Abstract
Tissues contain exquisite vascular microstructures, and patterns of chemical cues for directing cell migration, homing, and differentiation for organ development and function. 3D microfabrication by multi-photon photolithography is a flexible, high-resolution tool for generating 3D bioscaffolds. However, the combined fabrication of scaffold microstructure simultaneously with patterning of cues to create both geometrically and chemically defined microenvironments remains to be demonstrated. Here, we present a high-speed method for micron-resolution fabrication of scaffold microstructure and patterning of protein cues simultaneously using native scaffold materials. By the simultaneous microfabrication of arbitrary microvasculature geometries, and patterning selected regions of the microvasculature with the homing ligand P-selectin, we demonstrate adhesion, rolling, and selective homing of cells in defined 3D regions. This novel ability to rapidly generate high-resolution geometries replete with patterned cues at high speed enables the construction of biomimetic microenvironments for complex 3D assays of cell behavior.
Keywords: 3D Printing, engineered microvasculature, capillary network, cell trap, cell rolling
Introduction
Developing tissues display exquisite microarchitectures of blood vessels and patterns of growth factors that are essential for the development of viable tissues.[1–3] An emerging interest in tissue engineering is to recapitulate these biological structures using scaffolds with intricate in vivo-like 3D vascular microarchitectures with micropatterns of chemical cues. In particular, great promise has been shown by using decellularized scaffolds, formed by treating cadaver organs with chemicals to strip the tissue of its cells, leaving behind an extracellular matrix (ECM) containing native vasculature and patterns of protein cues.[4–7] However, supply, decellularization, recellularization, and safety issues limit the use of cadaveric tissues.[7]
Alternatively, a variety of microfabrication techniques are being investigated to engineer geometrically patterned scaffolds. Myocardial cells seeded onto a laser ablated scaffold with an accordion-like architecture have been shown to resemble native myocardial tissue function better than traditional isotropic scaffolds. [8] 3D printers have been used to generate blood vessel architectures by molding biomaterials around sacrificial materials fabricated via ink-jet printing of an organic phase,[9] fused-deposition modeled carbohydrate glass [10], or direct-ink written pluronic hydrogels.[11] Of existing methods, multi-photon photolithography (MPP) holds the greatest promise for generating arbitrarily shaped 3D microchannels and patterns of cues down to the single-micron resolutions exhibited by the smallest capillaries (Figure 1a,i). MPP has been used to microfabricate a variety of biomaterials, including collagen,[12,13] bovine serum albumin (BSA),[14] poly(ethylene-glycol) diacrylate (PEGDA),[15,16] fibrin,[17] and poly(lactic acid).[18] MPP has also been used to pattern chemical cues inside bulk scaffolds (Figure 1a,ii). West and colleagues have patterned short peptides within collagenase sensitive PEGDA scaffolds to direct endothelial cell migration.[19] Wylie et al. have patterned multiple cues inside photoactivatable agarose scaffolds to direct signalling pathways in retinal progenitor cells.[20] Seidlits et al. patterned peptides in hyaluronic acid and agarose hydrogels using a photosensitizer to crosslink biotinylated BSA.[21]
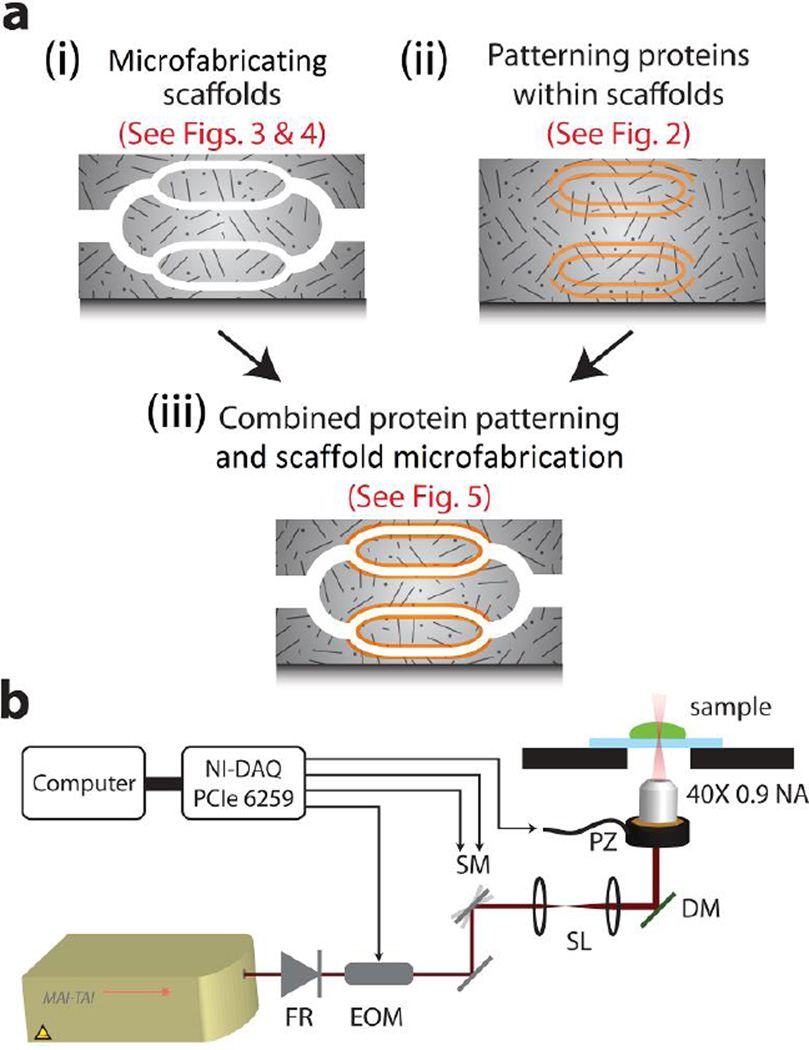
Figure 1. Multi-photon photolithography of scaffolds and internal cues.
(a) A schematic illustrating (i) microfabricating scaffolds, (ii) patterning of proteins inside scaffolds, and (iii) the combination of both scaffold microfabrication and protein patterning. (b) The optical setup used for performing MPP. FR = Faraday rotor, EOM = electro optic modulator, SM = x-y galvanometer scanning mirrors, SL = scan lenses, DM = dichroic mirror, PZ = piezo objective mount for z-axis motion.
However, MPP has not yet been used to rapidly pattern proteins inside a wide variety of native scaffold materials currently employed by tissue engineers, limiting its widespread adoption. Second, despite the fact that native ECMs comprise both a defined microarchitecture and micropatterns of protein cues, MPP has never been used to pattern both the scaffold and internal protein cues simultaneously (Figure 1a,iii). Third, MPP, like all 3D printing technologies, suffers from a cube-law in scaling: the number of voxels to be patterned or printed increases with the cube of the length-scale of the scaffold, significantly hindering its application at therapeutic scales. Rapid 3D laser microfabrication of collagen has proven particularly challenging. Most multi-photon photoinitiators function only in alkaline environments, in which collagen undergoes gelation.[12]
Here, we present a suite of novel techniques that overcome all these shortcomings of MPP, making it a uniquely powerful approach to rapidly microfabricate intricate tissue scaffolds simultaneously patterned with arbitrary protein cues at micron resolutions. Our approach makes use of the photobleaching of biotin-4-fluorescein (B4F), which has previously been used in conjunction with photomasks[22] or focused lasers[23,24] to produce 2D protein patterns in vitro, and we previously showed that multi-photon photobleaching of fluorescein or B4F can enable rapid laser micropatterning of proteins on a 2D, polymer monolayer coated surface.[25,26] We first show that multi-photon photobleaching of B4F can enable rapid 3D patterning of full length proteins in a wide range of natural, unmodified scaffold materials such as collagen, fibrin and agarose, or in modified materials such as gelatin methacrylate and agarose methacrylate, enabling its integration with a vast number of tissue engineering strategies. Next, we present a novel method for the rapid 3D microfabrication of collagen scaffolds by selectively photobleaching fluorescein inside a bulk collagen gel, followed by the development of the exposed structure in an acetic acid or tris buffer. We achieve rapid patterning speeds, rastering at up to 14 z-layers (100 × 100 × 2 µm) per second generating complex 3D shapes in collagen within seconds. Finally, we combine these two functionalities, showing for the first time MPP microfabricated scaffolds containing internal 3D patterns of proteins to direct cell behavior. Using our technique, we construct branched collagen microchannels containing micropatterns of P-selectin, a protein involved in recruiting leukocytes, hematopoietic stem cells and circulating tumor cells from blood vessels.[27] We show that HL-60 cells, when flowed through the microchannels, roll in the 3D patterned P-selectin regions, and can be captured in selected regions of the collagen gel. Taken together, this rapid method could enable the bottom-up construction of tissues with both a defined microarchitecture and internal patterns of protein cues for both basic science and drug screening assays in biomimetic environments, and also for targeted cell delivery for regenerative medicine applications.
Results
3D Patterning of Proteins in Scaffolds by Photobleaching of Biotin-4-Fluorescein
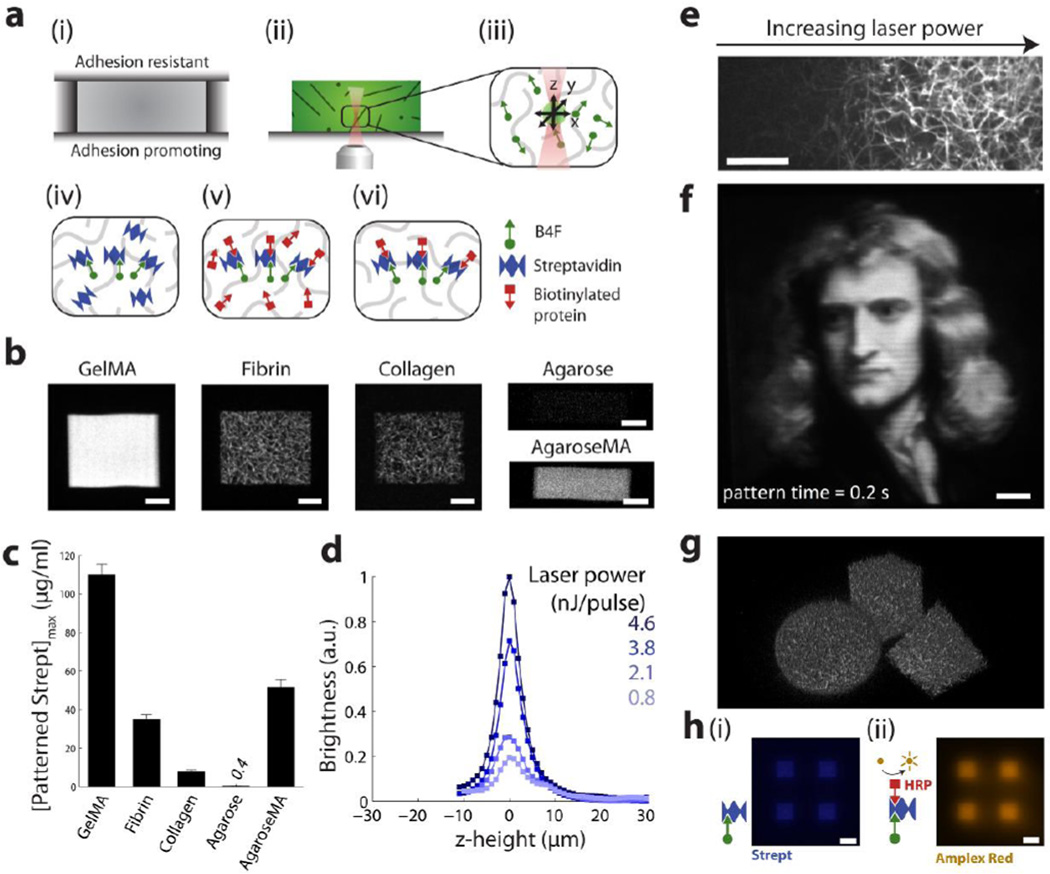
To perform MPP, a femtosecond laser 3D patterning system was assembled as presented in Figure 1b (See Methods). To create patterns of full-length proteins inside scaffolds (as illustrated in Figure 1a,ii), we first prepared collagen, gelatin methacrylate, fibrin and agarose scaffolds by sandwiching the prepolymer solution between adhesion promoting and adhesion resistant glass slides prior to the gelation of the scaffolds (Figure 2a, i). After gelation, the adhesion resistant coverslip was removed, B4F solution was added, and was allowed to diffuse into the scaffold (Figure 2a, ii). The laser was scanned in x, y and z to photocrosslink B4F to the scaffold backbone (Figure 2a, iii). Next, excess B4F was rinsed out of the scaffold, and streptavidin was added to bind to the patterned biotin moieties (Figure 2a, iv). After rinsing the scaffold to remove excess streptavidin, biotinylated protein was added to bind to the remaining streptavidin-biotin binding sites (Figure 2a, v), and a final rinse was performed (Figure 2a, vi). Using this process, we demonstrated patterns of fluorescent streptavidin in gelatin methacrylate (GelMA), fibrin, collagen, agarose, and agarose methacrylate scaffolds (Figure 2b). The patterns were formed using a scan speed of 250 mm/s and a scanline separation of 0.25 µm, patterning 100 × 100 µm regions in ~0.2 s. The various pore microarchitectures comprising the scaffolds are apparent from the fluorescent streptavidin patterns in Figure 2b, as the streptavidin binds to the B4F which, in turn, is bound to the scaffold backbone. We also showed that streptavidin patterns can be formed in collagen scaffolds with various porosities (Figure S1). Comparing the pattern brightness across the scaffold materials, exposure of B4F within gelatin methacrylate hydrogels resulted in the brightest streptavidin patterns, while patterns in agarose scaffolds were only 0.3% as bright (Figure 2c). We found that pattern brightness in agarose could be significantly boosted by pre-functionalization of the polysaccharide chains with methacrylate groups (forming agaroseMA) via a reaction with glycidyl methacrylate at neutral pH . The pattern brightness varies with laser power, and has a z-axis resolution of ~ 5 µm in collagen (Figure 2d), and ~ 10 µm in the other scaffold materials (Figure S2). The increased axial resolution of protein patterns in collagen is likely due to collagen shrinkage upon exposure to the laser pulses. Pattern brightness is affected by both laser power and laser scan speed (Figure S3). By continuously varying the laser power along a scanline using an electro optic modulator (EOM), we could generate smooth gradients of streptavidin patterns superposed on collagen fibrils (Figure 2e). By calibrating laser power to pattern brightness, complex micropatterns of streptavidin can be rapidly created, with ~1 µm resolution in the x-y plane, within the bulk of a gelatin methacrylate scaffold (Figure 2f). By patterning multiple z-layers, we generated arbitrary 3D patterns of streptavidin in fibrin scaffolds (Figure 2g, Movie S1a). Consisting of 40 z-layers, with a layer separation of 2 µm, each 3D shape was formed in ~ 8 s. The layer separation was chosen to be significantly below the axial resolution of ~10 µm to achieve a more uniform exposure throughout the patterned region. Finally, we demonstrated micropatterning of enzymatically active proteins by adding biotinylated horseradish peroxidase (HRP) to patterned streptavidin, whereupon patterns of fluorescence appear upon the addition of the HRP substrate amplex red (Figure 2h).
Figure 2. Patterning proteins inside scaffolds via biotin-4-fluorescein photobleaching.
(a) (i) Scaffold formation between two coverslips; (ii) Removal of lid and infusion of B4F; (iii) Laser scanning in x, y, and z to couple B4F to scaffold backbone; (iv) Addition of streptavidin to bind patterned biotin; (v) addition of biotinylated protein to bind to streptavidin; (vi) Final rinse to reveal patterns. (b) Various scaffold materials patterned with fluorescently labeled streptavidin. Images were brightened by differing degrees to highlight scaffold backbone microstructure. Scale bars = 20 µm. (c) Comparison of pattern brightness (defined as the maximum brightness of all z-crossections captured using two photon microscopy) across different scaffold materials. (d) Z-axis brightness profiles of streptavidin patterns in collagen. (e) Smooth gradients of fluorescent streptavidin patterned in a collagen scaffold by variation of laser power across scanlines. Scale bar = 20 µm. (f) A 100 µm × 100 µm painting, rendered by patterning fluorescent streptavidin in the bulk of a 3D gelatin methacrylate hydrogel. Scale bar = 10 µm. (g) 3D streptavidin shapes patterned in a fibrin scaffold. Patterning time = 8 s per shape. (h) Patterns of (i) streptavidin, and (ii) amplex red fluorescence to demonstrate patterns of enzymatically active biotinylated horseradish peroxidase. Scale bar = 50 µm.
3D Microfabrication of Collagen Scaffolds by Photobleaching of Fluorescein
Next, we discovered that collagen scaffolds can be 3D microfabricated at high speeds by exposing regions of a fluorescein infused collagen gel to the scanning femtosecond laser (Figure 3). After laser exposure, upon bathing the scaffold in acetic acid or tris-HCl, the unexposed regions of the collagen gel were dissolved away, leaving collagen only in the regions that were exposed to the laser (Figure 3a).
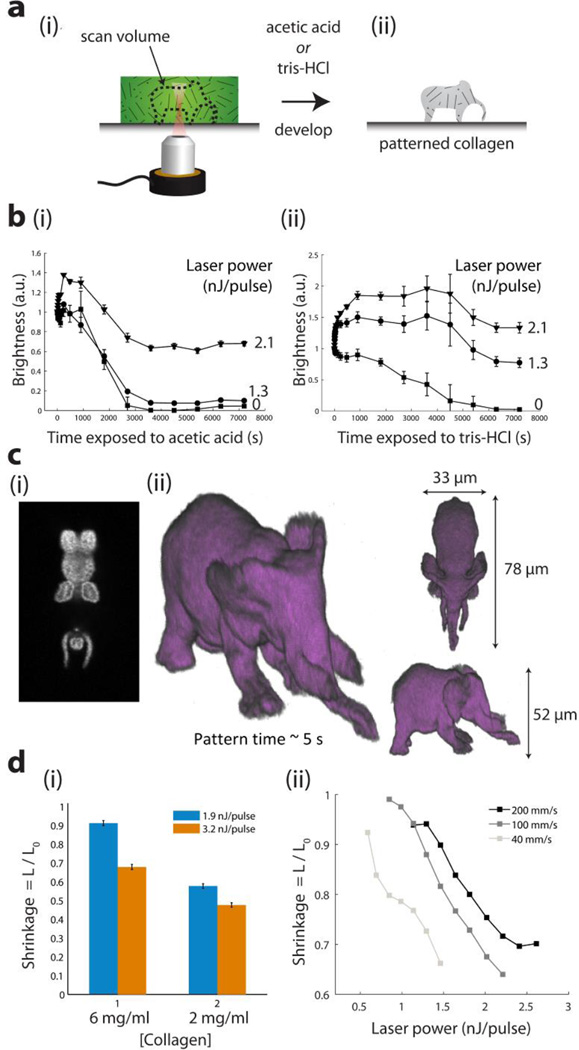
Figure 3. Multi-photon microfabricated collagen scaffolds.
(a) Method for multi-photon microfabricating collagen scaffolds by: (i) exposing a fluorescein infused collagen scaffold to the femtosecond laser; (ii) Addition of acetic acid or tris-HCl to dissolve unexposed regions. (b) Fluorescence decay curves showing dissolution and transport away of fluorescently labeled collagen scaffolds during development in (i) acetic acid; (ii) tris-HCl at different laser powers. (c) Arbitrary shapes exposed and developed in collagen. Laser scanning time ~ 5 sec. (i) Single z-slice imaged by 2-photon microscopy. (ii) Isosurface rendering by thresholding voxel dataset obtained from two-photon imaging of collagen scaffold (d) Dependence of collagen shrinkage on (i) collagen scaffold concentration and (ii) exposed laser power and scan speed.
To visualize the collagen development process, after exposing vertical columnar regions that span the thickness of a collagen scaffold to various laser powers, we fluorescently labeled the collagen molecules using an amine reactive dye (Figure S4a). Figure 3b and Figure S4b display the fluorescence intensity of the collagen regions exposed to varying laser powers during development in either acetic acid (Figure 3b, i) or tris-HCl (Figure 3b, ii). For each region, the fluorescence intensity was normalized to that measured at time t = 0. Fluorescence signatures of unexposed regions (0 nJ/pulse) decayed to zero, indicating complete collagen dissolution, while exposed regions retained a fluorescence signature, indicating undissolved collagen. The degradation kinetics of the collagen scaffold in acetic acid or tris-HCl were dependent upon the laser power that was incident on the collagen. Acetic acid acted as a more potent developer than tris-HCl, with exposed collagen scaffolds withstanding tris-HCl treatment, but dissolving in acetic acid at certain intermediate laser powers. After exposure, development of a ~30 µm thick scaffold was completed in approximately 2 h in both acetic acid and tris-HCl. The exposed collagen scaffolds remained sensitive to collagenase degradation, as both exposed and unexposed regions dissolved rapidly in the presence of collagenase I (Figure S4b). Furthermore, we show that blue light (one-photon) excitation of fluorescein is sufficient to stabilize collagen against acid dissolution, enabling a low-cost method for patterning regions of collagen scaffold using a collimated blue light-emitting diode and a printed transparency mask (Figure S5).
To demonstrate the ability to use MPP to rapidly microfabricate high-resolution, arbitrary collagen structures, we sliced an .stl file, and laser-rastered a structure at a speed of 14 z-layers per second, at a data rate of 0.8 Mvoxels/s, completing the structure in ~ 5 s (Figure 3c, Movie S1b). We found that the collagen structures shrink upon developing, with the degree of shrinkage varying with the collagen gel concentration (Figure 3d,i) and the incident laser power and laser scan speed used to crosslink the collagen (Figure 3d,ii). Note that this shrinkage is the likely cause of the initial increases in fluorescence intensity observed in Figure 3b, as the fluorescently labeled collagen structures increase in density, and hence the mean brightness also increases.
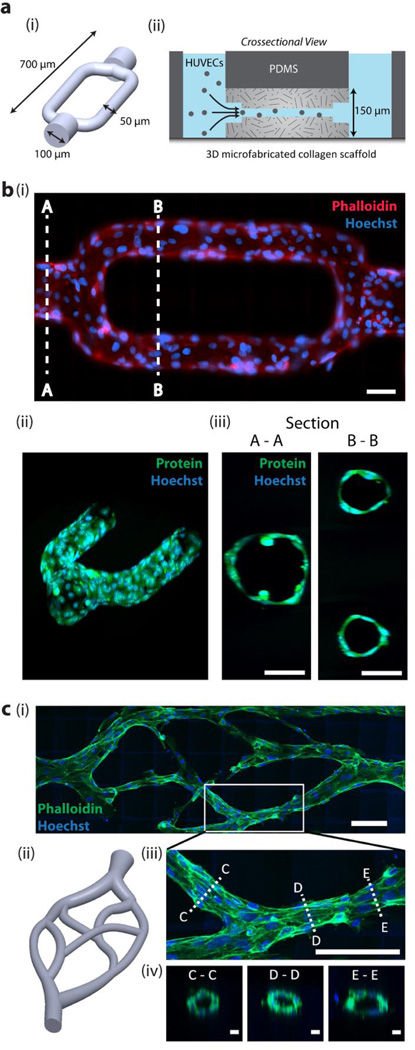
Next, to verify that the 3D microfabricated collagen scaffolds are biocompatible, we patterned microvascular channels in collagen (Figure 4a, i) which was gelled within a PDMS microchannel (Figure 4a, ii) to enable direct perfusion of the microfabricated channels. The channels required ~5 min of laser scanning time to generate. When human umbilical-vein endothelial cells (HUVECs) were seeded into the channels, they readily adhered along the collagen walls, and migrated down the length of the channels, forming a densely packed, confluent monolayer of cells that stain positive for VE cadherin (Figure 4b,i-ii and Figure S6). The resultant endothelialized vasculature maintained a patent lumen, ~50 µm in diameter, through which fluid could flow (Figure 4b,iii, and Movie S2). To the best of our knowledge, these vessels are the highest resolution perfusable endothelialized channels yet created. To further demonstrate microfabrication of high resolution microvasculature, we generated a series of biomimetic vascular channels (Figure 4c). When introduced into the larger channels at either side, HUVECs adhered to the walls and migrated along the channls towards the center of the scaffold. On crossection, the HUVECs formed patent lumens with inner diameters of ~ 20 µm (Figure 4c, iv).
Figure 4. Multi-photon microfabricated microvasculature.
(a) (i) 3D bifurcating microchannel design. The shape represents the unexposed regions in a block of collagen. (ii) Microchannels are microfabricated in a block of collagen within a microfluidic chip to enable addition of HUVECs via hydrostatic pressure driven flow. (b) (i) HUVECs fixed and stained at 12 h in vitro and imaged under (i) epifluorescence, and (ii-iii) 2-photon microscopy. Scale bars = 50 µm. (c) (i-ii) Biomimetic microvascular channels lined with HUVECs, fixed and phalloidin stained at 72 h in vitro. Image is acquired using two-photon microscopy and displayed as a maximum intensity projection. Scale bar = 100 µm. (iii) A maximum intensity image of a bifurcation. Scale bar = 100 µm, and (iv) crossections taken at three points along the vessel, showing a patent lumen. Scale bars = 10 µm.
Combined Microfabrication of Scaffolds and Patterning of Internal Protein Cues
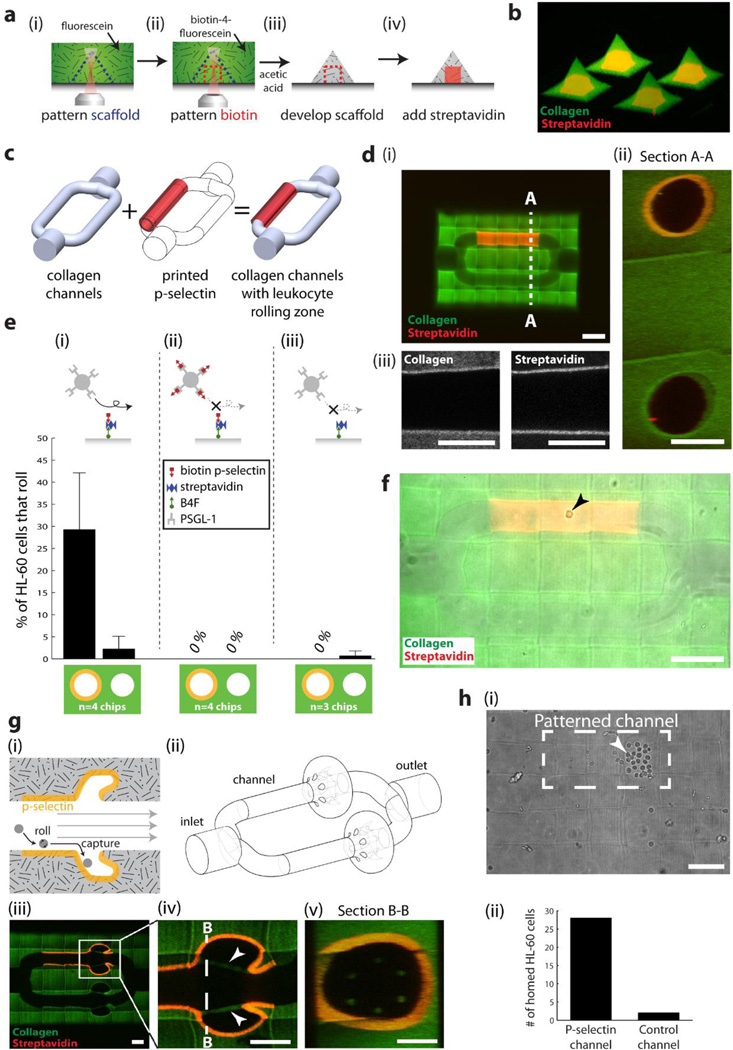
To generate microfabricated scaffolds with internal patterns of proteins, we combined our 3D protein patterning technique (Figure 2) with our method for 3D collagen scaffold microfabrication (Figures 3 & 4). This process can be performed by first exposing an arbitrary volume within a collagen scaffold that has been infused with fluorescein (Figure 5a,i), then an excess of B4F is added to replace the fluorescein via diffusion, followed by the patterning of a second arbitrary volume with biotin moieties by photobleaching the B4F (Figure 5a,ii). The gel is then removed from the laser setup, placed in a bath of acetic acid to develop the patterns (Figure 5a,iii), and streptavidin is added to bind to the internal patterns of biotin (Figure 5a,iv). The patterned streptavidin can then be used to bind a biotinylated protein, as in Figure 2. We used this to generate microfabricated collagen scaffolds, each containing internal patterns of streptavidin (Figure 5b, Movie S3). Crucially, by performing the steps in the order described here, and by carefully pipetting B4F such that the sample is not moved, no 3D realignment was necessary to align the internal patterns of protein to the patterned scaffolds.
Figure 5. Simultaneous microfabrication of collagen scaffold microarchitecture and patterning of internal protein cues.
(a) Method for microfabricating scaffold microarchitecture and internal patterns of proteins. (b) Microfabricated collagen shapes containing internal patterns of streptavidin. (c) Schematic showing the combination of collagen channels and patterned P-selectin. (d) Collagen channels containing a region patterned with streptavidin (i) Scale bar = 100 µm, (ii) Crossection of channels acquired from two-photon imaging. Scale bar = 50 µm. (iii) Two-photon images of microfabricated collagen and patterned streptavidin. Scale bar = 50 µm. (e) Percentage of HL-60 cells rolling on (i) P-selectin patterned, and non-patterned channels; (ii) the same channels, but the HL-60 cells are pre-incubated with soluble P-selectin; (iii) patterned channels coated with streptavidin only. (f) HL-60 cell rolling on patterned P-selectin (arrowhead). Scale bar = 100 µm. (g) (i) A device to selectively capture rolling cells; (ii) its corresponding 3D model; (iii-v) as microfabricated and imaged using two-photon microscopy. Arrowheads display collagen ‘tethers’ used to support overhanging collagen, and to prevent the overhangs from inverting in conditions of flow. Scale bars = 50 µm. (h) HL-60 cells preferentially home to the side patterned with P-selectin. Scale bar = 100 µm.
Next, we demonstrate the ability to generate complex biologically active scaffolds containing microarchitecture and patterned protein capable of directing cell behavior. Cell rolling on P-selectin is an essential step in the extravasation of immune cells, and is involved in the homing of mesenchymal stem cells to sites of injury, and in tumor metastasis.[28] Thus we produced a microfabricated construct for cell rolling using branched collagen channels with a patterned region of P-selectin (Figure 5c-d; Movie S4a). Upon addition of HL-60 human leukocytes, we found that cell rolling occurred on the P-selectin patterned branch, but not on the unpatterned branch (Figure 5e,i, Figure 5f; Movie S4a). This rolling on the patterned channel could be inhibited by pre-incubating HL-60 cells with soluble P-selectin to block the PSGL-1 receptor (Figure 5e,ii; Movie S4b). Furthermore, channels coated only with streptavidin (ie. without biotinylated P-selectin) did not elicit HL-60 cell rolling (Figure 5e,iii; Movie S4c).
Next, we tested multiple, iterative designs to generate a 3D cell niche that could selectively capture cells via receptor-P-selectin interaction (Figure 5f; Movie S4d-e). Using rapid prototyping, we tested numerous iterative designs to achieve our selective homing goal (Figure S7). The final model geometry consisted of a bulge with an overhanging flap to prevent cells from rolling out (Figure 5g). The flap was prevented from inverting during flow conditions by narrow (~10 µm) collagen tethers (arrowheads Figure 5g,iv) that were distributed radially (Figure 5g, v). Despite the geometries of both patterned and unpatterned channels being the same, HL-60 cells were only captured when traveling along the channel coated with P-selectin, demonstrating the ability of HL-60 cells to home to specific regions within this microfabricated geometry (Figure 5h, Movie S4d). Furthermore, cells that do not interact with P-selectin, for example human erythrocytes, were not trapped by either the P-selectin patterned, or the unpatterned branches (Movie S4e).
Discussion
We have demonstrated the fastest 3D biomaterial multi-photon photolithography (MPP) method to date, capable of patterning full-length proteins inside a range of tissue engineering scaffolds (Figure 2). Previously, MPP of full–length proteins in scaffolds had only been described in scaffolds composed of coumarin modified agarose,[20] unmodified agarose,[21] hyaluronic acid methacrylate[21] and collagenase sensitive poly(ethylene-glycol) diacrylate.[29] Our method, capable of patterning gradients of proteins inside off-the-shelf collagen, fibrin, gelatin methacrylate and agarose scaffolds, could readily be incorporated into many existing tissue engineering strategies, enabling the micropatterning of gradients of protein for directing cell development. This wide range of biomaterials can form scaffolds with cell-adhesive (collagen, fibrin and gelatin methacrylate), or non-adhesive properties (agarose and agarose methacrylate), enabling a wide range of possible cell behaviors on micropatterned scaffolds. Furthermore, these materials exhibit a wide range of scaffold stiffness, from soft collagen type I (~0.1 kPa[30]) and fibrin scaffolds(~ 0.1–4 kPa[31,32]), to stiff gelatin methacrylate (~ 10–30 kPa[33]) and agarose (~ 100–4,000 kPa[34]) scaffolds. This can enable selection of materials that are optimized for specific cell behaviors or differentiation pathways, for which scaffold stiffness is known to be a key factor.[35,36] The high patterning speeds are likely achieved by a combination of the rapid, oxygen independent multi-photon photobleaching of fluorescein,[25,37] using a high laser power, and the use of a high bath concentration of biotin-4-fluorescein. No biotinylated protein is ever directly exposed to the laser, mitigating any risk of protein denaturation upon irradiation. It should be noted that some proteins that avidly bind to the scaffold backbone cannot be patterned in that scaffold material using this method, as high-contrast patterning relies on a difference in binding affinities between the biotin patterned scaffold backbone and the scaffold itself.
We also discovered a new method for microfabricating collagen scaffolds in 3D at a rapid scan speed of 400 mm/s (Figure 3). Upon light exposure of the fluorescein, the collagen is rendererd resistant to dissolution in acid or tris buffer, likely due to the presence of additional cross-links that stabilize the collagen fibrils. Others have shown that both photo-generated singlet oxygen[38–40], generated by a reaction with singlet oxygen and fluorescein in its excited triplet state, or free-radicals[40] can react with histidine residues in collagen type I leading to the formation of stable cross-links. We also showed that one-photon (ie. blue light) exposure of fluorescein renders the collagen insoluble to acid, suggesting that singlet-oxygen driven cross-linking is sufficient to stabilize the collagen fibrils (Figure S5). Furthermore, the fact that the cross-linking occurs while the collagen is in the fibrillar phase likely enhances the rate of free-radical mediated fibril crosslinking, as the gel-phase collagen molecules are held in close apposition by hydrogen bonding. This is supported by our observations that collagen microfabrication does not occur if laser scanning is initiated upon neutralization of fluorescein infused collagen, but before bulk collagen gelation occurs. A major advantage of exposing a scaffold while it is in its gel phase is that it enables the facile layer-by-layer exposure of structures with negative overhangs (such as the ‘tusks’ in Figure 3c), as the surrounding unexposed scaffold regions can support the laser-exposed overhang until the entire structure is exposed and developed. This renders support material unnecessary for producing layer-by-layer 3D shapes via this method, while the need for support material remains a significant challenge in producing complex biomimetic architectures via many alternative 3D printing strategies.
Finally, we showed that both collagen scaffolds, and internal patterns of proteins can be microfabricated together to produce scaffolds with defined microarchitectures and internal micropatterns of protein cues. We microfabricated branched channels in collagen, and micropatterned a region with P-selectin, a protein known to cause homing of leukocytes, hematopoietic stem cells and circulating tumor cells. By careful design of geometry and patterned cues, we were able to selectively guide and capture specific cell types (HL-60) within the scaffold. When compared with alternative cell-capture scaffolds, such as a graphene-oxide sponge globally modified with anti-EpCAM coated struts[41], our MPP fabricated scaffolds can exhibit arbitrary, assay-specific geometries and can pattern selective regions for cell capture, albeit at the expense of manufacturing time and scale limitations. MPP fabricated scaffolds could, in the future, be geometrically and chemically designed to capture multiple different cell types in distinct regions based on cell-size and surface markers. Furthermore, as the scaffold is fabricated entirely from proteins, it likely exhibits a high level of biocompatibility and degradability.
Compared with many extrusion-based 3D bioprinting approaches, and in particular compared with methods for bioprinting endothelial cell-lined vascular structures[42], our MPP approach can achieve support-free fabrication of ECM, at a superior resolution (~ 1–10 µm), albeit at the expense of scalability; reasonable fabrication times require our MPP fabricated volumes to be limited to less than ~ 1 mm3. Existing approaches for generating precisely architected, endothelial cell-lined channels, including pin-casting[43,44], photolithographic molding[45] and 3D printing of sacrificial scaffolds[10,11,46,47], have not been able to construct microvasculature with patent lumen diameters of 20 µm, close to those of native capillaries (Table 1). Crucially, our approach is capable of generating truly three dimensional, non-linear channel geometries, without relying on support material considerations for structural stability during fabrication. Furthermore, the in-vitro modeling applications of our MPP approach extend beyond microvasculature, as many other tubular structures have inner diameters close to 20 µm, including proximal tubules (~ 20 µm),[48] microlymphatic vessels (10–60 µm),[49] and terminal bile ducts (< 15 µm).[50]
Table 1.
Methods for generating geometrically defined, endothelial-lined channels embedded in bulk hydrogels.
| Method | Min. Vessel Diameter |
Geometry | Refs |
|---|---|---|---|
| Pin casting | 55 µm | Linear channels | [43][44] |
| Photolithographic templating | 120 µm | Planar network | [45] |
| Printing sacrificial filaments | |||
| Carbohydrate glass template | 180 µm | 3D network | [10] |
| Pluronic-F127 template | 100 μm | 3D network | [11] |
| Agarose template | 150 µm | 3D network | [46] |
| Gelatin template | 500 µm | Linear channels | [47] |
| Multi-photon microfabrication | 20 µm | Freeform 3D network | This paper |
However, applying MPP to therapeutic scale scaffold production remains a daunting task, as a 1 cm3 scaffold with 1 µm3 resolution structures is composed of a trillion voxels. Microfabrication at such a scale would require some additional modifications to our system, including the use of an upright microscope fitted with a long working-distance, water immersion objective lens. The laser scanning needs to be accelerated by replacing our mechanically slow galvanometer scanning system with high speed resonant galvonometers, as 400 mm/s represented the maximum speed that our system could scan, not the maximum speed of the patterning chemistry itself. Our previous work[25], and the work of others[40], suggests that the protein patterning speed should be able to significantly increase in speed if the fluorescein solution is deoxygenated. Scanning at such an accelerated speed should enable the microfabrication of a scaffold at a physiologically relevant scale, ~ 1 cm3, with micron scale resolution in approximately 1 day.
We believe that this methodology of combined scaffold microfabrication and patterning of internal protein cues could enable highly engineered therapeutic scaffolds for regenerative medicine applications.
Methods
Monolayer Formation
All scaffolds were formed by sandwiching a precursor solution between two monolayer-coated surfaces: one surface was chosen to bind to the scaffold for the purpose of handling and washing, while the other surface was to act as a ‘lid’, and as such was designed to resist scaffold adhesion to facilitate its removal prior to patterning the scaffold.
For agarose, the adhesion promoting substrate was Gel-Fix coated plastic (SERVA Electrophoresis, Heidelberg, Germany). For collagen, fibrin and GelMA scaffolds, the adhesion promoting substrate was an aldehyde functionalized glass slide that was prepared via a two-step process. First, the surface was amine-functionalized by immersion of a plasma treated glass slide (5 min plasma treatment duration) in amino-propyltri(ethoxy-silane) (APTES; Sigma-Aldrich, St. Louis, MO) (10% v/v in ethanol) for 30 min. The amine coated slide was then rinsed in an ethanol bath, sonicated in DI water for 5 min, and finally rinsed under running DI water. The slide was then dried under a stream of nitrogen gas. Next, the aminated glass slide was aldehyde functionalized by addition of glutaraldehyde in DI water (6% v/v) for 10 minutes, in a fume hood. Then the glass slide was washed in DI and dried under nitrogen. The functionalized glass slides were stored in a desiccator prior to use.
The adhesion resistant ‘lid’ for all scaffolds was a PEG monolayer coated coverslip, prepared similarly to a previous protocol.[26] Briefly, a borosilicate glass coverslip was treated with plasma for 5 min, then placed in a bath of toluene containing 2-[methoxy(polyethyleneoxy)propyl]trichlorosilane (Gelest, Morrisville, PA) (1% v/v), under a nitrogen atmosphere for 1 h. Next, the glass slide was rinsed in fresh toluene, DI water, and dried under a stream of nitrogen. The PEG coated coverslips were stored in a dessicator prior to use.
Scaffold Preparation
Collagen scaffolds were prepared by neutralization of a high concentration stock solution of rat tail collagen I (BD Biosciences, San Jose, California) on ice with pre-chilled phsosphate buffered saline (PBS) containing HEPES (125 mM, pH 7.4) to form a prepolymer solution containing collagen (2 mg/ml, for all protein patterning experiments; or 2, 4, 6 or 7.5 mg/ml for microfabricating collagen scaffolds). The neutralized collagen was pipetted onto an aldehyde functionalized coverslip and covered by a ~ 0.5 cm2 PEG coated coverslip, and then gelled in an incubator at 37°C for 1 h (for low porosity scaffolds) or at 10°C overnight (for the high porosity scaffold patterned in Figure S1).
Fibrin scaffolds were prepared as previously described.[51] Briefly, the prepolymer solution is formed by dissolving fibrinogen in PBS (11 mg/ml) at 37°C, without mechanical mixing or agitation, to form an prepolymer solution (dissolution takes approximately 1 h). Next, fibrinogen solution (100 µl) was mixed on ice with CaCl2 solution in DI water (5 µl, 50 mM) and thrombin stock solution (5 µl, 100 U/ml), prior to immediately pipetting onto an aldehyde functionalized coverslip, which was then covered by a 0.5 cm2 PEG coated coverslip, and then gelled in an incubator at 37°C for 1 h.
GelMA scaffolds were formed by dissolving lyophilized gelatin methacrylate (50 mg, prepared as previously described [52]) in PBS (1 ml), at 60°C in an oven. Irgacure 2959 (I-2959) (100 mg) was dissolved in methanol (200 µl) and added to the prepolymer solution at a final I-2959 concentration of 0.5% wt/v of I-2959. Next, molten GelMA solution (10 µl) was pipetted onto an aldehyde functionalized coverslip, covered with a PEG-coated coverslip, and exposed to a UV lamp (5 mW/cm2, 2 min) to form a polymerized GelMA scaffold.
Agarose scaffolds (1% wt/v) were prepared by adding agarose (0.5 g) to PBS (50 ml), and dissolving by repeatedly boiling in a microwave and swirling gently. A small sample of the agarose solution (10 µl) was pipetted onto a Gel-Fix surface, covered with a PEG-coated coverslip and allowed to gel at room temperature for 30 min. Agarose scaffolds were modified with methacrylate groups (forming AgaroseMA) by bathing an agarose gel (1% wt/v) overnight in a solution of glycidyl methacrylate (3.6% v/v) in DI water, with triethylamine (TEA) (3.6% v/v) as a catalyst and tetrabutyl ammonium bromide (TBAB) (3.6% wt/v) as a phase catalyst. After methacrylation, agaroseMA scaffolds were rinsed thoroughly in PBS and rocked in a PBS bath for 1 h to remove unincorporated methacrylate groups before use.
Laser Patterning of Proteins Inside Scaffolds
Prior to patterning, PEG coverslips were gently removed from the tops of all scaffolds, and a solution of B4F in PBS (200 µg/ml, pH 7.4) was pipetted on top of the scaffold and incubated for 10 min to allow the B4F to diffuse into the scaffold. A MAI-TAI femtosecond laser (Newport Corp., Irvine, CA), with a 80 MHz pulse repetition rate and a ~100 fs pulse duration, was tuned to 780 nm, and was focused by a 40X 0.9 NA air objective mounted on a z-axis piezo actuator (PI, Auburn, MA) to enable rapid stepping along the z-axis. The laser was scanned in x-y by means of a pair of galvanometer scan mirrors (Cambridge Technology, Lexington, MA). Voltage signals to control x-y-z scanning signals and laser power were output from a NI-DAQ 6259 PCIe card (National Instruments, Austin, TX) at a sample rate of 400 kSamples/s. Laser power was modulated via an electro-optic modulator (Newport Corp.). Unless otherwise stated, the laser was scanned with a scanline separation and scan speed of 0.25 µm and 200–400 mm/s respectively for protein patterns inside scaffolds. For fabricating collagen scaffolds shown in Figure 3c, a scanline separation and scan speed of 0.5 µm and 400 mm/s, respectively, were used. Laser power was measured at the sample using an IR power meter (Newport Corp.).
After B4F patterning, scaffolds are placed in a bath of PBS and rocked for > 1 h to remove excess B4F. Next, scaffolds are bathed for 30 min in PBS containing BSA (3% w/v) to block non-specific adhesion, prior to bathing in a PBS solution containing Alexa-Fluor 594 conjugated streptavidin (10 µg/ml) (Life Technologies, Carlsbad, CA) and BSA (3% w/v) for 1 h. Next, excess unbound streptavidin is removed by placing the scaffold in a PBS bath, and rocking for > 1 h.
For patterning horseradish peroxidase (HRP), a solution of PBS, containing biotinylated HRP (10 µg/ml) (Thermo Fisher Scientific, Rockford, IL) and BSA (3% w/v), was added to the streptavidin patterned scaffold and was incubated for 1 h prior to washing by rocking in a bath of PBS. Next, a fluorescent HRP substrate Amplex Red (Thermo Fisher Scientific) was added (1:1000 dilution) to a solution of PBS (50% v/v) and glycerol (50% v/v) containing hydrogen peroxide (10 µM). This fluorescent HRP substrate solution was then pipetted onto the HRP patterned scaffold. Glycerol was used to limit the diffusion and convection of fluorescent Amplex Red away from the patterned HRP, thus enabling the visualization of the patterns of HRP. The scaffold was imaged using epifluorescence ~1 s after adding the Amplex Red solution to the HRP patterned scaffold.
Microfabricating Collagen Scaffolds
A .stl file was generated in SolidWorks (Dassault Systèmes SolidWorks Corporation, Waltham, MA), and passed to a z-section slicing program which outputted the .stl file as a series of bitmap pictures of z-sections (Freesteel Slicer). These pictures were sent to a custom built MATLAB interpreter which translated them into a series of x-, y-, z-coordinates, and EOM voltage signals for patterning.
For microfabricating a collagen scaffold, a collagen gel was bathed in PBS containing fluorescein (200 µg/ml). It was essential to keep the sample dark to prevent unwanted global photobleaching and crosslinking of collagen from ambient light. Next, the MAI-TAI laser was scanned in x-y-z to expose defined regions of the collagen scaffold.
Developing Patterned Collagen Scaffolds
After exposure to the laser, the collagen is washed in a bath of PBS to remove excess fluorescein, then developed in either acetic acid (0.2 M, pH 3.5), or if specified, tris-HCl buffer (1 M, pH 7.4). The developing solution is very gently agitated periodically. After development, the sample is neutralized by gently bathing in a bath of PBS. To visualize patterned collagen scaffolds for two photon imaging, the sample is fluorescently conjugated by placing the sample in a bath of borate buffer (pH 8) containing either N-hydroxysuccinimide (NHS) conjugated DyLight 488 (50 µg/ml) or DyLight 594 (65 µg/ml) for 5 min, then washed in PBS to remove unconjugated dye.
For the timelapse imaging of scaffold development in Figure 3b and Figure S4, collagen scaffolds were labeled, post laser exposure, with DyLight 594 (as described above) prior to development. The labeled scaffold was placed on an automated microscope stage (Prior Scientific, Rockland, MA), bathed in solutions of acetic acid (0.2 M, pH 3.5), tris-HCl (1M, pH 7.4) or PBS containing collagenase I (1 U/ml), and was imaged at various time intervals. Illumination was performed by briefly unshuttering an epifluorescence source and reshuttering within 0.5 s in an automated fashion. It was essential to minimize light exposure, as the excitation light exposed to the collagen fibrils was sufficient to render the collagen scaffold insoluble within ~ 1 min of light exposure due to singlet oxygen generated by excitation of conjugated fluorescent dye. To limit this singlet oxygen driven crosslinking of collagen under the epifluorescent light, L-ascorbic acid (10 mM) was added to the developing solution during the imaging process. Throughout timelapse imaging, the motorized stage was translated left and right with a frequency of ~ 0.5 Hz to gently rock the developer solution to aid the flow of solubilized collagen away from the field of view.
Combined Scaffold Microfabrication and Protein Patterning
To form microfabricated scaffolds with internal protein patterns (as shown in Figure 5), fluorescein-infused collagen scaffolds are first exposed to the scanning laser (without developing), while ensuring that all excess fluorescein solution surrounding the collagen gel is removed prior to exposure by gently dabbing a paper towel around the perimeter of the gel. After exposing the collagen scaffold shape, a few drops of B4F solution are gently added on top of the scaffold without directly touching it. In this manner, a realignment step between microfabrication of collagen and micropatterning of protein cues is not necessary, as the sample has not been moved on the microscope slide holder. After allowing 30 mins for the B4F to diffuse into the scaffold, a new region is scanned to generate patterns of biotin inside the collagen. Next, the scaffold is developed in acetic acid as previously described, switched into borate buffer and, to aid visualization, fluorescently conjugated using NHS-DyLight-488 (50 µg/ml). After 10 min, the scaffold is rinsed with PBS, and then the scaffold is blocked for 30 mins with PBS containing BSA (3% w/v), before bathing with streptavidin (10 µg/ml) in PBS containing BSA (3% w/v) for 5 min. Finally the scaffold is bathed in PBS for 30 mins to remove excess streptavidin prior to imaging.
Formation of Microvascular Structures
A simple microfluidic device (Figure 4a) consisting of two wells connected by a single channel ( > 1 mm × 500 µm × 150 µm LxWxH) was fabricated using standard soft lithography techniques. Briefly, SU-8 2050 (MicroChem, Newton, MA) was spun at 1250 rpm for 30 s to generate a 150 µm film, softbaked at 65°C for 5 min followed by 95 °C for 25 min, exposed to UV through a printed transparency, postbaked at 65°C for 5 min and 95°C for 10 min. The patterns were developed in PM acetate, and hard baked for 15 min at 150°C. The wafer was then silanized by placing the wafer in a vacuum chamber containing a few drops of (Tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane for 10 min. Sylgard 184 was mixed with its crosslinker (10:1 ratio), poured over the SU-8 patterned wafer, degassed for ~ 1 h in a vacuum chamber, and cured at 70°C for 1 h. The cured PDMS was removed from the wafer, and wells were punched out at either side of the channel using a 7 mm diameter biopsy punch. The chip and a clean glass coverslip were plasma bonded, and collagen precursor solution (7.5 mg/ml) in PBS containing fluorescein (200 µg/ml ) was pipetted directly into the microchannel, and gelled for 30 min in an incubator at 37°C.
To generate microvascular channels, a collagen structure (700 µm × 500 µm × 150 µm LxWxH for Figure 4b or 1200 µm × 500 µm × 150 µm LxWxH for Figure 4c) containing branched channels was microfabricated in 5 rows and either 7 or 12 columns of 100 × 100 × 150 µm sections that comprise the complete structure. Each section was exposed in ~ 8 s, finishing the structure shown in Figure 4b in ~ 5 min. After laser scanning is complete, fluorescein was washed out of the wells using PBS, and acetic acid (0.2 M, pH 3.5) was added for ~ 30 min with 100 µl in one well and 110 µl in the other to drive a gentle flow to aid the dissolving of the collagen channels. Next, the acetic acid is replaced 5x with fresh acetic acid to remove the dissolved collagen, and neutralized with 10x rinses in PBS. The collagen is then crosslinked by adding glutaraldehyde (6% v/v) in DI in each well for 2 min. As with the acetic acid, a 10 µl excess of glutaraldehyde solution is added to one well to ensure perfusion through the channels. The glutaraldehyde is then removed by 10 rinses with PBS.
Human umbilical-vein endothelial cells (HUVECs) were obtained from Lonza (Basel, Switzerland) and cultured in Endothelial Basal Media supplemented with an EGM-1 Bulletkit (Lonza). The PBS in the wells was replaced with media, and a 10 µl solution of passage 3–5 cells at a high density (> 1M cells/mL) was added to one well of the chip, and the cells entered the channel under hydrostatic flow and attached to the walls. The addition of cells was repeated until a high density of cells coated the walls of the channels. The cells were maintained at 37°C, 5% CO2. After 12 h (Figure 4b) or 72 h (Figure 4c), cells were fixed in aqueous paraformaldehyde (4% v/v) for 10 mins. The cells were permeabilized for 3 min in PBS containing Triton X-100 (0.1% v/v), blocked in PBS containing casein (1% w/v) for 30 mins, and actin was labeled by incubation with PBS containing phalloidin-Alexa Flour 594 (Life Technologies) (2 U/ml) and casein (1% w/v) for 30 mins. Nuclei were stained by a 3 min incubation in Hoescht solution in PBS (1:5000). To obtain a bright staining for high-contrast two photon imaging of 3D microchannels for Figure 4b, (ii)-(iii), cellular protein was stained using a solution of Alexa-Flour 488-NHS (50 µg/ml) in PBS through the channels for 5 min, followed by 10× rinses in PBS.
HL-60 Rolling Assays
A collagen channel (500 × 700 × 150 µm) was microfabricated in a microfluidic channel as described above, with all incubations or washes being performed with 10 µl excess fluid in one well to drive flow through the collagen. Before developing the exposed collagen channel structure in acetic acid, PBS containing B4F (200 µg/ml) was added to the PDMS wells, and allowed to diffuse into the collagen scaffold for 1 h. Care was taken not to displace the chip at this stage, such that alignment with the exposed collagen regions would not be disrupted. Next, annular patterns of B4F were patterned around one of the branches (see Figure 5c-d). B4F was then rinsed out of the wells with excess PBS, which was then replaced by acetic acid to develop the collagen channels. After developing for 1 h, acetic acid was removed from the wells by washing with excess PBS, and the channels were blocked for > 1 h in PBS containing BSA (3% w/v). Next, streptavidin (10 µg/ml) in PBS containing BSA (3% w/v) was added to the wells . After 1 h, the streptavidin was rinsed out with excess PBS and subsequently incubated in PBS for 1 h to remove unbound streptavidin. Next, biotinylated P-selectin (R&D Systems) (10 µg/ml) in PBS containing BSA (3% w/v) was added for 1 h, and was subsequently washed in PBS for 1 h to remove unbound P-selectin. Before use, HL-60 cells were cultured in flasks containing IMDM with FBS (10% v/v) containing penicillin/streptomycin (100 U/ml). Once microfabricated, the collagen channels were bathed in the HL-60 cell culture media described above, and 2 µl of HL-60 cells were added to one well to drive a flow of cells through the chip at a speed of ~200 µm/s. Cells were counted as rolling if they remained adhered to the wall for > 100 µm along the straight section of the P-selectin coated or uncoated branch. From this count, a cell rolling ratio was calculated as the ratio of the number of rolling cells in a branch to the total number of cells that flowed through the branch. The HL-60 homing assay was prepared as the rolling assay, except for the use of a modified geometry. As a negative control for the homing geometry, human erythrocytes (Zen-bio, Inc., NC) were introduced to the bulged channels in the same manner as the HL-60 cells.
Image Analysis
Three-dimensional images of patterned proteins and microfabricated scaffolds were obtained using a two-photon fluorescence microscope (Prairie Technologies, Middleton, WI) using a MAI-TAI laser. Pattern brightness was calculated, using ImageJ, as the difference between the brightness of patterned regions and the background. For the quantitation of patterned fluorescent streptavidin concentration in Figure 2c, the pattern brightness for each material was compared to the brightness of a 10 µg/ml solution of streptavidin. All error bars display standard deviations.
Supplementary Material
Acknowledgments
MAS was supported by a Kennedy Memorial Trust fellowship and a Martinos Scholarship. AD was supported by the National Space Biomedical Research Institute through NASA NCC 9-58. This project was funded by an NIH R01 EUREKA Award (R01-NS066352), a Packard Award in Science and Engineering, and an NIH Director’s Pioneer Award (DP1-NS082101).
Footnotes
Conflict of Interest Statement
The authors report no conflict of interest associated with this work.
References
- 1.Ashe HL, Briscoe J. Development. 2006;133:385. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- 2.Tessier-Lavigne M, Goodman CS. Science. 1996;274:1123. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 3.Conover JC, Notti RQ. Cell Tissue Res. 2008;331:211. doi: 10.1007/s00441-007-0503-6. [DOI] [PubMed] [Google Scholar]
- 4.Ott HC, Matthiesen TS, Goh S-K, Black LD, Kren SM, Netoff TI, Taylor DA. Nat. Med. 2008;14:213. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 5.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Nat. Med. 2010;16:927. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 6.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, Herzog E, Niklason LE. Science. 2010;329:538. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crapo PM, Gilbert TW, Badylak SF. Biomaterials. 2011;32:3233. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat. Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachlos E, Reis N, Ainsley C, Derby B, Czernuszka JT. Biomaterials. 2003;24:1487. doi: 10.1016/s0142-9612(02)00528-8. [DOI] [PubMed] [Google Scholar]
- 10.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat. Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. Adv. Mater. 2014;26:3124. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 12.Basu S, Cunningham LP, Pins GD, Bush KA, Taboada R, Howell AR, Wang J, Campagnola PJ. Biomacromolecules. 2005;6:1465. doi: 10.1021/bm049258y. [DOI] [PubMed] [Google Scholar]
- 13.Bell A, Kofron M, Nistor V. Biofabrication. 2015;7:035007. doi: 10.1088/1758-5090/7/3/035007. [DOI] [PubMed] [Google Scholar]
- 14.Harper JC, Brozik SM, Brinker CJ, Kaehr B. Anal. Chem. 2012;84:8985. doi: 10.1021/ac301816c. [DOI] [PubMed] [Google Scholar]
- 15.Ovsianikov A, Malinauskas M, Schlie S, Chichkov B, Gittard S, Narayan R, Löbler M, Sternberg K, Schmitz K-P, Haverich A. Acta Biomater. 2011;7:967. doi: 10.1016/j.actbio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honegger T, Elmberg T, Berton K, Peyrade D. Microelectron. Eng. 2011;88:2725. [Google Scholar]
- 17.Koroleva A, Gittard S, Schlie S, Deiwick A, Jockenhoevel S, Chichkov B. Biofabrication. 2012;4:015001. doi: 10.1088/1758-5082/4/1/015001. [DOI] [PubMed] [Google Scholar]
- 18.Melissinaki V, Gill AA, Ortega I, Vamvakaki M, Ranella A, Haycock JW, Fotakis C, Farsari M, Claeyssens F. Biofabrication. 2011;3:045005. doi: 10.1088/1758-5082/3/4/045005. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Moon JJ, West JL. Biomaterials. 2008;29:2962. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. Nat. Mater. 2011;10:799. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 21.Seidlits SK, Schmidt CE, Shear JB. Adv. Funct. Mater. 2009;19:3543. [Google Scholar]
- 22.Holden M, Cremer P. J. Am. Chem. Soc. 2003;125:8074. doi: 10.1021/ja035390e. [DOI] [PubMed] [Google Scholar]
- 23.Bélisle JM, Correia JP, Wiseman PW, Kennedy TE, Costantino S. Lab Chip. 2008;8:2164. doi: 10.1039/b813897d. [DOI] [PubMed] [Google Scholar]
- 24.Bélisle JM, Kunik D, Costantino S. Lab Chip. 2009;9:3580. doi: 10.1039/b911967a. [DOI] [PubMed] [Google Scholar]
- 25.Scott MA, Wissner-Gross Z, Yanik M. Lab Chip. 2012;12:2265. doi: 10.1039/c2lc21105j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wissner-Gross ZD, Scott MA, Ku D, Ramaswamy P, Fatih Yanik M. Integr. Biol. 2011;3:65. doi: 10.1039/c0ib00058b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEver RP. Curr. Opin. Cell Biol. 2002;14:581. doi: 10.1016/s0955-0674(02)00367-8. [DOI] [PubMed] [Google Scholar]
- 28.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Nat. Rev. Immunol. 2007;7:678. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 29.Mosiewicz KA, Kolb L, van der Vlies AJ, Martino MM, Lienemann PS, Hubbell JA, Ehrbar M, Lutolf MP. Nat. Mater. 2013;12:1072. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 30.Leung LY, Tian D, Brangwynne CP, Weitz DA, Tschumperlin DJ. FASEB J. 2007;21:2064. doi: 10.1096/fj.06-7510com. [DOI] [PubMed] [Google Scholar]
- 31.Duong H, Wu B, Tawil B. Tissue Eng. Part A. 2009;15:1865. doi: 10.1089/ten.tea.2008.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiron S, Tomczak C, Duperray A, Lainé J, Bonne G, Eder A, Hansen A, Eschenhagen T, Verdier C, Coirault C. PLoS One. 2012;7:e36173. doi: 10.1371/journal.pone.0036173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Normand V, Lootens DL, Amici E, Plucknett KP, Aymard P. Biomacromolecules. 2000;1:730. doi: 10.1021/bm005583j. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ. Nat. Mater. 2014;13:970. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 36.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Nat. Mater. 2014;13:979. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson GH, Piston DW. Biophys. J. 2000;78:2159. doi: 10.1016/S0006-3495(00)76762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu A, Naru E, Arakane K, Masunaga T, Shinmoto K, Nagano T, Hirobe M, Mashiko S. Chem. Pharm. Bull. (Tokyo) 1997;45:1243. doi: 10.1248/cpb.45.1243. [DOI] [PubMed] [Google Scholar]
- 39.Au V, Madison SA. Arch. Biochem. Biophys. 2000;384:133. doi: 10.1006/abbi.2000.2070. [DOI] [PubMed] [Google Scholar]
- 40.Webster A, Britton D, Apap-Bologna A, Kemp G. Anal. Biochem. 1989;179:154. doi: 10.1016/0003-2697(89)90216-9. [DOI] [PubMed] [Google Scholar]
- 41.Yin S, Wu Y-L, Hu B, Wang Y, Cai P, Tan CK, Qi D, Zheng L, Leow WR, Tan NS, Wang S, Chen X. Adv. Mater. Interfaces. 2014;1 n/a. [Google Scholar]
- 42.Bogorad MI, DeStefano J, Karlsson J, Wong AD, Gerechtc S, Searson PC. Lab Chip. 2015;15:4242. doi: 10.1039/c5lc00832h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price GM, K. Wong KH, Truslow JG, Leung AD, Acharya C, Tien J. Biomaterials. 2010;31:6182. doi: 10.1016/j.biomaterials.2010.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chrobak KM, Potter DR, Tien J. Microvasc. Res. 2006;71:185. doi: 10.1016/j.mvr.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9342. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Khademhosseini A. Lab Chip. 2014;14:2202. doi: 10.1039/c4lc00030g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee VK, Lanzi AM, Haygan N, Yoo S-S, Vincent PA, Dai G. Cell. Mol. Bioeng. 2014;7:460. doi: 10.1007/s12195-014-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhai XY. J. Am. Soc. Nephrol. 2003;14:611. doi: 10.1097/01.asn.0000051725.00406.0c. [DOI] [PubMed] [Google Scholar]
- 49.Scallan J, Huxley VH, Korthuis RJ. The Lymphatic Vasculature. Morgan & Claypool Life Sciences; 2010. [PubMed] [Google Scholar]
- 50.Strazzabosco M, Fabris L. Anat. Rec. (Hoboken) 2008;291:653. doi: 10.1002/ar.20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kathleen K, WSM J. Vis. Exp. 2012 [Google Scholar]
- 52.Chen Y-C, Lin R-Z, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Adv. Funct. Mater. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.