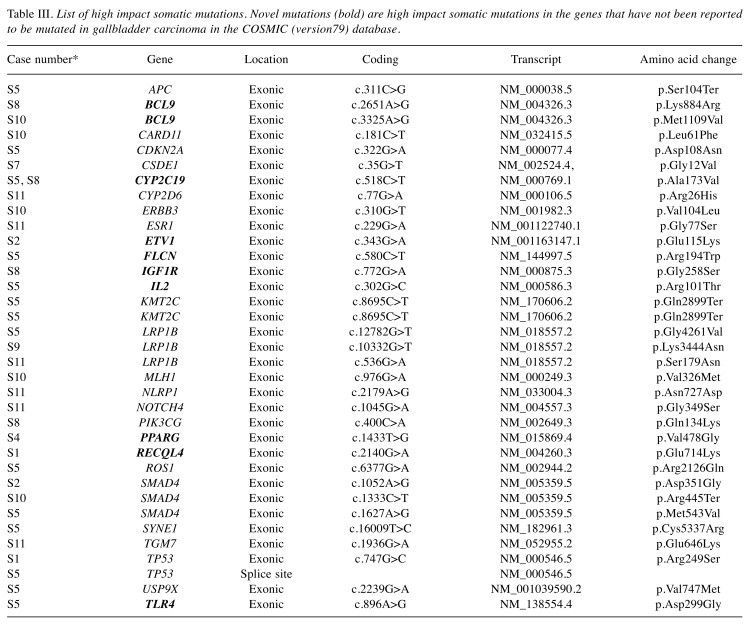
Biological relevance of somatic mutation profiles. We selected 409 genes implicated in various cancer types. Instead of using statistical pathway enrichment, we looked for pathway annotations of different mutated genes using Ion Reporter’s pathway annotation track and cross-validated them using independent annotation tools. Based on our results, we assessed 39 high-impact mutations in 28 genes. The list of high-impact variants is presented in Figure 3, and Table III. High-impact mutations are either present in strategic regions of functional domains, or affect accessibility of functional domains. We found somatic mutations in genes mostly belonging to cell cycle, apoptosis [i.e. B-cell CLL/lymphoma 9 (BCL9), cyclin-dependent kinase inhibitor 2A (CDKN2A), DNA damage and repair (i.e. PARP1, XRCC2, MLH1), and chromatin binding and remodeling [i.e. KDM5C, PR/SET domain 1 (PRDM1] pathways. Some of the cancer predisposition genes [i.e. FLCN in S5, FANCA in S5 and S10, RecQ Like Helicase 4 (RECQL4) in S1 and S8] were also found to be somatically mutated in a few samples. We also detected somatic mutations in key genes from WNT [e.g. adenomatosis polyposis coli tumor suppressor (APC), transcription factor 7 like 1 (TCF7L1)] and NOTCH signaling (NOTCH2, NOTCH4) pathways. Although we adopted a universal cancer gene panel for sequencing, we observed enrichment of a few key cancer-associated biological pathways even in our set of nine GBC tumors, indicating relevance of these pathways in GBC pathogenesis.
Figure 3. Spectrum of high-impact somatic mutations and their sample-wise distribution. High-impact somatic mutations of low-density lipoprotein receptor-related protein 1 (LRP1B) and SMAD family member 4 (SMAD4) were mutated in the highest proportion of samples (3/9 samples). We were unable to detect any actionable mutation in S6. Right color bar panel represents actionability or druggability status of these genes. Numbers of mutations in a gene of a tumor are presented according to the color key. The bar plot in the top panel presents the total number of high-impact somatic mutations detected.
Table III. List of high impact somatic mutations. Novel mutations (bold) are high impact somatic mutations in the genes that have not been reported to be mutated in gallbladder carcinoma in the COSMIC (version79) database.
Targetable mutated genes. Like several other mutation or variant excavation studies, our goal was to assess the gene mutation architectures of these nine GBC cases to identify nodal molecular switches that might be considered as effective potential therapeutic targets. Thorough understanding of the altered molecular hierarchy would theoretically help in defining case-specific effective therapeutic strategies. To accomplish this, we intersected the existing targetable list with our functionally relevant observations (mutations). We considered several different databases including ClinicalTrials.gov, My Cancer Genome Clinical Trials, DrugBank, My Cancer Genome, and TDG clinical trial, which aggregate information on clinically approved drugs as well as several novel agents under clinical trial. In this study, we identified 16 potential druggable genes and 17 clinically actionable genes in GBC (Figure 3). However, in the case of sample S3, we failed to find any high-impact actionable mutation.
Spectrum of deleterious germline variants. In the light of Knudson’s two-hit hypothesis and recent cancer genomics studies, the importance of inherited germline mutations in carcinogenesis has become clearer (27-29). Instead of entire profile of germline variants, we restricted our study only to rare and deleterious germline mutations. Population allelic frequencies were extracted from dbSNP (version 138), ExAC and 1000 Genomes database. We also looked for known deleterious missense mutations in ClinVar. Initially, a total 102 high quality variants were detected in 51 unique genes. Information from several databases including Pfam was also consulted followed by lenient adoption of ACMG recommendations to narrow down to 60 candidate unique germline mutations in 46 different genes (Table IV) (30). In this study, we tagged mutations as deleterious or likely deleterious, instead of pathogenic or likely pathogenic mutations. Most of the detected deleterious or likely deleterious mutations were never observed to have a second hit in any of the studied tumor tissues. An exception was noticed with Tet methylcytosine dioxygenase 1 (TET1) gene, which is associated with the myeloproliferative phenotype of different hematologic cancers. Loss of TET1 is known to be associated with aberrant hypermethylation in liver tumors leading to disrupted expression homeostasis (31). The impact of two TET1 mutations in a sample (two hits) was reflected through early age of disease onset of this patient (S6, age at diagnosis: 46 years). In sample S12, two genes [mucin 1, cell surface associated (MUC1) and protein kinase, DNA-activated, catalytic polypeptide (PRKDC)] had mutations in two loci each. Interestingly, the age at diagnosis of this patient was 38 years (Table I).
Table IV. Genes harboring rare germline variants in gallbladder carcinoma cases.
Discussion
This is one of the first studies from Northern-India (a GBC incidence hotspot) performing a high-throughput sequencing based exploration of the GBC mutation profile. Instead of performing exome- or genome-wide horizontal mutation excavation, we restricted our study only to the regions/genes of common genomic aberrations to take advantage of targeted sequencing approaches including higher depth of coverage to rarer mutation detection with greater confidence. Many of the driver events, as well as several tumor progression events, were easily tagged with biological pathways associated with 409 cancer genes analyzed in the present study. The somatic mutation spectrum in our study is dominated by C>T/G>A transition substitutions, which is consistent with three other GBC exome sequencing studies (9-11). Transitions are also common in all other common cancer types (6).
With our focused ultra-deep sequencing, we have identified several novel mutations in GBC shedding light on molecular pathobiological understanding of this disease (Table I). We identified loss of function (LOF) mutations in some of the genes such as SMAD4 and APC that were already known for their implications as drivers in other cancer types (32,33). On the other hand, we also predicted a gain of function due to hypermethylation of the ETS variant 1 (ETV1) gene. Being a transcription factor, such hypermethylation of ETV1 may impact its downstream transcriptome profile in tumor tissue. ETV1 hypermethylation in prostate cancer, for example, is suggested to have a link with disease progression and aggressiveness of the cancer (34). All detected novel mutations were nonrecurring (appeared once only) in our studied set of tumor samples and all of them were assessed as high-impact mutations in genes that are already well known for their implications in other cancer types. Although the limited number of samples in our study prevents us from making any definitive inference to enlist these mutations as causal to GBC carcinogenesis; considering their significance in other cancer types these gene sets are candidates for further investigation in this specific cancer type.
Like many other studies with common cancer types, we observed high-impact somatic mutations in several well-known oncogenes such as BCL11B, ETV1 and tumor suppressor genes like TP53 and SMAD4 in GBC (35). Considering the low sample size and heterogeneous nature of carcinoma samples, we kept our focus on the mutation load of every individual tumor sample. The observed somatic mutation frequency per megabase of covered targeted genome seems to be exceptionally high in some samples. The definitive reason behind this observation may not be clear. One reason could be the higher sequencing depth of the cancer-related genes and thus greater sensitivity in detecting rarer mutations. Evidently three out of the nine samples qualified as hyper-mutated cases with a high burden of somatic mutations. As a general finding, there was a large difference in the number of mutations across the studied tumors. Such a wide and highly variable range of mutation load may partly be attributed to differences in the coverage between fresh frozen and FFPE samples. FFPE samples tend have lower number of mutations detected in these targeted portions of the genome. On the other hand, even within the fresh frozen tumor tissue set, we observed quite a large variability in numbers of detected mutations. It may be considered as a hint to the heterogeneous molecular phenotype of the included GBC tumors. Our targeted sequencing approach was able to reliably detect several somatic mutations in well-known cancer driver genes in most of the samples. In sample S4, we detected only two somatic mutations in PPARG and NSD1 genes. Although PPARG gene mutation has a crucial supportive role in tumor metabolism, these two mutations alone are not sufficient to explain GBC carcinogenesis. TP53 one of the most frequently mutated genes in GBC and was detected in ~25% of all studied GBC cases (9-11). Our study is not an exception to that. We observed high-impact TP53 mutation in two out of the 11 studied GBC cases.
Interestingly, the majority of the mutated genes in our study were not reported by two earlier exome sequencing studies with 9 and 32 GBC samples (9,10). However, we noted a remarkable overlap with the recently published study (28 samples) on GBC mutation profile with three exceptions (11). Three genes (KMT2C, phosphodiesterase 4D interacting protein (PDE4DIP) and dihydropyrimidine dehydrogenase (DYPD) were detected in more than one tumor in our study but never appeared in the mutation profile reported by Nakamura et al. (11). Some of the observed mutations are novel in GBC (Table III). As per the cosmic database (version 79) mutations in these genes have not previously been reported in GBC (24). These mutations might have been missed by previous sequencing studies because of their relatively low depth of sequencing and/or due to recruitment of patients from a totally different lifestyle exposure or these events may be too rare in GBC. Nik-Zainal et al. demonstrated that sub-clonal variants can be readily detected with ultra-high coverage (188x) genome sequencing (36). Our ultra-deep sequencing approach gave us an edge in detecting subclonal variants with confidence.
In the early NGS era, the focus of most studies was to identify and understand the spectrum of somatic mutations in cancer (7,35). Several cancer drivers and progression markers were established in this way. More recently, the cancer research field has begun to understand the importance of rare deleterious germline variants in carcinogenesis and progression (28,29,37). GBC is not among the most heritable types of human cancer, although variability in its ethnic and geographical incidence rates has been partly associated with cancer predisposition (38-40). Recently, we reported a genome-wide association study recruiting cases of GBC, it outlined the contribution of common variants to GBC predisposition. Northern Indians, especially women, are one population with very high incidence rates (21.5/100,000) and a very high variability in age of onset (40). In this study, we excavated the rare germline variant profile of these cases and detected deleterious or likely deleterious rare germline point mutations following several stringent filtering protocols. We observed the existence of such deleterious mutations even in the two samples (S3 and S6) whose somatic mutation profiles were filtered out of this study due to certain unavoidable qualitative issues (such as low and uneven sequencing coverage).
GBC has an incidence bias towards female gender (1). This study had only two males and nine females. Sample S4 was from one of the two males, harboring a germline deleterious mutation in genes ADAM metallopeptidase with thrombospondin type 1 motif 2 (ADAMTS2) and fibroblast growth factor receptor 1 (FGFR1). ADAMTS2 is a key player in angiogenesis and cancer development (41). Somatic mutations in FGFR1 have been reported in common types of cancer, including its fusion genes in biliary tract cancer (11,42). Thus, altered function of these genes might facilitate tumor survival and may confer an increased metastatic potential to this tumor. Among the other detected germline mutated genes one notable gene was KMT2D also known as MLL2, which encodes lysine-specific methyltransferase 2D, and is also known for its role in neoplastic cell proliferation (43,44). The Sample S8 was also found to have double germline hits in the onco-protein coding gene MUC1. We identified one TP53 germline mutation in sample S9. Interestingly the age at diagnosis of this patient was 80 years (Table I). S9 tumor is one of most frequently mutated tumors as per the detected somatic mutation profile. In addition, this tumor harbored several high-impact somatic mutations in genes such as fibronectin 1 (FN1), KMT2C, nuclear mitotic apparatus protein 1 (NUMA1), and RECQL4. Possibly, the advanced age of this patient together with the deleterious mutation in TP53 gene contributed to the tumor’s hyper-mutator phenotype.
It is known that deleterious mutations are extremely rare in the normal population and even if it is present in an individual, the most likely observed genotypes are heterozygous. Our observations across germline targeted genes are not an exception. Sample S11 was found to have two germline heterozygous likely deleterious mutations located in DNA repair-associated Fanconi anemia-related genes FANCG and homologous recombination (HR)-associated gene RAD50. RAD50 has been reported earlier in breast cancer for its possible haploinsufficient impact in HR-mediated DNA repair phenotype double-stranded break repair (45). Since we did not notice any second hit on FANCG in the tumor, this mutation likely conferred a disease risk rather than acting as a driver. An exception to that is the germline mutation of Iow-density lipoprotein receptor related protein 1b (LRP1B) in S11 which had a second hit in the tumor, thus making it a possible candidate for LRP1B target therapy, which is currently being studied (46).
S6 was another interesting sample harboring two germline mutations in TET1 gene, which is linked with hyper-methylation-dependent breast cancer carcinogenesis. The other two early onset samples (S8 and S9) were found to harbor deleterious mutations in genes with high impact such as ERCC2, FLT3, KMT2C, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB), SYNE1 and transglutaminase 7 (TGM7). One of these two samples (S8) harbored hazard ratio-associated MRE11A germline heterozygous mutation and double hits in the PRKDC gene, which performs the role of non-homologous end joining-mediated DNA repair (47). This consistent observation of mutations in DNA-repair pathway-associated genes in all four early onset cases hints at the relevance of DNA repair pathway in age of onset of GBC. Interestingly, the same set of samples had also acquired a few somatic mutations, which would directly or indirectly impact DNA repair pathway homeostasis. Sample S10 also had mutations in E1A binding protein P300 (EP300) and Erb-B2 receptor tyrosine kinase 3 (ERBB3), whereas sample S11 was observed to have mutations in FN1, NOTCH4, KMT2C (histone methylation), NLRP1 (inflammasome suppression) and S8 tumor was somatically mutated in genes such as RECQL4 and PIK3CG. All these detected mutations in DNA repair-associated genes point towards the significance of DNA-repair pathways in GBC carcinogenesis.
In this first NGS study recruiting North-Indian patients with GBC, we have shed light on the importance of deleterious DNA-repair gene mutations in GB carcinogenesis. Because of the nature of generated data, we restricted our study to analyzing SNVs (somatic and germline) and therefore this study unravels only one aspect of the molecular alterations in gallbladder tumors. Other exome-sequencing studies have, however suggested that somatic single nucleotide mutations are major players in GB carcinogenesis (9-11). Considering all somatic and germline mutations together, we established a few highly aberrant biological pathways including DNA double-strand break repair, extracellular matrix maintenance, cellular homeostasis-associated FGF signaling, NOTCH signaling, loss of innate immunity or altered immunity-based immune evasion, altered glucose metabolism, altered demethylase activity and several growth signaling switches such as erythropoietin-producing hepatoma (EPH) as some of the most significantly disrupted pathways in GBC. We were able to capture actionable molecular stratification even within such a small number of cases. We predicted actionability in about eight out of nine cases. Accounting for only three molecular nodes, namely PIK3CG, TP53 and LRP1B, five out of the nine cases could be considered as being clinical actionable (Figure 3). This small-scale study puts forwards the likely applicability of clinical targeted sequencing in practice that could impact the treatment decisions for a large number of patients with GBC. However, any definitive inference would be possible only after large-scale studies.
In conclusion, it could be perceived that for future precision medicine practice in GBC, not only the somatic mutation profile, but the typing of germline mutations could be highly informative both for a comprehensive understanding of the disease biology and for recommendation of effective therapeutic regimes. Despite our small sample size, this study will assist the field to move one step forward and guide us to design future studies to realize the extent to which germline pathogenic mutations and DNA-repair gene impairment impact global GBC pathobiology.
Acknowledgments
The Authors thank the patients with GBC for giving their consent to provide their samples for the study and Department of Biotechnology (DBT), Government of India for providing the funds for the study. The Authors acknowledge financial help from the Indian Council of Medical Research, and Council of Scientific and Industrial Research in the form of Emeritus Medical Scientist award (BM) and research fellowship (SY), respectively.
References
- 1.Kanthan R, Senger JL, Ahmed S, Kanthan SC. Gallbladder cancer in the 21st century. J Oncol. 2015;2015:967472. doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor VK, McMichael AJ. Gallbladder cancer: An 'indian' disease. Natl Med J India. 2003;16(4):209–213. [PubMed] [Google Scholar]
- 3.Roa I, Garcia H, Game A, de Toro G, de Aretxabala X, Javle M. Somatic mutations of PI3K in early and advanced gallbladder cancer: Additional options for an orphan cancer. J Mol Diagn. 2016;18(3):388–394. doi: 10.1016/j.jmoldx.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Roa JC, Roa I, de Aretxabala X, Melo A, Faria G, Tapia O. [K-RAS gene mutation in gallbladder carcinoma] Rev Med Chil. 2004;132(8):955–960. [PubMed] [Google Scholar]
- 5.Wistuba II, Gazdar AF. Gallbladder cancer: Lessons from a rare tumour. Nat Rev Cancer. 2004;4(9):695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 6.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27(1):15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJ, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R, Velculescu VE, Hruban RH, Vogelstein B, Kinzler KW, Papadopoulos N, Wood LD. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Zhang Z, Li X, Ye J, Wu X, Tan Z, Liu C, Shen B, Wang XA, Wu W, Zhou D, Zhang D, Wang T, Liu B, Qu K, Ding Q, Weng H, Mu J, Shu Y, Bao R, Cao Y, Chen P, Liu T, Jiang L, Hu Y, Dong P, Gu J, Lu W, Shi W, Lu J, Gong W, Tang Z, Zhang Y, Wang X, Chin YE, Weng X, Zhang H, Tang W, Zheng Y, He L, Wang H, Liu Y. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ERBB pathway. Nat Genet. 2014;46(8):872–876. doi: 10.1038/ng.3030. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, Hiraoka N, Ojima H, Shimada K, Okusaka T, Kosuge T, Miyagawa S, Shibata T. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47(9):1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 12.Zhu AX, Hong TS, Hezel AF, Kooby DA. Current management of gallbladder carcinoma. Oncologist. 2010;15(2):168–181. doi: 10.1634/theoncologist.2009-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edge SB, Compton CC. The american joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of tnm. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 14.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. Sift web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using polyphen-2. Curr Protoc Hum Genet. 2013;Chapter 7, Unit 7:20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The human gene mutation database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison SM, Riggs ER, Maglott DR, Lee JM, Azzariti DR, Niehaus A, Ramos EM, Martin CL, Landrum MJ, Rehm HL. Using ClinVar as a resource to support variant interpretation. Curr Protoc Hum Genet. 2016;89:11–18. doi: 10.1002/0471142905.hg0816s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. Panther version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44(D1):D336–342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mort M, Sterne-Weiler T, Li B, Ball EV, Cooper DN, Radivojac P, Sanford JR, Mooney SD. Mutpred splice: Machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol. 2014;15(1):R19. doi: 10.1186/gb-2014-15-1-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The catalogue of somatic mutations in cancer (COSMIC) Curr Protoc Hum Genet Chapter. 2008;10:Unit 10.11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatachalam S, Tyner SD, Pickering CR, Boley S, Recio L, French JE, Donehower LA. Is p53 haploinsufficient for tumor suppression? Implications for the p53+/− mouse model in carcinogenicity testing. Toxicol Pathol. 2001;29 Suppl:147–154. doi: 10.1080/019262301753178555. [DOI] [PubMed] [Google Scholar]
- 26.An O, Dall'Olio GM, Mourikis TP, Ciccarelli FD. NCG 5.0: Updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic Acids Res. 2016;44(D1):D992–999. doi: 10.1093/nar/gkv1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knudson AG Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cereda M, Gambardella G, Benedetti L, Iannelli F, Patel D, Basso G, Guerra RF, Mourikis TP, Puccio I, Sinha S, Laghi L, Spencer J, Rodriguez-Justo M, Ciccarelli FD. Patients with genetically heterogeneous synchronous colorectal cancer carry rare damaging germline mutations in immune-related genes. Nat Commun. 2016;7:12072. doi: 10.1038/ncomms12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Xie M, Wendl MC, Wang J, McLellan MD, Leiserson MD, Huang KL, Wyczalkowski MA, Jayasinghe R, Banerjee T, Ning J, Tripathi P, Zhang Q, Niu B, Ye K, Schmidt HK, Fulton RS, McMichael JF, Batra P, Kandoth C, Bharadwaj M, Koboldt DC, Miller CA, Kanchi KL, Eldred JM, Larson DE, Welch JS, You M, Ozenberger BA, Govindan R, Walter MJ, Ellis MJ, Mardis ER, Graubert TA, Dipersio JF, Ley TJ, Wilson RK, Goodfellow PJ, Raphael BJ, Chen F, Johnson KJ, Parvin JD, Ding L. Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson JP, Ottaviano R, Unterberger EB, Lempiainen H, Muller A, Terranova R, Illingworth RS, Webb S, Kerr AR, Lyall MJ, Drake AJ, Wolf CR, Moggs JG, Schwarz M, Meehan RR. Loss of tet1-associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 2016;76(10):3097–3108. doi: 10.1158/0008-5472.CAN-15-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, Zhou JR, Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71(3):998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fodde R. The apc gene in colorectal cancer. Eur J Cancer. 2002;38(7):867–871. doi: 10.1016/s0959-8049(02)00040-0. [DOI] [PubMed] [Google Scholar]
- 34.Angulo JC, Andres G, Ashour N, Sanchez-Chapado M, Lopez JI, Ropero S. Development of castration resistant prostate cancer can be predicted by a DNA hypermethylation profile. J Urol. 2016;195(3):619–626. doi: 10.1016/j.juro.2015.10.172. [DOI] [PubMed] [Google Scholar]
- 35.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ, Stephens PJ, McLaren S, Tarpey PS, Papaemmanuil E, Davies HR, Varela I, McBride DJ, Bignell GR, Leung K, Butler AP, Teague JW, Martin S, Jonsson G, Mariani O, Boyault S, Miron P, Fatima A, Langerod A, Aparicio SA, Tutt A, Sieuwerts AM, Borg A, Thomas G, Salomon AV, Richardson AL, Borresen-Dale AL, Futreal PA, Stratton MR, Campbell PJ. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R, Elemento O, Rubin MA, Robinson D, Lonigro R, Hussain M, Chinnaiyan A, Vinson J, Filipenko J, Garraway L, Taplin ME, AlDubayan S, Han GC, Beightol M, Morrissey C, Nghiem B, Cheng HH, Montgomery B, Walsh T, Casadei S, Berger M, Zhang L, Zehir A, Vijai J, Scher HI, Sawyers C, Schultz N, Kantoff PW, Solit D, Robson M, Van Allen EM, Offit K, de Bono J, Nelson PS. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava K, Srivastava A, Sharma KL, Mittal B. Candidate gene studies in gallbladder cancer: A systematic review and meta-analysis. Mutat Res. 2011;728(1-2):67–79. doi: 10.1016/j.mrrev.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mhatre S, Wang Z, Nagrani R, Badwe R, Chiplunkar S, Mittal B, Yadav S, Zhang H, Chung CC, Patil P, Chanock S, Dikshit R, Chatterjee N, Rajaraman P. Common genetic variation and risk of gallbladder cancer in India: a case–control genome-wide association study. Lancet Oncol. 2017;18(4):535–544. doi: 10.1016/S1470-2045(17)30167-5. [DOI] [PubMed] [Google Scholar]
- 40.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: Geographical distribution and risk factors. Int J Cancer. 2006;118(7):1591–1602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Rao N, Ge R. Emerging roles of ADAMTSS in angiogenesis and cancer. Cancers. 2012;4(4):1252–1299. doi: 10.3390/cancers4041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner N, Grose R. Fibroblast growth factor signalling: From development to cancer. Nat Rev Cancer. 2010;10(2):116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 43.Guo C, Chen LH, Huang Y, Chang CC, Wang P, Pirozzi CJ, Qin X, Bao X, Greer PK, McLendon RE, Yan H, Keir ST, Bigner DD, He Y. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4(11):2144–2153. doi: 10.18632/oncotarget.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao RC, Dou Y. Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15(6):334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heikkinen K, Rapakko K, Karppinen SM, Erkko H, Knuutila S, Lundan T, Mannermaa A, Borresen-Dale AL, Borg A, Barkardottir RB, Petrini J, Winqvist R. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27(8):1593–1599. doi: 10.1093/carcin/bgi360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beer AG, Zenzmaier C, Schreinlechner M, Haas J, Dietrich MF, Herz J, Marschang P. Expression of a recombinant full-length LRP1B receptor in human non-small cell lung cancer cells confirms the postulated growth-suppressing function of this large LDL receptor family member. Oncotarget. 2016;7(42):68721–68733. doi: 10.18632/oncotarget.11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee R, Russo N, Liu M, Basrur V, Bellile E, Palanisamy N, Scanlon CS, van Tubergen E, Inglehart RC, Metwally T, Mani RS, Yocum A, Nyati MK, Castilho RM, Varambally S, Chinnaiyan AM, D'Silva NJ. Trip13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat Commun. 2014;5:4527. doi: 10.1038/ncomms5527. [DOI] [PMC free article] [PubMed] [Google Scholar]