Abstract
A range of acyl-lysine (acyl-Lys) modifications on histones and other proteins have been mapped over the past decade but for most, their functional and structural significance remains poorly characterized. One limitation in the study of acyl-Lys containing proteins is the challenge of producing them or their mimics in site-specifically modified forms. We describe a cysteine alkylation-based method to install hydrazide mimics of acyl-Lys post-translational modifications (PTMs) on proteins. We have applied this method to install mimics of acetyl-Lys, 2-hydroxyisobutyryl-Lys, and ubiquityl-Lys that could be recognized selectively by relevant acyl-Lys modification antibodies. The acyl-Lys modified histone H3 proteins were reconstituted into nucleosomes to study nucleosome dynamics and stability as a function of modification type and site. We also installed a ubiquityl-Lys mimic in histone H2B and generated a diubiquitin analog, both of which could be cleaved by deubiquitinating enzymes. Nucleosomes containing the H2B ubiquityl-Lys mimic were used to study the SAGA deubiquitinating module’s molecular recognition. These results suggest that acyl-Lys mimics offer a relatively simple and promising strategy to study the role of acyl-Lys modifications in the function, structure, and regulation of proteins and protein complexes.
Graphical Abstract
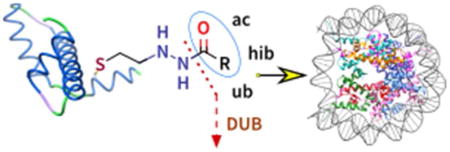
INTRODUCTION
Lysine sidechain modifications on histones were discovered about 50 years ago.1a Since then, these reversible covalent marks have been shown to be important for the regulation of genome processing including gene expression and DNA repair.1 The last decade has seen the discovery of a plethora of novel post-translational modifications (PTMs) involving the acylation of the lysine sidechain (acyl-Lys).2 Thousands of lysine acetylation and ubiquitylation sites have been mapped and increasing numbers of novel acyl-Lys modifications such as 2-hydroxyisobutyrylation (hib) have been identified (Figure 1A).3 The biological functions of the vast majority of these lysine modifications, however, have remained elusive. To determine the functions of such acyl-Lys PTMs, it is important to have proteins containing stoichiometric and site-specific modifications or faithful mimics. Standard site-directed mutagenesis has been used to investigate the roles of specific acetyl-Lys modifications by introducing glutamine as a mimic, but the results can be difficult to interpret.4
Figure 1.
Lysine sidechain modifications. (A) Naturally occurring acyl-Lys PTMs. (B) Previously reported acetyl-Lys mimics.9a,8a,8b (C) Three-step protocol presented in this manuscript to install acyl-Lys mimics into proteins.
Several elegant strategies have been developed for installing acyl-Lys modifications and their mimics into proteins, including total chemical synthesis,5 chemical ligation strategies,6 nonsense suppression mutagenesis,7 and cysteine (Cys) modifications8a,9,10 but the technical complexity, scope, or yields have so far limited their applications. Total chemical synthesis of proteins containing ubiquitylation and acetylation has been shown to be effective for small proteins11 but because of technical challenges, this method is out of reach for most biochemistry labs. Expressed protein ligation and related semisynthetic methods have been used to introduce acetyl-6c,12 and ubiquityl-Lys13 modifications into proteins, but these methods have typically been employed to install modifications at the termini of a protein of interest.14 The use of nonsense codon suppression to incorporate acyl-Lys has shown promise, but has largely been used in prokaryotic expression systems, the yields can be variable, and enzymatic deacylation during isolation is a concern.7c The unique reactivity of the Cys sulfhydryl group in a protein has been harnessed before to install good mimics of methyl-arginine,15a methyl-lysine,10b,15b and acyl-lysine8a,9,10 (Figure 1B) modifications. The key step in the mimic installation in these methods is either a chemoselective alkylation of the thiol8a,15 or a thiol–ene reaction9,10 of the Cys side-chain, where the latter also includes a variation of Cys to dehydroalanine conversion followed by thiol–ene reaction with a modification bearing thiol.10b Thus, all these mimics contain a sulfur atom instead of the methylene, but that minor difference has not proven detrimental to the mimicry.
Ubiquitylation, a PTM that results in an appendage of 8.5 kDa, can be regarded as an especially challenging target among acyl-Lys modifications. Cys modification methods,10,13 disulfide linkage,16 azide-alkyne cycloaddition,17 total chemical synthesis,5a–c and nonsense suppression mutagenesis followed by traceless ligation18 have all been shown to have utility in the production of ubiquityl-protein conjugates. Perhaps because of either technically challenging chemical manipulations in some cases or the significant departure from the native structures in others, however, these strategies have not yet been widely adopted. As there are tens of thousands of acyl-Lys sites that have been mapped, simple and efficient methods are needed to more rapidly interrogate the impact of these PTMs.
Here we investigate a new Cys modification strategy that can install hydrazide analogs of acyl-Lys mimics. We show that this relatively simple semisynthetic method can be used to install small modifications into proteins such as an acetyl-Lys mimic, as well as larger modifications such as a ubiquityl-Lys (Figure 1C). We examine the recognition of these acyl-Lys mimics using antibodies and their macromolecular effects regarding nucleosome dynamics and recognition by deubiquitinases (DUBs) and describe these studies below.
EXPERIMENTAL METHODS
Acid hydrazides
Acetohydrazide was purchased from Sigma-Aldrich. 2-Hydroxy-isobutyric acid hydrazide19a and ubiquitin hydrazide19b have been reported previously and our method of preparation is delineated in the Supporting Information.
Expression and purification of histones
Protocols for bacterial overexpression and purification of H3-K56C, H3-K122C (these K→C mutants are derived from human H3.1 harboring C96S, G102A, and C110A mutations), H2B-K120C, H2B-K116A,K120C (these mutants originated from truncated, aa4–125, Xenopus laevis H2B-1.1) and other core histones are detailed in the Supporting Information.
Installation of hydrazide mimics into histones and ubiquitin
Lyophilized powder of H3-K56C or K122C (10 mg, final concentration: ~1.0 mM) was dissolved in 100 mM HEPES (added from 1 M stock, pH 8.0) containing 50 mM reducing agent, tris(2-carboxyethyl)phosphine (TCEP; added from 0.5 M stock, pH 7.0) and 4.5 M guanidinium hydrochloride. Chloroacetaldehyde (50% w/v soln.; final concentration: 15 mM) was then added and stirred vigorously at room temperature for 30–45 min. The reaction mixture was transferred to a 15 mL centrifugal filter-unit (Amicon Ultra-15, 3 kDa cutoff, MilliporeSigma) and filled to the brim with dH2 O and spun at 4,000 rpm until the volume was reduced to about 3 mL. This operation was repeated twice by filling the filter unit with acetate buffer (1 M AcOH + 1 M NaOAc, pH 4.5). At this stage the filter unit was filled with acetate buffer containing acyl-hydrazide (20 mM in the case of acetohydrazide and 2-hydroxyisobutyrohydrazide, and 10 mM for ubiquitin hydrazide [Ub-Hz]) and the procedure was repeated. The final volume of about 1 mL was then transferred to a 5 mL tube and supplemented with 15 mM acyl-hydrazide (this is not needed in the case of Ub-Hz, because, it is retained during Amicon ultrafiltration) and the mixture was vigorously stirred at room temperature for 14–18 h. The final step in this three-step protocol is the reduction of hydrazone to hydrazide which is achieved by adding sodium cyanoborohydride (final concentration: 50 mM) in 4 M urea and stirring vigorously at room temperature for 2 h. This reaction mixture can be directly injected (if not ready for HPLC right away, the sample should be rid of urea by Amicon ultrafiltration) onto a PROTO 300 C18 column on HPLC (4.6×250 mm, 53, 300 Å; Higgins Analytical) and eluted with acetonitrile/H2 O (gradient ramp of 25% to 85% acetonitrile over 40 min) containing 0.05% trifluoroacetic acid (TFA). The HPLC fractions were examined by ESI-MS, purified fractions containing desired protein species were pooled, concentrated and lyophilized to obtain the desired histone-PTM mimics as powders and stored at -80 °C. It should be noted that we installed ubiquitin hydrazide mimics on H2B-K120C, H2B-K116A,K120C, and ubiquitin-K48C (makes K48-diubiqutin mimic) using the procedure described above. Starting from 10 mg of histone, yields of pure histone PTM mimics ranged between 2–6 mg.
Note: Henceforth, we will use shorthand designations for acyl-Lys mimics. For example, H3-Kc56ac will stand for histone H3 bearing acetyl-Lys hydrazide mimic at position 56, where the mimic descended from Cys.
H3-Kc56ac
Average Mass (Mav), calculated: 15,313.8; measured, ESI-MS: 15,315.5 ± 4.6.
H3-Kc56hib
Mav, calculated: 15,357.9; measured, MALDI-TOF: 15,357.5, ESI-MS: 15,359.5 ± 3.0.
H3-Kc56ub
Mav, calculated: 23,810.5; measured, ESI-MS: 23,814.5 ± 4.7.
H3-Kc122ac
Mav, calculated: 15,313.8; measured, ESI-MS: 15,316.4 ± 4.1.
H3-Kc122hib
Mav, calculated: 15,357.9; measured, MALDI-TOF: 15,356.6, ESI-MS: 15,357.2 ± 3.8.
H3-Kc122ub
Mav, calculated: 23,810.5; measured, ESI-MS: 23,813.5 ± 3.9.
H2B-Kc120ub
Mav, calculated: 22,065.5; measured, ESI-MS: 22,067.5 ± 3.1.
H2B-K116A,Kc120ub
Mav, calculated: 22,008.4; measured, ESI-MS: 22,012.5 ± 4.2
Kc48-diUbiquitin
Mav, calculated: 17,128.6; measured, MALDI-TOF: 17,132.4.
Immunoblotting, nucleosome reconstitutions, and subsequent assays including deubiquitination
These experiments were conducted according to known methods and are described in the Supporting Information.
RESULTS AND DISCUSSION
Synthetic approach to acyl-Lys mimic incorporation
The goal of this work was to generate acyl-Lys mimics in a chemically simple way that would be compatible with recapitulating at least some of the properties of acyl-Lys modifications. In this regard, we set our sights on thioether-containing acyl-hydrazide mimics of acyl-Lys (Figure 1C, 6). Such acyl-Lys mimics substitute a sulfur atom for the -methylene in the Lys side chain and contain an additional nitrogen atom in the connection between the e-amino group and the carbonyl. Prior studies on Lys thioether derivatives8a,9,10b and on hydrazide8b analogs (Figure 1B) indicated that such functionality could preserve at least some of the structural and biochemical features of the corresponding acyl-Lys.
Figure 6.
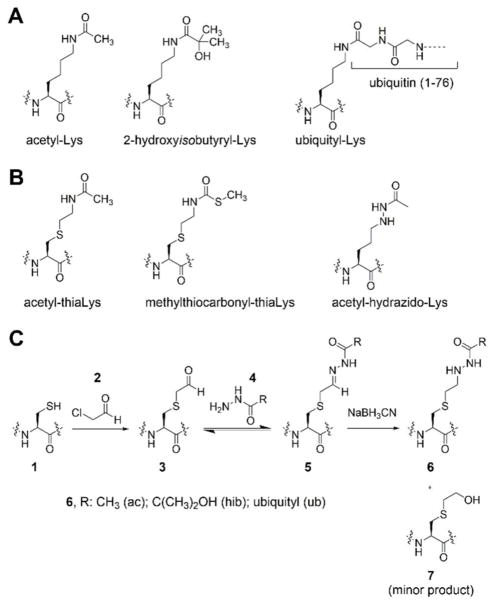
Effect of H3-K122 modifications on nucleosome stability. (A) Normalized FRET response due to a titration of NaCl in presence of nucleosomes with and without acyl-Lys mimics at H3-K122. (B) Decay of FRET acceptor response through donor excitation on nucleosomes with and without acyl-Lys modifications at H3-K122 at a fixed concentration of salt (1,125 mM). (C) Tabulation of results from salt titration and kinetics.
The general synthetic approach toward these acyl-Lys analogs pursued here involves a three-step chemical sequence starting with a Cys-containing peptide (model peptide reaction detailed in Supporting Information) or protein: thiol alkylation with chloroacetaldehyde followed by reaction with the corresponding acyl-hydrazide and then sodium cyanoborohydride reduction (Figure 1C). During optimization of chloroacetaldehyde alkylation of cysteines on model peptides and proteins, we observed that the presence of up to 50 mM TCEP is tolerated and necessary to drive the reaction to completion. TCEP can theoretically undergo alkylation20 to deplete chloroacetaldehyde but the near complete modification of the cysteine sulfhydryl group suggests that this side reaction is inconsequential under our reaction conditions. We performed these reactions on histones and ubiquitin engineered to have a single Cys at the Lys modification site of interest. Moving from peptides to histones and ubiquitin, the use of 4.5 M guanidine hydrochloride as chaotrope was found to be beneficial in the alkylation step. However, the chaotrope impeded the hydrazone formation, and we found acetate buffer at pH 4.5 to be conducive to this process, and hence a buffer exchange was incorporated at this step.
It should be noted that hydrazone formation is likely a slower and reversible step (requires ~14 hours to reach near completion). This would explain that after cyanoborohydride reduction besides the desired PTM mimic we do observe as a minor side product, separable on HPLC, hydroxyethyl modified proteins presumably resulting from aldehyde to alcohol reduction (Figure 1C, 7). This approach was used to incorporate acetyl, 2-hydroxyisobutyryl, and ubiquityl mimics into proteins. In general, the three-step process proceeds to ~50–70% conversion and final modified protein products are purified by reversed-phase chromatography and structural confirmation performed using mass spectrometry (Figure 2)
Figure 7.
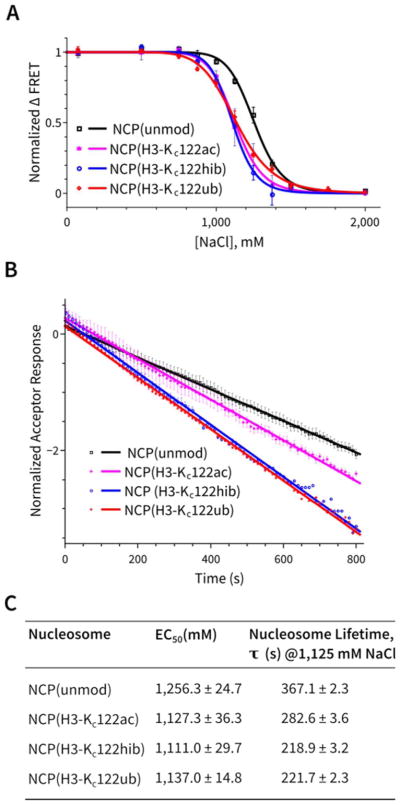
Deubiquitinase reactions of (A) Ubp10, (B) Ubp8/SAGA DUB module, and (C) hOTUB1 with natural and hydrazide mimic versions of K48-diubiquitin.
Figure 2.
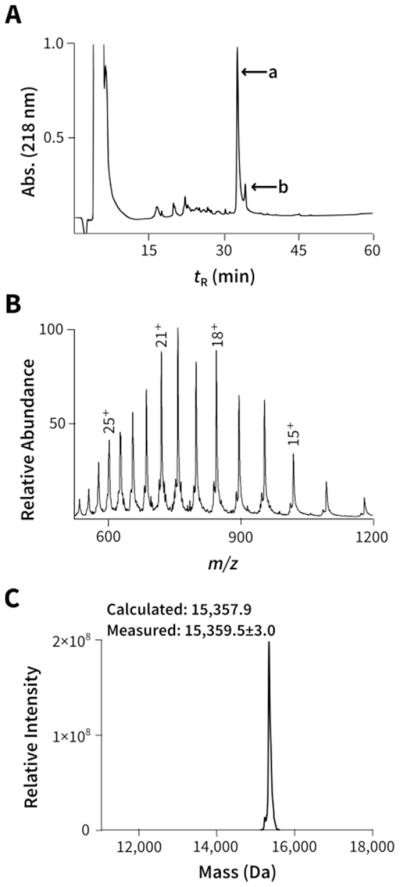
Representative characterization data for hydrazide mimics (data shown for H3-Kc56hib, remainder of the hydrazide mimics are in Supplementary Figures). (A) HPLC trace, peak ‘a’ corresponds to H3-Kc56hib and after ESI-MS analysis ‘b’ was attributed to the minor side product 7, shown in Fig. 1C. (B) ESI-MS of HPLC fractions corresponding to peak ‘a’. (C) Deconvolution of ESI-MS spectra shown in Fig. 2B above. Reported here are the Average Masses (Mav).
Effects of acyl-Lys on nucleosome dynamics and stability
Given the frequency and diversity of histone Lys modifications and their potential importance in influencing nucleosome dynamics, we decided to investigate the effects of acyl-Lys mimics in the context of histones. We incorporated three acyl-Lys mimics for acetyl (ac), hydroxyisobutyryl (hib), and ubiquityl (ub) at sites of histone modifications that were identified in proteomics studies.1–3 The sites studied here in histone H3 include H3-K56 (ac, hib, and ub) and H3-K122 (ac, hib, and ub). These sites were chosen in part because they occur in the globular region of histone H3, where they are in proximity to the DNA and therefore may be more likely to affect nucleosome stability or dynamics. H3-K56 and H3-K122 acetylation have been shown previously to influence nucleosome biophysical properties.21
As one test of whether the acyl-Lys mimics recapitulated properties of native acyl-Lys modifications, we used Western blotting to test whether antibodies specific for particular acyl-Lys modifications would recognize the acyl-Lys mimics. As shown in Figure 3, the hydrazide mimics installed on histone H3 (H3-Kc56ac, hib, and ub) were selectively recognized by commercial antibodies that correspond to the appropriate acyl-Lys modification. These results are consistent with the designed structural similarity between the acyl-Lys mimics and the genuine modifications.
Figure 3.
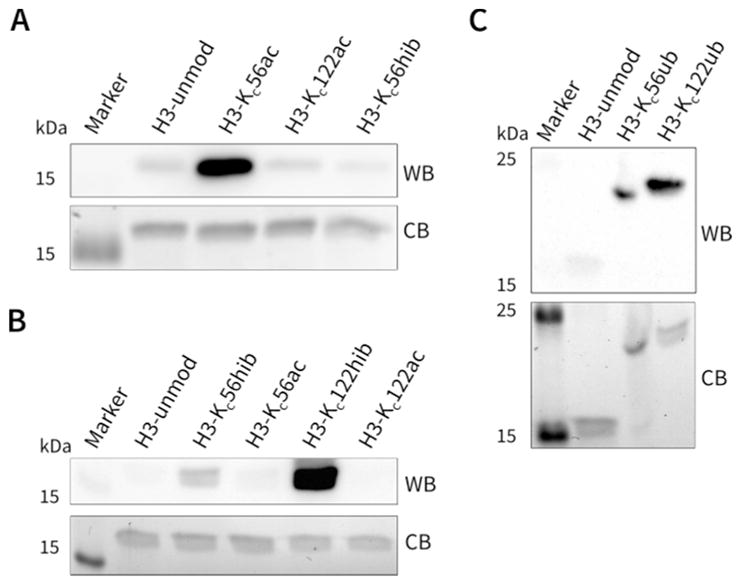
Selective antibody recognition of hydrazide mimics. (A) Antibody against H3-K56ac (WB: immunoblot; CB: Colloidal Blue visualization of the loading control). (B) Pan 2-hydroxyisobutyryl-Lys antibody. (C) Ubiquitin antibody. H3-unmod refers to the unmodified human H3.1 harboring C96S, G102A, and C110A mutations. Full gels of loading controls stained with Colloidal Blue corresponding to Figure 3A–C are shown in Supplementary Figure 10A–C, respectively. Pan-2-hydroxyisobutyryl-Lys antibody recognizes the modification in H3 better at Kc122 than at Kc56, and that same observation was made in each replicate.
One potential consequence of acyl-Lys modifications in the context of histones is that they may loosen nucleosome structure. Indeed, prior work on acetylation of histone H3 on Lys56, a modification seen in DNA damage in yeast, has shown that this modification can enhance nucleosome DNA unwrapping in the entry/exit region of the nucleosome.5d We therefore proceeded to compare the effects of the various histone H3 modifications on nucleosome DNA unwrapping. We assembled nucleosomes containing a LexA protein binding site in a Widom 601 DNA nucleosome positioning sequence (LexA site bp 8–27) that also included Cy3 and Cy5 fluorescent tags at DNA bp 1 and histone H2A-K119C, respectively (Figure 4A and 4C). Cy3 is a FRET donor to Cy5, so when LexA binds to its canonical site in the nucleosome, it traps the nucleosome in a partially unwrapped state increasing the Cy3–Cy5 distance and decreasing the FRET signal. The affinity of LexA for these nucleosomes can therefore be measured by monitoring the FRET change (Figure 5A).
Figure 4.
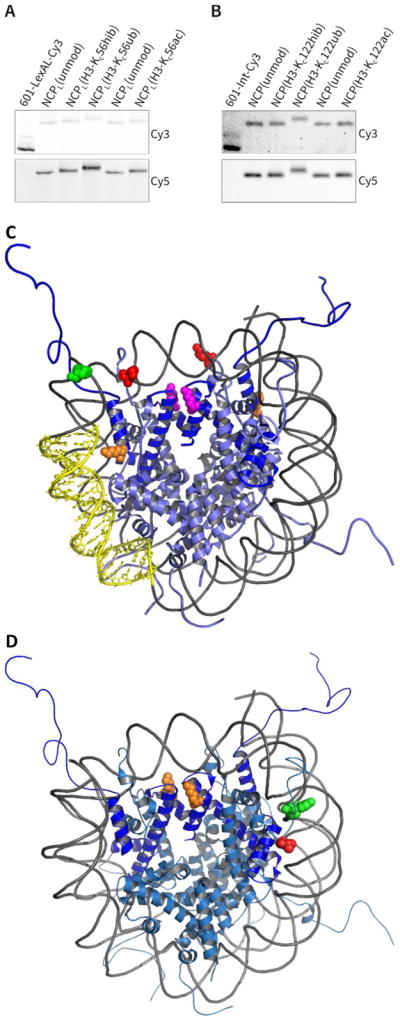
Native PAGE of reconstituted nucleosomes bearing hydrazide mimics (NCP stands for nucleosome core particle). Two types of nucleosomes we prepared to study their dynamics: NCPL with LexA target sequence in the dsDNA and standard NCP which does not include LexA dsDNA. (A) Nucleosomes with H3-Kc56 modifications. (B) Nucleosomes containing H3-Kc122 modifications. (C and D) Structures of NCPs (PDB ID:22 1KX5), depicting the location of fluorophores and PTM mimics. Both copies of H3 are rendered in blue. Locations of Cy3 (green), Cy5 (red), and PTM mimics are in shown as ‘spheres’. (C) Nucleosomes used in the LexA binding studies show LexA target sequence (yellow ‘sticks’), H3-K56 (orange), and H3-K122 (magenta). (D) NCPs used in salt titrations and kinetics show H3-K122 residues in orange. H3-unmod (as described in Figure 3 legend above) was used to assemble both NCPL(unmod) and NCP(unmod) in Figures 4A and 4B, respectively. Also, these NCPs were loaded on every third lane to help reveal the gel shifts due to PTMs. This observation has been noted in previous reports (see Fig. 3c–d in reference 5d or Fig. 2A in reference 21c).
Figure 5.
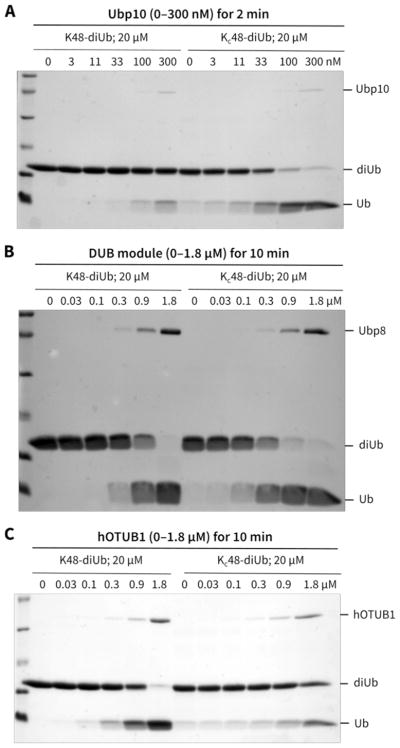
Effect of H3-K56 acyl modifications on nucleosome dynamics. (A) Titration of LexA against unmodified (comprising H3-unmod) and modified nucleosomes bearing hydrazide mimics on H3-K56. (B) Tabulation of S½ values (concentration at which half of the nucleosomes are bound by LexA) and relative DNA accessibilities of nucleosomes for LexA binding with different H3-K56 acyl mimics. *Prior data on natural acyl-Lys5d; **previous5d S½ was normalized relative to the current S½ of NCPL(unmod).
It had previously been shown that H3-K56 acetylation induces a ~3-fold enhanced affinity for LexA in this assay, correlating with enhanced nucleosome DNA unwrapping with this PTM.5d Here we show that the acetyl-Lys hydrazide mimic at the H3-K56 position increases LexA affinity by 2-fold, closely agreeing with the effect of the natural acetyl-Lys.5d In contrast to the effects of H3-K56 acetyl-Lys mimic, H3-K122 modification by the acetyl-Lys mimic had essentially no influence on nucleosome DNA unwrapping (Supplementary Figure 11). These results are consistent with the expectation that H3 PTMs distal from the nucleosome entry/exit sites do not enhance unwrapping and that the chemical manipulation of H3 per se is not a source of non-specific effects. Moreover, the H3-K56hib and H3-K56ub hydrazide mimics increased LexA binding by 12-fold and 6-fold, respectively (Figure 5B). These findings suggest that both the hydroxyisobutyrylation and ubiquitylation of H3-K56 show a heightened ability to enhance nucleosome DNA unwrapping, perhaps because of their larger size as compared to acetylation. That the significantly bigger ubiquitylation does not perturb unwrapping more than hydroxyisobutyrylation indicates that PTM size per se is not the only factor to influence nucleosome DNA unwrapping at the histone H3-K56 position. Rather, a combination of polarity, hydrogen bonding, and other atomic features are apparently important in mediating the effects of H3-K56 modifications on nucleosome DNA unwrapping.
We also explored the effects of H3-K122 acyl-Lys mimics on nucleosome stability. For these studies, FRET of the intact nucleosome was assessed using DNA labeled with Cy3 at bp 58 and histone H4 tagged at the 21 position (V21C) with Cy5 which can report on the nucleosome dyad symmetry axis. This dyad region shows dense histone-DNA interactions and acetylation of H3-K122 can enhance remodeling by SWI/SNF.23 Here we found that incorporation of the acetyl, hydroxyisobutyryl and ubiquityl hydrazide-Lys modifications each showed similar destabilizing effects of the nucleosome as a function of increasing salt concentration (Figure 6A). Corroborating these findings, the rate of nucleosome denaturation was faster at a fixed ionic strength (Figure 6B). These results suggest that acyl modification of the natural Lys side chain at H3-K122, regardless of bulk of the substitution, is sufficient to destabilize nucleosomes in this region (Figure 6C).
Ubiquityl-Hydrazide Lys mimics as deubiquitinase substrates
The molecular recognition of ubiquityl hydrazide analogs was probed with three purified deubiquitinase (DUB) enzymes: yeast Ubp10, yeast Ubp8, and human OTUB1. Ubp10 and Ubp8 have been implicated in the removal of ubiquitin from Lys123 in histone H2B24 whereas OTUB1 is reported to cleave K48-linked polyubiquitin25 and stabilize the transcription factor FOXM1.
Ubp8 functions as part of a DUB module in the yeast SAGA complex which is a multi-protein complex comprising the proteins, Ubp8, Sgf11, Sus1, and Sgf73.26 An X-ray crystal structure has revealed the architecture of the complex formed by the DUB module with a ubiquitinated nucleosome.27 Like many DUBs, Ubp10, Ubp8, and OTUB1 are Cys hydrolase enzymes in which the active Cys serves as a nucleophile that attacks the isopeptide (amide) carbonyl carbon of the ubiquitin attached to a Lys sidechain in protein substrates. Subsequent hydrolysis of the Cys-Ub thioester affords free ubiquitin and regenerates the unmodified DUB catalyst.
For the initial experiments, we prepared the corresponding diubiquitin substrates linked at the Lys48 position in ubiquitin via the Cys modification strategy described above. The ability of Ubp10, Ubp8/SAGA DUB module, and OTUB1 to cleave the diubiquitin hydrazide analogs was compared with cleavage of K48-linked diubiquitin containing a native linkage. In these experiments, we used fixed reaction times and diubiquitin protein substrate along with various concentrations of DUB. The reaction conditions were based on the specific reactivity of the particular DUB with natural K48-linked diubiquitin. We found that the diubiquitin hydrazide analog was readily cleaved by each of the three DUBs analyzed, suggesting that the diubiquitin hydrazides were recognized in an enzymatically relevant way by the enzymes. Interestingly, Ubp10 cleaved the diubiquitin hydrazide substrate about 10-fold more efficiently than the natural diubiquitin substrate (Figure 7A) and the Ubp8 SAGA DUB module processed diubiquitin hydrazide about 3-fold more efficiently than natural diubiquitin (Figure 7B). By contrast, OTUB1 cleaved diubiquitin hydrazide about 3-fold less efficiently than natural diubiquitin substrate (Figure 7C).
These results indicate that, for these three DUBs, the hydrazide mimic is well-recognized by the enzymes in terms of binding and catalysis. However, there appear to be unique features for each of the enzymes that influence the precise rate of processing. Since cleavage rates on the hydrazide mimic were higher for the two USP class DUBs, Ubp8 and Ubp10, and lower for OTUB1, a member of the OTU class of DUBs, it is possible that unique features near the active site influence reactivity of the hydrazide mimic. We speculate that the increased cleavage observed with Ubp10 and Ubp8 on the diubiquitin hydrazide substrate relative to the natural substrate may be because of the lower pKa of the hydrazine leaving group relative to an amine leaving group, although further mechanistic studies will be needed to evaluate this.
We next explored the well-established Ubp8/SAGA DUB module's enzymatic function by analyzing the H2B-K120-ubiquitylated (H2B-monoUb at K123 in yeast is equivalent to K120 in Xenopus laevis or human) hydrazide nucleosome substrate. Prior elucidation of the Ubp8/SAGA DUB module's structural interactions with an H2B K120-ubiquitylated nucleosome used a histone H2B that contained a K116A mutation that was introduced to simplify H2B-K120Ub synthesis.27 It was formally possible that Lys116 might influence Ubp8/SAGA DUB module recognition of the K120-ubiquitylated nucleosome substrate, which was not investigated previously. We therefore prepared two forms of Lys120-ubiquitin hydrazide histone, H2B containing either Lys116 or Ala116 and then incorporated these modified H2Bs into nucleosomes. Reaction of these modified nucleosomes with Ubp8/SAGA DUB module showed that both substrates were rapidly processed with approximately equal rates (Figure 8). Overall, these studies suggest that Lys116 in ubiquitylated H2B does not make critical contacts for Ubp8/SAGA DUB module binding and turnover.
Figure 8.
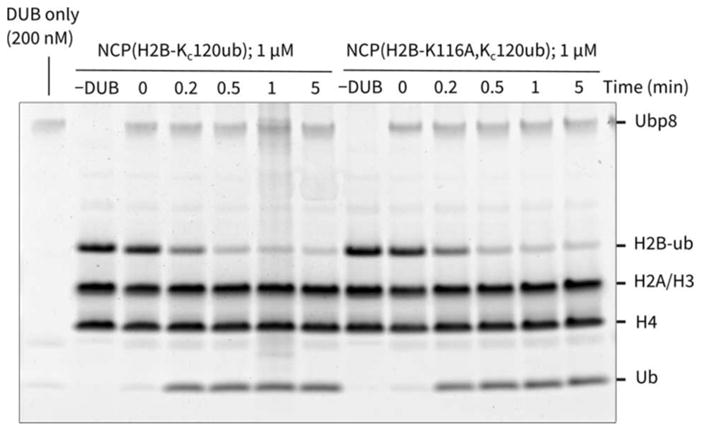
Deubiquitination of nucleosomes bearing H2B-Kc120ub and H2B-K116A-Kc120ub by Ubp8/SAGA DUB module.
CONCLUSION
Here we have developed a new approach to installing acyl-Lys mimics at specific sites in proteins by Cys modification. The chemical steps are relatively simple to execute and can readily be performed on histones and ubiquitin. Like other Cys alkylation approaches, it is necessary to replace natural Cys in a protein of interest to avoid non-targeted mutations. Although we have thus far applied this method to relatively small proteins that can be denatured and refolded, the lack of a requirement for organic solvent suggests that it should be possible to adapt this method to a wide range of larger proteins of interest without the need for denaturation.
The sulfur for methylene substitution and the extra nitrogen in the acyl-Lys mimics used here seem to be tolerated in antibody recognition and in the case of ubiquitin, DUB processing. The latter feature might make them particularly useful mechanistic tools for the study of DUB substrate selectivity and structural interactions. As with all mimics, there will undoubtedly be cases where the differences of these mimics from the natural functionalities will alter biochemical behaviors. Nevertheless, we expect that the chemical ease for generating these mimics will allow them to be investigated in a range of settings to explore the structure and function of acyl-Lys protein modifications.
Supplementary Material
Acknowledgments
We thank the NIGMS GM62437 (P.A.C.), GM095822 (C.W.) and GM121966 (M.G.P.) and FAMRI Foundation (P.A.C.) for financial support.
Footnotes
Notes
The authors declare no competing financial interest.
Preparation of 2-hydroxyisobutyric hydrazide and its installation as a 2-hydroxyisobutyrl-Lys mimic onto a model peptide.
Preparation of ubiquitin hydrazide, K→C mutant H3 and H2B histones, core histones, other proteins used in the assays, and DNA constructs.
Details of immunoblotting experiments.
Histone octamer refolding and nucleosome reconstitutions. Site accessibility and stability assays with nucleosomes. Production and purification of Ubp10, hOTUB1, Ubp8/SAGA DUB module and deubiquitination assays.
Selected HPLC traces, MALDI-TOF, ESI-MS, and the corresponding deconvolution spectra of hydrazide mimics of model peptide, H3, H2B, and diubiquitin.
Full gels of the loading controls corresponding to Figures 3A–C.
LexA—nucleosome binding isotherms comprising a negative control (NCP with H3-Kc122ac).
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Allfrey VG, Mirsky AE. Science. 1964;144(3618):559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]; (b) Li B, Carey M, Workman JL. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]; (c) Cole PA. Nature Chem Biol. 2008;4(10):590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bannister AJ, Kouzarides T. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Lin H, Su X, He B. ACS Chem Biol. 2012;7(6):947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Olsen CA. ChemMedChem. 2014;9(3):434–437. doi: 10.1002/cmdc.201300421. [DOI] [PubMed] [Google Scholar]; (c) Rousseaux S, Khochbin S. Cell J. 2015;17(1):1–6. doi: 10.22074/cellj.2015.506. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Huang H, Lin S, Garcia BA, Zhao Y. Chem Rev. 2015;115(6):2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hirschey MD, Zhao Y. Mol Cell Proteomics. 2015;14(9):2308–315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) PhosphoSitePlus® Online Systems Biology Resource. https://www.phosphosite.org/
- 3.(a) Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, Sabari BR, Deng Z, Allis CD, Ren B, Khochbin S, Zhao Y. Nat Chem Biol. 2014;10(5):365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]; (b) Huang H, Luo Z, Qi S, Huang J, Xu P, Wang X, Gao L, Li F, Wang J, Zhao W, Gu W, Chen Z, Dai L, Dai J, Zhao Y. Cell Res. 2018;28(1):111–125. doi: 10.1038/cr.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Wang YH, Tsay YG, Tan BCM, Lo WY, Lee SC. J Biol Chem. 2003;278(28):25568–576. doi: 10.1074/jbc.M212574200. [DOI] [PubMed] [Google Scholar]; (b) Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. J Biol Chem. 2009;284(35):23312–321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wollebo HS, Bellizzi A, Cossari DH, Salkind J, Safak M, White MK. J Neurovirol. 2016;22(5):615–625. doi: 10.1007/s13365-016-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Kumar KSA, Spasser L, Erlich LA, Bavikar SN, Brik A. Angew Chem Int Ed. 2010;49(48):9126–131. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]; (b) El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Angew Chem Int Ed. 2010;49(52):10149–153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kumar KSA, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A. Angew Chem Int Ed. 2011;50(27):6137–141. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]; (d) Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. J Mol Biol. 2011;408(2):187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Dawson P, Muir T, Clark-Lewis I, Kent S. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]; (b) Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci USA. 1998;95(12):6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311(5762):844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]; (d) McGinty RK, Chatterjee C, Muir TW. Methods Enzymol. 2009;462:225–243. doi: 10.1016/S0076-6879(09)62011-5. [DOI] [PubMed] [Google Scholar]
- 7.(a) Neumann H, Peak-Chew SY, Chin JW. Nat Chem Biol. 2008;4(4):232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]; (b) Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. Angew Chem Int Ed. 2010;49(18):3211–214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]; (c) Gattner MJ, Vrabel M, Carell T. Chem Commun. 2013;49:379–381. doi: 10.1039/c2cc37836a. [DOI] [PubMed] [Google Scholar]; (d) Wang WW, Zeng Y, Wu B, Deiters A, Liu WR. ACS Chem Biol. 2016;11(7):1973–981. doi: 10.1021/acschembio.6b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wang ZA, Kurra Y, Wang X, Zeng Y, Lee YJ, Sharma V, Lin H, Dai SY, Liu WR. Angew Chem Int Ed. 2017;129(6):1665–669. doi: 10.1002/anie.201611415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, Marmorstein R, Cole RN, Khochbin S, Cole PA. J Am Chem Soc. 2010;132(29):9986–987. doi: 10.1021/ja103954u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dancy BCR, Ming SA, Papazyan R, Jelinek CA, Majumdar A, Sun Y, Dancy BM, Drury WJ, III, Cotter RJ, Taverna SD, Cole PA. J Am Chem Soc. 2012;134(11):5138–148. doi: 10.1021/ja209574z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Li F, Allahverdi A, Yang R, Lua GBJ, Zhang X, Cao Y, Korolev N, Nordenskiöld L, Liu CF. Angew Chem Int Ed. 2011;50(41):9611–14. doi: 10.1002/anie.201103754. [DOI] [PubMed] [Google Scholar]; (b) Jing Y, Liu Z, Tian G, Bao X, Ishibashi T, Li XD. Cell Chem Biol. 2018;25(2):166–174. doi: 10.1016/j.chembiol.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 10.(a) Valkevich EM, Guenette RG, Sanchez NA, Chen YC, Ge Y, Strieter ER. J Am Chem Soc. 2012;134(16):6916–919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chalker JM, Lercher L, Rose NR, Schofield CJ, Davis BG. Angew Chem Int Ed. 2012;51(8):1835–839. doi: 10.1002/anie.201106432. [DOI] [PubMed] [Google Scholar]
- 11.Kent SBH. Chem Soc Rev. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 12.(a) Chiang KP, Jensen MS, McGinty RK, Muir TW. ChemBioChem. 2009;10(13):2182–187. doi: 10.1002/cbic.200900238. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Karukurichi KR, Wang L, Uzasci L, Manlandro CM, Wang Q, Cole PA. J Am Chem Soc. 2010;132(14):1222–223. doi: 10.1021/ja909466d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Nguyen UTT, Bittova L, Muller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW. Nat Methods. 2014;11(8):834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Miller TCR, Simon B, Rybin V, Grotsch H, Curtet S, Khochbin S, Carlomagno T, Muller CW. Nat Commun. 2016;7:13855. doi: 10.1038/ncomms13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453(7196):812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fierz B, Kilic S, Hieb AR, Luger K, Muir TW. J Am Chem Soc. 2012;134(48):19548–551. doi: 10.1021/ja308908p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarzer D, Cole PA. Curr Opin Chem Biol. 2005;9(6):561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 15.(a) Le DD, Cortesi AT, Myers SA, Burlingame AL, Fujimori DG. J Am Chem Soc. 2013;135(8):2879–82. doi: 10.1021/ja3108214. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narliker GT, Shokat KM. Cell. 2007;128(5):1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee C, McGinty RK, Fierz B, Muir TW. Nat Chem Biol. 2010;6(4):267–69. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 17.(a) Rösner D, Schneider T, Schneider D, Scheffner M, Marx A. Nat Protoc. 2015;10(10):1594–611. doi: 10.1038/nprot.2015.106. [DOI] [PubMed] [Google Scholar]; (b) Zhao X, Lutz J, Hollmuller E, Scheffner M, Marx A, Stengel F. Angew Chem Int Ed. 2017;56(49):15764–768. doi: 10.1002/anie.201705898. [DOI] [PubMed] [Google Scholar]; (c) Zhang F, Smits AH, van Tilburg GBA, Jansen PWTC, Makowski MM, Ovaa H, Vermeulen M. Mol Cell. 2017;65(5):941–55. doi: 10.1016/j.molcel.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 18.(a) Yang R, Pasunooti KK, Li F, Liu X-W, Liu C-F. J Am Chem Soc. 2009;131(38):13592–593. doi: 10.1021/ja905491p. [DOI] [PubMed] [Google Scholar]; (b) Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. J Am Chem Soc. 2011;133(28):10708–711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Nat Chem Biol. 2010;6(10):750–57. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 19.(a) Galons H, Cave C, Miocque M, Rinjard P, Tran G, Binet P. Eur J Med Chem. 1990;25(9):785–788. [Google Scholar]; (b) Li YT, Liang J, Li JB, Fang GM, Huang Y, Liu L. J Pept Sci. 2014;20(2):102–107. doi: 10.1002/psc.2568. [DOI] [PubMed] [Google Scholar]
- 20.Bos J, Muir TW. J Am Chem Soc. 2018;140(14):4757–4760. doi: 10.1021/jacs.7b13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. Mol Cell. 2009;36(1):153–63. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Proc Natl Acad Sci USA. 2011;108(31):12711–716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) North JA, Shimko JC, Javaid S, Mooney AM, Shoffner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. Nucleic Acids Res. 2012;40(20):10215–227. doi: 10.1093/nar/gks747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee N, North JA, Dechassa ML, Manohar M, Prasad R, Luger K, Ottesen JJ, Poirier MG, Bartholomew B. Mol Cell Biol. 2015;35(23):4083–4092. doi: 10.1128/MCB.00441-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Genes Dev. 2011;25(21):2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. J Mol Biol. 2009;386(4):1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Genes Dev. 2003;17(21):2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Cell. 2010;141(4):606–17. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Science. 2010;328(5981):1025–29. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. Science. 2016;351(6274):725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.