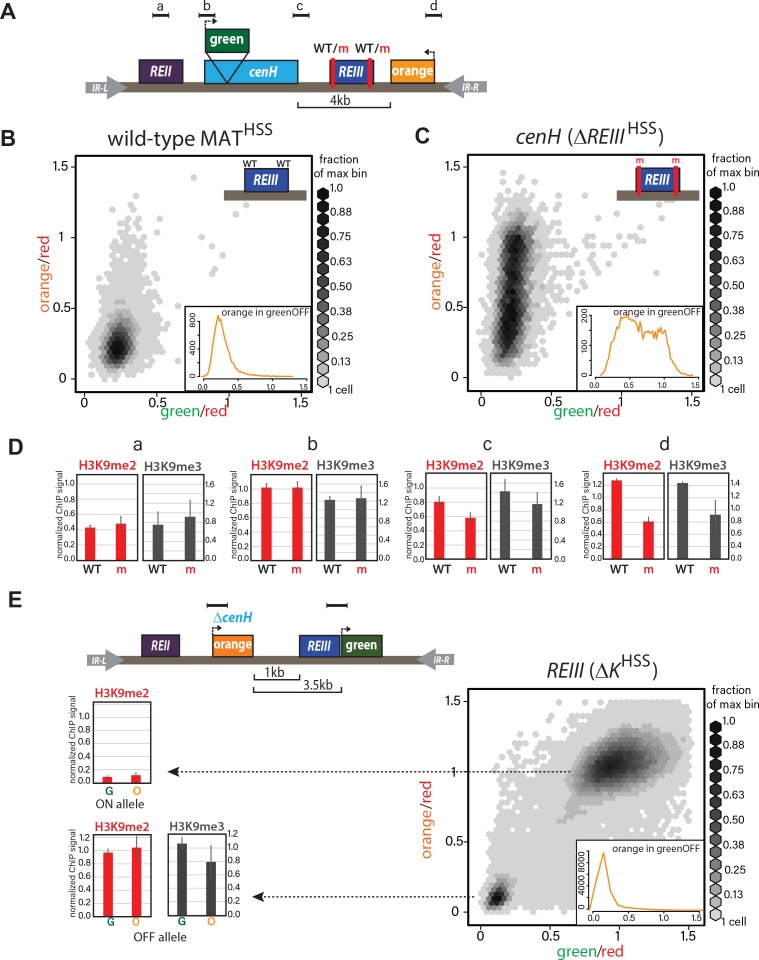
Figure 2. ncRNA-dependent and independent nucleation yields qualitatively different spreading reactions in the MAT locus.
(A) Diagram of the reporters within MATHSS and ΔREIIIHSS. WT and m for REIII indicate the presence or deletion of the Atf1/Pcr1 binding sites, respectively. (B) 2D-density hexbin plot showing the ‘red’-normalized ‘green’ and ‘orange’ fluorescence for wild-type MATHSS cells. Scale bar shows every other bin cutoff as a fraction of the bin with the most cells. Inset: histogram of the ‘red’-normalized ‘orange’ fluorescence distribution of ‘green'OFF cells. (C) 2D-density hexbin plot and inset as above for ΔREIIIHSS, which contains two 7 bp Atf1/Pcr1-binding site deletions (m) within the REIII element. (D) ChIP for H3K9me2 (red) and H3K9me3 (grey) for amplicons indicated in (A). normalized to dh. WT, wild-type MATHSS, m, ΔREIIIHSS. (E) TOP: diagram of the reporters within ΔKHSS. The cenH nucleator and additional 5’ sequence is deleted and replaced by ‘orange’. ‘green’ is located directly proximal to REIII and serves as the nucleation clamp. ChIP amplicons are indicated as black bars. BOTTOM: 2D- density hexbin plot and inset as above. LEFT: ChIP for H3K9me2 (red) and H3K9me3 (grey) for ‘green’ and ‘orange’ in isolated ΔKHSS-ON or ΔKHSS-OFF alleles. In hexbin plots, the Δclr4 derivative of each strain was used to normalize the X- and Y-axes to = 1. Error bars indicate standard deviation of technical replicates.