Abstract
Heme is a cofactor that is essential for cellular respiration and for the function of many enzymes. If heme levels become too low within the cell, S. aureus switches from producing energy via respiration to producing energy by fermentation. S. aureus encodes two heme oxygenases, IsdI and IsdG, which cleave the porphyrin heme ring releasing iron for use as a nutrient source. Both isdI and isdG are only expressed under low iron conditions and are regulated by the canonical Ferric Uptake Regulator (Fur). Here we demonstrate that unregulated expression of isdI and isdG within S. aureus, leads to reduced growth under low iron conditions. Additionally, the constitutive expression of these enzymes leads to decreased heme abundance in S. aureus, an increase in the fermentation product lactate, and increased resistance to gentamicin. This work demonstrates that S. aureus has developed tuning mechanisms, such as Fur regulation, to ensure that the cell has sufficient quantities of heme for efficient ATP production through aerobic respiration.
Keywords: Heme oxygenase, IsdG family, Staphylococcus aureus, heme homeostasis, iron
Introduction
Heme is an important small molecule and an essential cofactor for a variety of enzymes (1–4). During cellular respiration, heme populates cytochromes and serves as an electron acceptor in the electron transport chain (2, 5). Heme-dependent respiration is critical for many organisms (6). If heme is unable to populate the cytochromes, either due to genetic inactivation of the cytochromes or a lack of cellular heme, cells are unable to respire and must switch to a fermentative state (7). Fermentation through glycolysis results in the production of 2 ATP molecules, compared to respiration that can generate up to 38 molecules of ATP per molecule of glucose.
Staphylococcus aureus is a Gram-positive coccoid bacterium and is the leading cause of skin and soft tissue infections (8). In order to meet the cellular requirements for heme, S. aureus both biosynthesizes heme and imports heme from the extracellular milieu (9, 10). Heme import is mediated through the high-affinity iron-regulated surface determinant (Isd) heme acquisition system (9, 11). The genes in the isd operons are regulated by the Ferric Uptake Regulator (Fur) (9, 12). Fur dimerizes when iron is present to bind Fur boxes in the promoter regions of target genes and repress transcription. This repression is alleviated under iron deplete conditions, when there is insufficient intracellular iron to allow Fur dimerization (13). Regulation by Fur is widely conserved throughout bacterial species, and Fur regulates a variety of transcripts associated with pathogenesis (14–18).
In addition to being an important enzymatic cofactor, heme can also be used as a source of iron. Vertebrate-associated microorganisms, especially pathogenic bacteria, exploit host heme as a nutrient source. Aerobically, heme degradation is performed by heme oxygenases, while anaerobic bacteria use enzymes that rely on radical catabolism (19). Once heme is imported into the cell through the Isd proteins, heme is used to populate heme-binding proteins or heme is degraded by heme degrading enzymes. S. aureus encodes two such heme degrading enzymes, the heme oxygenases IsdI and IsdG. IsdG and IsdI facilitate the degradation of heme to produce the secondary catabolites staphylobilin and formaldehyde (20, 21). This degraded heme also results in the release of iron, which the bacteria can use to meet their iron requirements (22).
Here we describe work initiated to understand the role of the heme degradation products in the context of S. aureus. Through an RNA-Sequencing experiment comparing a S. aureus strain containing constitutive heme oxygenase activity to one lacking heme degradation, we found a significant increase in a number of transcripts from genes associated with oxygen-independent energy production. This led to the hypothesis that dysregulation of isdI causes aberrant degradation of the intracellular heme pool. Further analysis comparing constitutively and endogenously expressed isdI and isdG showed a significant decrease in intracellular heme levels and heightened fermentation. This work demonstrates the importance of Fur regulation for optimal bacterial growth in low iron conditions.
Results
Expression of heme oxygenases in S. aureus is essential for growth with heme as the sole source of iron
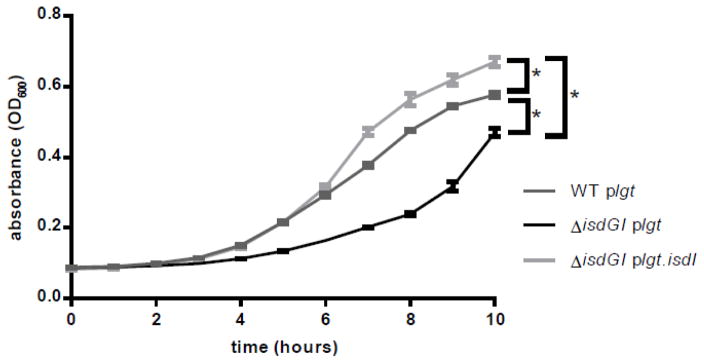
The function of the heme degradation products in S. aureus are unclear. To try to elucidate the roles of these catabolites in the cell, we created two strains of S. aureus; one lacking heme oxygenase activity (ΔisdGI plgt) and one containing the endogenous heme oxygenase isdI added back in trans under the constitutive expression of the promoter for lipoprotein diacylglycerol transferase (lgt) (23) (ΔisdGI plgt.isdI). Removing both isdI and isdG from S. aureus results in a decrease in growth compared to the wildtype strain when grown in the presence of heme as the sole source of iron. Providing isdI in trans complemented the growth defect of ΔisdGI plgt (Fig 1). These data indicate that the expression of IsdI is necessary for growth when heme is the sole source of iron, and provide an experimental condition where heme is actively degraded in the presence of the heme oxygenases but it is not degraded in their absence.
Fig. 1. Expression of isdI is required for growth of S. aureus with heme as the sole source of iron.
Growth of S. aureus strains in Chelex treated RPMI with 0.75 mM EDDHA and 1 μM heme. 2-way ANOVA analysis was applied to the 10 hour time point (p<0.05), error bars represent standard error of the mean (S.E.M.).
Constitutive expression of isdI leads to an increase in expression of transcripts associated with oxygen-independent energy production
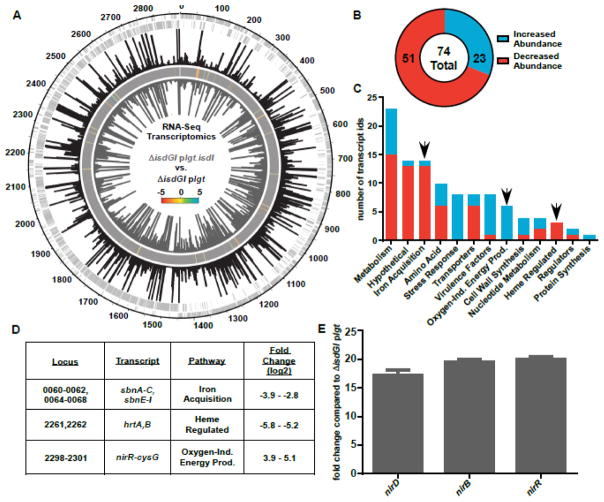
In order to elucidate the role of heme catabolites in S. aureus, ΔisdGI plgt and ΔisdGI plgt.isdI were grown with heme as the sole iron source. Samples were collected for RNA isolation at mid-log phase growth and RNA from these samples was submitted for RNA-Sequencing analysis. In total, the abundance of 74 transcripts were significantly different between ΔisdGI plgt.isdI and ΔisdGI plgt (q≤0.05) (Fig 2A). Of these, 23 were increased in abundance and 51 were decreased in abundance when ΔisdGI plgt.isdI transcripts were compared to ΔisdGI plgt (Fig 2B). As expected, there were significant changes in abundance of transcripts for genes associated with iron acquisition (sbnA-C (NWMN_0060-_0062), sbnE-I (NWMN_0064-_0068)) and genes known to be regulated by heme, such as the heme regulated transporter (hrtAB (NWMN_2261, _2262)) genes, which are regulated by the heme sensing two component system (HssRS) (14, 24) (Fig 2C, D). However, there was also a significant increase in the expression of genes associated with oxygen-independent energy production (Fig 2C, D, Table 1). This result was surprising because heme oxygenases require oxygen to function (25) and the cultures were grown aerobically.
Fig. 2. Constitutive expression of isdI leads to an increase in transcripts associated with non-aerobic growth.
RNA-Sequencing analysis comparing RNA from ΔisdGI plgt.isdI to ΔisdGI plgt. A, RPKM mapping of RNA-Seq data for ΔisdGI plgt (black bars, fourth strip) and ΔisdGI plgt.isdI (grey bars, sixth/inner strip) mapped relative to the chromosomal location (outer strip) for each of the positive strand (second strip) and negative strand (third strip) genes. A heat map (fifth strip) is included showing the log2-fold changes for transcripts with significant q-values. B, Pie chart showing the number of total differentially abundant transcripts, as well as the number with increased (blue) and decreased (red) abundance. C, Pathway analysis was performed for the differentially abundant transcripts with KEGG Pathway. The number of transcript identifications which fall into each pathway bin is shown, as well as whether the transcript has increased (blue) or decreased (red) abundance. D, Fold change values for a subset of transcripts identified in the RNA-Seq analysis from the iron acquisition, heme regulated, and oxygen-independent energy production pathways. E, qRT-PCR analysis of the nitrite reductase operon transcripts confirming their increased abundance in ΔisdGI plgt.isdI compared to ΔisdGI plgt (Student’s T-test, p<0.05, error bars represent S.E.M.).
To confirm the results of the RNA-Seq, quantitative Real Time Polymerase Chain Reaction (qRT-PCR) was performed on transcripts from a subset of the genes linked with oxygen-independent energy production. The nitrite reductase operon contains three genes (nirD (NWMN_2299), nirB (NWMN_2300), and nirR (NWMN_2301) known to be upregulated under anaerobic conditions in various bacteria, including the closely related Staphylococcus carnosus (26–28). NirBD form the nitrite reductase, which acts to reduce nitrate to nitrite. Nitrite can be used as an alternative electron acceptor during growth under anaerobic conditions. While nitrite is likely not being used as the terminal electron acceptor since the cultures are grown aerobically, the reduction of nitrate to nitrite performed by NirBD consumes NADH, and therefor may be involved in recycling reduction equivalents. Analysis by qRT-PCR confirmed that these transcripts had a 17–20 fold increase in transcript abundance in the ΔisdGI plgt.isdI strain compared to ΔisdGI plgt strain.
Due to this increased expression of genes associated with oxygen-independent energy production, we hypothesized that the over expression of isdI under the control of the lgt promoter may lead to unregulated production of the heme oxygenase, unregulated heme degradation, and disruption of heme homeostasis.
Constitutive expression of S. aureus heme oxygenases leads to heme-dependent growth inhibition
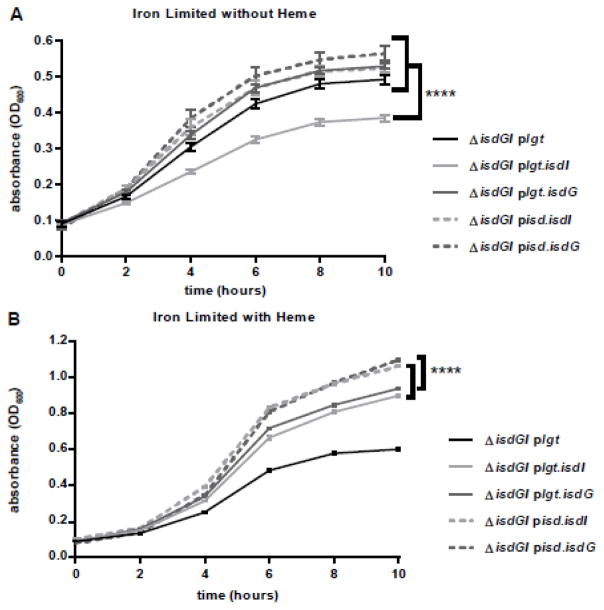
Since the heme oxygenases of S. aureus are regulated by Fur, additional strains were created expressing both of the heme oxygenases, isdI and isdG, on the pOS1 plasmid controlled by either the lgt promoter (plgt) or their endogenous isdI/isdG promoters (pisd). The strains were grown in heme as the sole source of iron. Under these conditions, the strains expressing the heme oxygenases from their native promoters (pisd.isdI and pisd.isdG) grew to slightly higher optical densities than the strains containing the constitutively expressed heme oxygenases (plgt.isdI and plgt.isdG) (Fig 3B). Additionally, when the strains were grown under iron limiting media lacking heme, the strain expressing the constitutively expressed isdI grew significantly worse than all of the other strains (Fig 3A). Unlike the strain constitutively expressing isdI, the strain constitutively expressing isdG did not exhibit reduced growth under these conditions. This is consistent with the fact that IsdG requires heme for protein stability, and in the absence of heme, IsdG is rapidly degraded post-translationally (29). These data indicate that while heme oxygenases are active under iron limited conditions, constitutive isdI expression results in reduced growth.
Fig. 3. Misregulation of isdI expression leads to heme-dependent growth inhibition.
Growth of S. aureus strains in Chelex treated RPMI with 0.75 mM EDDHA either in the absence (A) or presence (B) of 1 μM heme. A, Under iron deplete conditions lacking heme, ΔisdGI plgt.isdI (dark grey) exhibits reduced growth compared to any of the other strains. B, When grown in the iron limited conditions in the presence of heme, the endogenously expressed strains (ΔisdGI pisd.isdI and ΔisdGI pisd.isdG) exhibit enhanced growth as compared to constitutively expressed strains (ΔisdGI plgt.isdI and ΔisdGI plgt.isdG). 2-way ANOVA analysis was applied to the 10 hour time point for each growth curve (p<0.05), error bars represent S.E.M..
Heme levels are decreased in strains with constitutively expressed heme oxygenases
We hypothesized that the decreased growth in the constitutively expressed heme oxygenase-containing strains is due to unregulated heme degradation. This hypothesis predicts that these strains contain lower levels of cellular heme. However, the levels of heme in these strains under the low iron conditions were below the limit of detection of our assays. Therefore, the strains were grown under iron replete conditions. Since the heme oxygenases are expressed from pOS1, a high copy number plasmid, we performed immunoblot analysis to ensure that the enzymes expressed from their endogenous promoters were suppressed by Fur under these iron replete conditions. There is a significant increase in IsdI (2.4 fold) and IsdG (2.5 fold) abundance in ΔisdGI plgt.isdI and ΔisdGI plgt.isdG compared to ΔisdGI pisd.isdI and ΔisdGI pisd.isdG, respectively, suggesting that Fur is actively repressing in these conditions (Fig S1). However, since we did detect protein expression of the heme oxygenases expressed from their endogenous promoters, the amount of Fur in the cell may be insufficient to fully repress transcription.
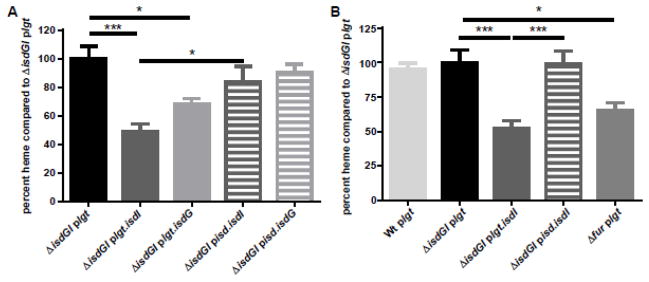
Using the iron replete conditions established above, we performed heme quantification on each of the strains harvested at mid-log phase growth. Both ΔisdGI plgt.isdI and ΔisdGI plgt.isdG contained significantly less heme than ΔisdGI plgt (~32.1–51.2%), as well as ΔisdGI pisd.isdI and ΔisdGI pisd.isdG, respectively (Fig 4A). We hypothesize that we see these differences between the strains containing the two unregulated heme oxygenases because of the post-translational regulation of heme on IsdG. Interestingly, while both of the constitutively expressed heme oxygenases have lower levels of cellular heme than the strain lacking heme oxygenase activity, the strain containing isdG contains more heme than the isdI containing strains. Once IsdG degrades a certain amount of cytoplasmic heme, there is likely not a sufficient amount of heme in the cytoplasm to stabilize the protein leading to its degradation. These fluctuations in IsdG proteins levels in the cell allow for a higher level of total heme in the cell compared to the heme levels in a cell constitutively expressing the stable IsdI protein.
Fig. 4. Heme levels are decreased in strains with constitutive heme oxygenase expression.
Quantification of cellular heme levels of the heme oxygenase strains compared to the strain lacking heme oxygenase activity (ΔisdGI plgt). The ΔisdGI plgt.isdI (dark grey, solid) strains contain significantly less heme than ΔisdGI plgt (black) and ΔisdGI pisd.isdI (dark grey, dashed) strains. Additionally, the ΔisdGI plgt.isdG (light grey, solid) strain contains significantly less heme than ΔisdGI plgt (1-way ANOVA, p<0.05), error bars represent S.E.M..
Additionally, the heme levels in a fur mutant are also significantly less (60.9%) than that of the ΔisdGI plgt, and not significantly different from levels present in ΔisdGI plgt.isdI (Fig 4B). These data indicate that dysregulation of heme oxygenase results in significantly less heme in S. aureus, potentially leading to alterations in cell physiology.
Strains with constitutively expressed heme oxygenases undergo fermentation
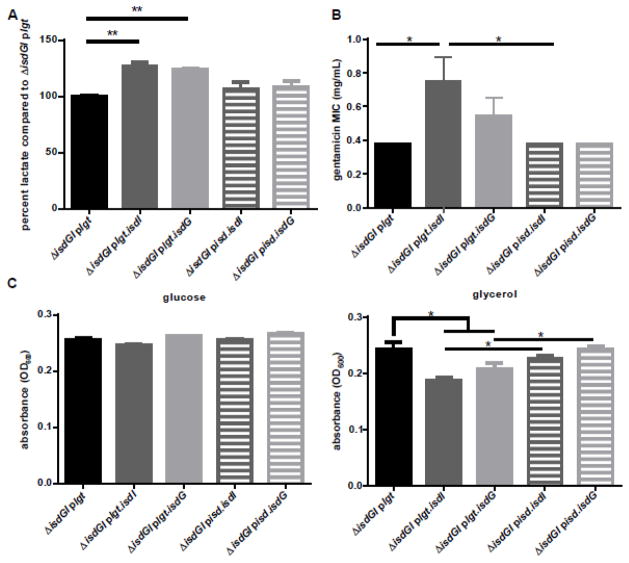
Since cells lacking heme are unable to respire, and there were lower levels of heme in ΔisdGI plgt.isdI and ΔisdGI plgt.isdG, we hypothesized that these strains may be forced to rely on fermentation for energy production. To determine if the strains containing the constitutively expressed heme oxygenases generate energy through fermentation, the fermentation product lactate was measured from the supernatant of cells grown under iron replete conditions. ΔisdGI plgt.isdI and ΔisdGI plgt.isdG produce higher levels of lactate (23.7–26.5% increase) than ΔisdGI plgt, whereas ΔisdGI pisd.isdI and ΔisdGI pisd.isdG do not have significant increases in lactate production (Fig 5A).
Fig. 5. Strains with constitutively expressed heme oxygenase activity undergo fermentation.
A, Supernatant lactate levels were quantified and graphed as percent lactate compared to ΔisdGI plgt. Both ΔisdGI plgt.isdI (dark grey, solid) and ΔisdGI plgt.isdG (light grey, solid) supernatants contain significantly more lactate than the ΔisdGI plgt (black) supernatant (2-way ANOVA, p<0.05), error bars represent S.E.M.. B, Strains were plated onto TSA containing 10 g/mL chloramphenicol, then a gentamicin E-test strip was added. The minimum inhibitory concentration (MIC) was then determined the next day. The gentamicin MIC was significantly greater for ΔisdGI plgt.isdI (dark grey, solid) than either ΔisdGI plgt (black) or ΔisdGI pisd.isdI (dark grey, dashed) (Student’s t-test, p<0.05, error bars represent S.E.M.). C, S. aureus strains were grown in RPMI without glucose containing 22 mM glycerol as the carbon source. Both the ΔisdGI plgt.isdI (dark grey, solid) and ΔisdGI plgt.isdG (light grey, solid) had significantly less growth at 6 hours than ΔisdGI plgt (black) or ΔisdGI pisd.isdI (dark grey, dashed) and ΔisdGI pisd.isdG (light grey, dashed), respectively (1-way ANOVA, p<0.05, error bars represent S.E.M.).
In addition to lactate production, fermenting cells exhibit decreased proton motive force generation (30). Gentamicin requires functional respiration within S. aureus to be bactericidal (30). When exposed to gentamicin, ΔisdGI plgt.isdI had a 2 fold increased minimum inhibitory concentration (MIC) than ΔisdGI plgt and ΔisdGI pisd.isdI (Fig 5B), suggesting constitutive expression of isdI reduces the proton motive force within the cell. However, while we see a slight increase in the gentamicin MIC for the isdGI plgt.isdG strain, this increase in not statistically significant. This may be due to the instability of IsdG once heme levels drop in the cell from IsdG degradation.
If the strains which are constitutively expressing heme oxygenases are less able to respire to produce energy, they would have reduced growth using a carbon source that is unable to be fermented. When grown in the non-fermentable carbon source glycerol, there is a significant decrease in growth of ΔisdGI plgt.isdI and ΔisdGI plgt.isdG strains after 6 hours (Fig 5C, Fig S2). However, when these same strains were grown with glucose as the primary carbon source, there is no decrease in growth (Fig 5C, Fig S2). This indicates that the constitutively expressed heme oxygenase strains grow less well in glycerol because they are not able to ferment. Taken together, these data are consistent with a model where unregulated heme oxygenase activity decreases respiration and increases reliance on fermentation for energy production in S. aureus.
Discussion
Heme is a cofactor for a variety of cellular processes, including cytochromes where it facilitates respiration by acting as an electron acceptor (2, 5, 31). If S. aureus is unable to make or acquire heme, then the cell must depend on fermentation for energy production, which generates less ATP than respiration (32). The production of less energy leads to significantly less growth and to changes in cellular physiology.
In addition to the importance of heme as an enzymatic cofactor, heme also acts as an important iron source. In order to access the iron within heme, S. aureus encodes heme degrading enzymes known as heme oxygenases (33, 34). The two heme oxygenases encoded by S. aureus, IsdG and IsdI, are essential for growth under iron deplete conditions where heme is the sole source of iron (Fig 1). We initially sought to investigate the role of heme degradation products in S. aureus. However, upon performing RNA-Sequencing analysis, we identified that expressing isdI under the constitutively expressed lgt promoter led to an increase in expression of genes linked to oxygen-independent energy production (Fig 2). Since the cultures for these experiments had been grown aerobically, we hypothesized that the overproduction of isdI led to excess heme degradation, resulting in an insufficient amount of heme present to populate the cytochromes required for efficient aerobic respiration. To test this hypothesis, we developed strains of S. aureus expressing each heme oxygenase either under the endogenous promoters (ΔisdGI pisd.) or the constitutively expressed lgt promoter (ΔisdGI plgt.). When these strains were grown under iron limitation with heme as the sole source of iron, the endogenously expressed heme oxygenases grew better than the constitutively expressed heme oxygenases (Fig 3B). We next investigated the effect of heme oxygenase transcriptional regulation on cellular heme abundance and found that the strains constitutively expressing isdI and isdG contained less heme (Fig 4). Additionally, when the fur gene is removed from S. aureus, there is also a significant decrease in cellular heme levels compared to the strain lacking heme oxygenase activity. As a result of this decrease in cellular heme, the strains constitutively expressing the heme oxygenases produce higher levels of the fermentation end-product lactate. Additionally, the constitutively isdI expressing strain is also more resistant to gentamicin, which requires active electron transport for entry into the cell. Finally, the constitutively expressing heme oxygenase strains had a slight growth defect compared to a strain lacking heme oxygenase activity when grown on the non-fermentable carbon source glycerol.
S. aureus strains that are unable to respire, due to lack of either heme or menadione, exhibit a small colony variant (SCV) phenotype (35, 36). These SCVs have increased resistance to aminoglycoside antibiotics that contributes to their ability to cause persistent infections (37, 38). While constitutively expressing heme oxygenases in strains of S. aureus do not have the same colony morphology as SCVs, the dysregulation of the heme oxygenases leads to a significant decrease in cellular heme. The reduced heme levels exhibited by the S. aureus strains constitutively expressing heme oxygenases lower overall fitness. Therefore, connecting heme oxygenase expression to iron availability through Fur may ensure optimal heme levels for bacterial fitness. This is especially important since removing fur leads to a significant decrease in heme levels compared to a strain lacking heme oxygenase activity. Additionally, all functionally identified IsdG family heme oxygenases are regulated by Fur, as are the heme acquisition systems encoded within these organisms. We predict that heme oxygenases are co-expressed with heme acquisition systems to prevent the heme oxygenases from degrading the endogenously biosynthesized heme. We anticipate that this work will be broadly applicable within bacteria that contain Fur-regulated heme oxygenases, since removing fur leads to a decrease in cellular heme. This could have extensive physiological implications on not only respiration, but also on a variety of other heme-associated processes, such as catalase functionality and bacterial nitric oxide synthase.
This work indicates that Fur regulation of the heme oxygenases is necessary to ensure that there is enough cellular heme to allow for respiration and optimal fitness under both iron limiting and iron replete conditions. Because a large number of organisms encode heme oxygenases, this may be a widely conserved paradigm that organisms must balance iron acquisition and heme homeostasis, or control oxygenase expression in the context of iron limitation or heme uptake.
Materials and methods
Bacterial Strains and Growth Conditions
The Staphylococcus aureus strain Newman was used for all experiments (39). The ΔisdGI strain was made previously, as well as the ΔisdGI strain containing plgt.isdI and plgt.isdG (29). The endogenous promoter for isdG was defined as the intergenic region between the isdA and isdCDEFsrtBisdG operons. The endogenous promoter for isdI was defined as the intergenic region between orfX (NWMN_0112) and aldA (NWMN_0113). However, the Fur box for isdI jis contained within the 3′ region of the orfX operon, and this region was also included in the constructed promoter for isdI. Each of these promoter regions were amplified from Newman and were combined with either the gene encoding isdG or isdI with the Fur box and 3′ region of orfX. These amplicons as well as pOS1 plasmid were combined using Gibson cloning. Bacteria were grown overnight at 37°C with shaking at 180 r.p.m. in tryptic soy broth (TSB; BD) and 10 μg/mL chloramphenicol unless otherwise stated.
RNA-Sequencing
The ΔisdGI plgt and ΔisdGI plgt.isdI were grown in biological triplicate in chelex (Sigma) treated Roswell Park Memorial Institute (RPMI) medium (Corning) with 0.75 mM Ethylenediamine-N,N-bis(2-hydroxyphenylacetic acid) (EDDHA: LGC Standards), 10 μg/mL chloramphenicol, and 1 μM porcine hemin (Sigma). EDDHA was resuspended in 100% ethanol to a stock concentration of 100 mM. The cultures were grown at 37°C shaking at 180 r.p.m. to an OD600 of 0.4. At the correct density, a 1:1 solution of Acetone:Ethanol was added at an equal volume to each sample. These samples were stored at −80°C until the RNA was extracted. Cells were resuspended in 750 mL of LETs buffer (1 M LiCl, 0.5 M EDTA, 1 M Tris pH 7.4) and added to tubes holding lysing matrix B (MP Biomedicals). A FastPrep-24 (MP Biomedicals) bead beater was used to lyse cells by bead beating for 45 seconds at 6 m/s. Samples were then heated at 55°C for 5 min then centrifuged for 10 min. The upper phase was transferred to a fresh tube and 1 mL TRIzol (Thermo Scientific) was added. A 200 uL aliquot of chloroform was added to each tube and vortexed. The mixed samples were then centrifuged for 15 minutes to separate the aqueous and organic layers. The aqueous (upper) layer was then aliquoted into a fresh tube. The RNA was precipitated by the addition of 1 mL of isopropyl alcohol and incubated for 10 min at room temperature. Samples were then centrifuged for 10 min and the supernatant was removed. The RNA was washed with 200 μL of 70% ethanol. The RNA pellet was dried for 1 minute, then resuspended in 100 μL of water. DNA contamination was eliminated through the addition of 8 μL RQ1 DNase, 12 μL of 10x RQ1 buffer, and 2 μL RNase inhibitor (Promega) to the purified RNA. After DNase treatment for 2 hours, the samples were further purified using the RNeasy miniprep RNA cleanup kit (Qiagen). The RNA concentration was determined by using the Synergy 2 with Gen 5 2.1 software (BioTek).
Library creation and RNA-Sequencing was performed by Vanderbilt Technologies for Advanced Genomics Core Facility (VANTAGE) similarly to previous descriptions (40). Briefly, 1.5 μg of RNA from each sample was submitted and an Agilent Bioanalyzer Nano RNA chip was used to determine the quality of the RNA. A Ribo-Zero kit was then used to remove ribosomal RNA by following the manufacturer’s protocol. After cDNA creation, samples were pooled for multiplexing on a flow cell then loaded on a Illumina HiSeq 2500 for a 50-bp paired-end run. Raw reads were processed for FASTQ conversion, and only reads passing the pass filter were used for further analysis.
RNA-Sequencing Analysis
Rockhopper was used to analyze RNA-seq data, including reads alignment, quantification of transcript abundance, identification of differential expression and characterization of operon structures (41). The genome alignment figure (Fig 2A) was created in Circos. The RPKM values were set to a max of 2000 being shown. The heat map shows the log2-fold changes ranging from −5 to 5. The pathway analysis was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) Mapper to search the KEGG pathway databases of annotated S. aureus genes. Additional annotations were made through literature searches.
qRT-PCR
Samples were grown and RNA was extracted as described for the RNA-Seq experiment. cDNA was synthesized and qRT-PCR was performed as previously described (40). The assay was repeated to test three biological replicates in triplicate.
Growth curves
S. aureus Newman strains were grown in RPMI with 1% Casamino acids, 0.5 mM EDDHA, and 10 μg/mL chloramphenicol. Overnight cultures were grown in biological triplicate diluted 1:50 in chelex treated RPMI with 100 μM CaCl2, 25 μM ZnCl2, 1 mM MgCl2, 25 μM MnCl2, 1% Casamino acids, 0.75 mM EDDHA, and 10 μg/mL chloramphenicol, either with or without 1 μM hemin. Growth curves were setup to a total volume of 200 μL 96 well flat bottom plates. The OD600 was monitored every 1 to 2 hours for up to 16 hours. For the glycerol growth curve, Newman strains were grown in biological triplicate overnight as above. Cultures were diluted 1:50 in RPMI without glucose with 22 mM glycerol and 10 μg/mL chloramphenicol. The assay was repeated to test three biological replicates in triplicate. For the glucose growth curve, Newman strains were grown in biological triplicate overnight as above. Cultures were diluted 1:50 in RPMI without glucose with 11 mM glucose and 10 μg/mL chloramphenicol. The assay was repeated to test three biological replicates in triplicate. Each of the growth curves are representative of three experiments.
Heme quantification
S. aureus Newman strains were grown overnight in TSB. Cultures were diluted 1:200 in TSB and grown aerobically at 37°C while shaking at 180 r.p.m. to an OD600 of 0.4 – 0.45. Heme was quantified as previously described (42). Briefly, the pellets were resuspended in 20 mM potassium phosphate containing 20 μL of lysostaphin. Samples were incubated at 37°C for 30 minutes, then lysed by sonication. Unbroken protoplasts were removed by centrifugation, the protein concentration of each sample was determined using a Pierce BCA Protein Assay Kit. A 450 μL aliquot of each sample was transferred to a cuvette, then heme was extracted by adding 0.2 M NaOH and 40% pyridine with 200 μM potassium ferricyanide in 450 μL. The absorbance was measured from 540–590 nm. Then 10 μL of 0.5 M DTT was added, then the absorbance was again measured from 540–590 nm. The heme quantity was then calculated using the coefficient of 324 mM−1 cm−1.
Lactate assay
S. aureus strains were grown to mid-log phase (OD600=0.400) in 20 mL TSB with 10 μg/mL chloramphenicol. Samples were centrifuged and the supernatant was collected. L-Lactate levels were determined using the r-Biopharm D-Lactic acid/L-Lactic acid kit. 13.4 μL of supernatant was added to 134 μL of glycylglycine buffer, 26.8 μL nicotinamide-adenine dinucleotide (NAD), and 2.68 μL of glutamate-pyruvate transaminase suspension to a final volume of 298 μL in a 96 well plate. These samples were incubated for 5 min then the absorbance at 340 nm was read (A1). Then 2.68 μL of L-lactacte dehydrogenase solution was added. The samples were then mixed and incubated for 30 min then the absorbance at 340 nm was read a second time (A2). The concentration of L-lactic acid was determined by subtracting A1 from A2 for each samples then multiplying the AΔ by the final volume times the molecular weight of L-lactic acid, then dividing by the extinction coefficient of NADH at 340 nm.
Gentamicin sensitivity assay
S. aureus strains were grown overnight in 5 mL TSB with 10 μg/mL chloramphenicol. 200 μL of overnight culture were plated onto Tryptic Soy Agar (TSA) plates containing 10 μg/mL chloramphenicol. One gentamicin Etest strip (Biomerieux) was applied to each plate. The plates were incubated for ~16 hours at 37°C, then the minimum inhibitory concentration was determined by the concentration at which there was no zone of clearing. Immunoblot-Cultures of S. aureus were grown in TSB to a final OD600 of 0.4 and pelleted by centrifugation. Pellets were resuspended in 100 mM Tris pH 7.0, 500 mM sucrose, 10 mM MgCl2 with 20 μL of lysostaphin and were incubated at 37°C for 1 hour. The samples were then pelleted to remove the cell wall, and resuspended in 500 μL phosphate buffered saline with proteinase inhibitor. Samples were sonicated for 10 seconds. The cell wall was removed by ultracentrifugation at 180,000 r.p.m. for 1 hour. A Pierce Bicinchoninic acid Protein Assay Kit was used to determine the protein concentration in the cytoplasmic fraction. 40 μg of total protein were analyzed by 12% SDS-PAGE. Samples were run in biological triplicate. Anti-IsdG and –IsdI sera was used as previously described (29). The raw intensities of the protein bands were quantified using ImageJ and expressed in arbitrary units.
Acknowledgments
We thank members of the Skaar laboratory for critical evaluation of the manuscript. This work was supported by grants from the National Institutes of Health, R01AI069233 (EPS), and T32GM008554-21 (LJL).
Footnotes
Author Contributions
Conception or design of study: LJL, AJF, EPS. Acquisition, analysis, or interpretation of data: LJL, AJF, AW. Writing of the manuscript: LJL, EPS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.George P. A comparison of the decomposition of hydrogen peroxide by catalase, ferrous and ferric ions, haemin and ferrous phthalocyanine. Biochem J. 1948;43:287–295. doi: 10.1042/bj0430287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison M, Stotz E. Partition chromatography of hemins. Separation of the prosthetic groups of cytochromes a and a3. J Biol Chem. 1954;213:373–8. [PubMed] [Google Scholar]
- 3.Maehly AC. Splitting of horseradish peroxidase into prosthetic group and protein as a means of studying the linkages between hemin and protein. Biochim Biophys Acta. 1952;8:1–17. doi: 10.1016/0006-3002(52)90002-4. [DOI] [PubMed] [Google Scholar]
- 4.Igo RP, Mackler B, Duncan H. Liver aldehyde oxidase: The nature of hematin component. Arch Biochem Biophys. 1961;93:435–9. doi: 10.1016/0003-9861(61)90290-9. [DOI] [PubMed] [Google Scholar]
- 5.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. MBio. 2013:4. doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musser SM, Chan SI. Evolution of the Cytochrome c Oxidase Proton Pump. Journal of Molecular Evolution. 1998;46:508–520. doi: 10.1007/pl00006332. [DOI] [PubMed] [Google Scholar]
- 7.Von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Gotz F. A Site-Directed Staphylococcus aureus hemB Mutant is a Small-Colony Variant Which Persists Intracellularly. J Bacteriol. 1997;179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK Investigators ftABCsAM. Invasive Methicillan-Resistant Staphylococcus aureus Infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakis DM, Schneewind O. Passage of Heme-Iron Across the Envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 10.Tien W, White DC. Linear Sequential Arrangement of Gene for the Biosynthetic Pathway of Protoheme in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1968;61:1392–1398. doi: 10.1073/pnas.61.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–9. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong A, Singh VK, Cabrera G, Jayaswal RK. Molecular characterization of the ferric-uptake regulator, Fur, from Staphlyococcus aureus. Microbio. 2000;146:659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]
- 13.Bagg A, Neilands JB. Ferric Uptake Regulation Protein Acts as a Repressor, Employing Iron(II) as a Cofactor To Bind the Operator of an Iron Transport Operon in Escherichia coli. Biochem J. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 14.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, Friedman DB, Heinrichs DE, Dunman PM, Skaar EP. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun. 2010;78:1618–28. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litwin CM, Boyko SA, Calderwood SB. Cloning, Sequencing, and Transcriptional Regulation of the Vibrio cholerae fur Gene. J Bacteriol. 1992;174:1897–1903. doi: 10.1128/jb.174.6.1897-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsolis RM, Baumler AJ, Stojiljkovic I, Heffron F. Fur Regulon of Salmonella typhimurium: Identification of New Iron-Regulated Genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanui CK, Shyntum DY, Priem SL, Theron J, Moleleki LN. Influence of the ferric uptake regulator (Fur) protein on pathogenicity in Pectobacterium carotovorum subsp. brasiliense. PLoS One. 2017;12:e0177647. doi: 10.1371/journal.pone.0177647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beall BW, Sanden GN. Cloning and Initial Characterization of the Bordetella pertussus fur Gene. Curr Microbiol. 1995;30:223–226. doi: 10.1007/BF00293637. [DOI] [PubMed] [Google Scholar]
- 19.LaMattina J, Nix D, Lanzilotta W. Radical new paradigm for heme degradation in Escherichia coli 0157:H7. Proc Natl Acad Sci U S A. 2016;113:12138–12143. doi: 10.1073/pnas.1603209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, Wright DW, Bachmann BO, Murphy ME, Skaar EP. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–38. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui T, Nambu S, Ono Y, Goulding CW, Tsumoto K, Ikeda-Saito M. Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry. 2013;52:3025–7. doi: 10.1021/bi400382p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-Source Preference of Staphylococcus aureus Infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 23.Selvan AT, Sankaran K. Localization and characterization of prolipoprotein diacylglyceryl transferase (Lgt) critical in bacterial lipoprotein biosynthesis. Biochimie. 2008;90:1647–55. doi: 10.1016/j.biochi.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–19. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streit BR, Kant R, Tokmina-Lukaszewska M, Celis AI, Machovina MM, Skaar EP, Bothner B, DuBois JL. Time-resolved Studies of IsdG Protein Identify Molecular Signposts along the Non-canonical Heme Oxygenase Pathway. J Biol Chem. 2016;291:862–71. doi: 10.1074/jbc.M115.666560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neubauer H, Pantel I, Gotz F. Molecular characterization of the Nitrite-Reducing System of Staphylococcus carnosus. J Bacteriol. 1998;181:1481–1488. doi: 10.1128/jb.181.5.1481-1488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayaraman PS, Peakman TC, Busby SJW, Quincey RV, Cole JA. Location and Sequence of the Promoter of the Gene for the NADH-dependent Nitrite Reductase of Escherichia coli and its Regulation by Oxygen, the Fnr Protein and Nitrite. J Mol Biol. 1987;196:781–788. doi: 10.1016/0022-2836(87)90404-9. [DOI] [PubMed] [Google Scholar]
- 28.Goldman BS, Roth JR. Genetic Structure and Regulation of the cysG Gene in Salmonella typhimurium. J Bacteriol. 1992;175:1457–1466. doi: 10.1128/jb.175.5.1457-1466.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–15. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryan LE, Kwan S. Roles of Ribosomal Binding, Membrane Potential, and Electron Transport in Bacterial Uptake of Streptomycin and Gentamicin. Antimicrob Agents Chemother. 1983;23:835–845. doi: 10.1128/aac.23.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurt E, Hauska G. A Cytochrome f/b6 Complex of Five Polypeptides with Plastoquinol-Plastocyanin-Oxidoreductase Activity from Spinach Chloroplasts. Eur J Biochem. 1981;117:591–599. doi: 10.1111/j.1432-1033.1981.tb06379.x. [DOI] [PubMed] [Google Scholar]
- 32.Jurtshuk P., Jr . Bacterial Metabolism. 4. University of Texas Medical Branch at Galveston; Galveston (TX): 1996. [PubMed] [Google Scholar]
- 33.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–43. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 34.Wu R, Skaar EP, Zhang R, Joachimiak G, Gornicki P, Schneewind O, Joachimiak A. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–6. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates D, von Eiff C, McNamara P, Peters G, Yeaman M, Bayer A, Proctor R. Staphylococcus aureus menD and hemB Mutants Are as Infective as the Parent Strains, but the Menadione Biosynthetic Mutant Persists within the Kidney. JID. 2003;187:1654–1661. doi: 10.1086/374642. [DOI] [PubMed] [Google Scholar]
- 36.von Eiff C, McNamara P, Becker K, Bates D, Lei XH, Ziman M, Bochner BR, Peters G, Proctor RA. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J Bacteriol. 2006;188:687–93. doi: 10.1128/JB.188.2.687-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musher DM, Baughn RE, Merrell GE. Selection of Small-Colony Variants of Enterobacteriaceae by in vitro Exposure to Aminoglycosides: Pathogenecity for Experimental Animals. JID. 1979;140:209–214. doi: 10.1093/infdis/140.2.209. [DOI] [PubMed] [Google Scholar]
- 38.Kahl B, Herrmann M, Everding AS, Koch HG, Becker K, Harms E, Proctor RA, Peters G. Persistent Infection with Small Colony Variant Strains of Staphylococcus aureus in Pateints with Cystic Fibrosis. JID. 1998;177:1023–1029. doi: 10.1086/515238. [DOI] [PubMed] [Google Scholar]
- 39.Duthie ES, Lorenz LL. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 40.Mortensen BL, Rathi S, Chazin WJ, Skaar EP. Acinetobacter baumannii Response to Host-Mediated Zinc Limitation Requires the Transcriptional Regulator Zur. J Bacteriol. 2014;196:2616–2626. doi: 10.1128/JB.01650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mike LA, Dutter BF, Stauff DL, Moore JL, Vitko NP, Aranmolate O, Kehl-Fie TE, Sullivan S, Reid PR, DuBois JL, Richardson AR, Caprioli RM, Sulikowski GA, Skaar EP. Activation of heme biosynthesis by a small molecule that is toxic to fermenting Staphylococcus aureus. Proc Natl Acad Sci U S A. 2013;110:8206–8211. doi: 10.1073/pnas.1303674110. [DOI] [PMC free article] [PubMed] [Google Scholar]