Abstract
Diabetes leads to markedly accelerated rates of many associated macrovascular complications like hypertension and atherosclerosis, and microvascular complications like nephropathy and retinopathy. High glucose, the hallmark of diabetes, drives changes in vascular and inflammatory cells that promote the development of these complications. Understanding the molecular processes involved in the development of diabetes and its debilitating complications can lead to much needed newer clinical therapies. Recently, long-noncoding RNAs (lncRNAs) have been shown to be important in the biology of vascular cells and there is growing evidence that lncRNAs are also involved in the cell biology relevant to diabetic vascular complications. In this review, we provide an overview of lncRNAs that function in vascular cells, and those that have been linked to diabetic complications.
Keywords: Diabetic vascular complications, lncRNAs, endothelial, macrophage, vascular smooth muscle cells
Graphical Abstract
INTRODUCTION
Diabetes is a metabolic disorder defined by hyperglycemia that currently affects 400 million patients worldwide [1]. Diabetes results in numerous vascular complications such as cardiovascular diseases, retinopathy, neuropathy and nephropathy leading to the damage of corresponding target organs [1–5]. The chronic hyperglycemic state in diabetes causes vascular abnormalities of both microvessels and macrovessels characterized by microangiopathy and macroangiopathy [2–5]. Irreversible glycation of proteins, increased oxidative stress and inflammation, as well as endothelial and smooth muscle dysfunction are considered to be the main factors for vascular damage [3–5]. These long-term complications result in high mortality rates of diabetic patients, and approaches to treat vascular complications have been shown to reduce this burden [1, 2].
The mammalian genome is widely transcribed by RNA pol II and produces large number of long and small non-coding RNAs (e.g. long non-coding RNAs (lncRNAs), microRNAs (miRNAs), tRNA fragment RNAs (tRFs)) along with protein coding mRNAs [6, 7]. LncRNAs are defined as transcripts with more than 200 nucleotides. These transcripts can possess structural similarity to mRNAs, but have functionally distinct features from mRNAs such as cis-regulatory capacity and absence of an open reading frame [8]. Increasing evidence shows that lncRNAs are not mere transcriptional noise, but can be involved in various cellular mechanisms such as gene regulation ranging from transcription and splicing to translation and providing structural integrity to a cell [9]. LncRNA expression patterns are also associated with diverse physiologic processes including pluripotency of stem cells, differentiation, and organismal development [10]. Many lncRNAs have now been shown to be dysregulated in human diseases, and furthermore, many genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in lncRNA loci implicating their role in multiple diseases, including cardiometabolic diseases [11, 12]. One of the best examples is the ANRIL (antisense noncoding RNA in the INK4 locus) lncRNA locus, which is associated with coronary disease and also type 2 diabetes [13–15]. The role of ANRIL in disease is related to its role in regulating the entire p15/CDKN2B-p16/CDKN2A locus, impacting both proliferation and senescence of cells [16]. LncRNAs have also been mapped to diabetes susceptibility loci and it is likely there are also additional lncRNA loci that are associated with diabetic vascular complications [17, 18].
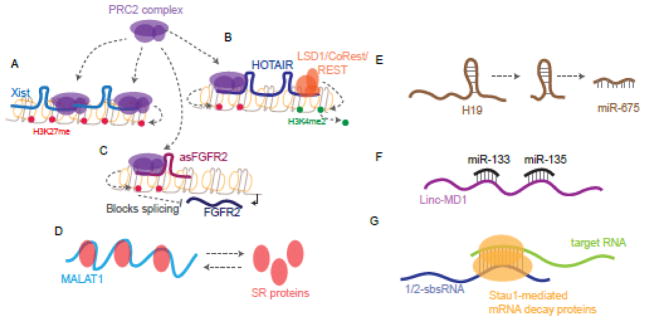
LncRNAs have many functional roles in the cell [9] (Figure 1). In the nucleus, lncRNAs can impact gene expression in a number of ways that include epigenetic mechanisms. At the chromatin level, lncRNAs, including H19, which regulates embryonic development and growth, and Xist, which impacts X-inactivation in females, act through binding to the repressive chromatin modifier, polycomb repressive complex 2 (PRC2) and regulate the activity of the complex at target genomic locations [19–24]. This affinity for the PRC2 complex is not unique to these two lncRNAs as additional lncRNAs, which are involved in a myriad of biological processes, such as HOTAIR, RepA, Kcnq1ot1, and NEAT1 are also shown to interact with PRC2 complexes indicating that regulation of this complex is a common function of a number of lncRNAs [25]. Recent sequence composition analysis of these PRC2-binding lncRNAs shows that they possess distinct, highly conserved sequence features [26]. Other chromatin modifying complexes can also be regulated by lncRNAs. For example, HOTAIR, a regulator of the HOXC locus, acts as a modular scaffold for both PRC2 complex (which affects H3K27me3 levels) and lysine-specific demethylase-1A/REST corepressor/RE-1 silencing transcription factor (LSD1/CoREST/REST) complex (which affects H3K4 methylation levels). Therefore, HOTAIR impacts both lysine 27 methylation and lysine 4 demethylation of histone H3 at target sites [27]. Clearly, these examples show how lncRNAs can be involved intimately with chromatin to impact levels of gene expression through modulating the transcriptional machinery. LncRNAs have also been implicated in regulating co-transcriptional processes including alternative splicing of pre-mRNAs. The lncRNA MALAT1, localized to nuclear speckles, controls the levels and distribution of serine/arginine (SR) splicing factors [28]. Additionally, lncRNAs can directly impact alternative splicing at their transcribed locus as shown for the splicing of FGFR2 transcripts [29]. Studies have also indicated that lncRNAs have the potential to function as host genes for microRNAs (miRNAs) and other small RNAs [9]. For example, H19, one of the conserved lncRNAs involved in genomic imprinting also functions as a primary miRNA precursor for miR-675 that targets insulin-like growth factor 1 receptor (Igf1r) gene [30, 31]. Beyond impacting transcription, lncRNAs are also known to sequester miRNAs due to the presence of miRNA response element (MREs). This function impacts the levels of mRNAs targeted by the miRNAs. Such lncRNAs are referred to as competing endogenous RNAs (ceRNAs) [32]. For example, the muscle specific lncRNA, Linc-MD1 is a ceRNA that sequesters miR-133 and miR-135 and regulates the expression of transcription factors MAML1 and MEF2C respectively to influence muscle differentiation [33]. In addition to impacting miRNA binding to target sites, lncRNAs themselves can directly interact with mRNAs. The lncRNA, 1/2-sbsRNA (half STAU1-binding site RNA), binds to target mRNA 3′UTRs which results in mRNA degradation through the Staufen 1(STAU1)-mediated messenger RNA decay (SMD) pathway [34]. TINCR lncRNA, which controls somatic tissue differentiation, like 1/2-sbsRNA, can also bind to target mRNAs which are also recognized by the SMD pathway [35]. Thus, another common theme of post-transcriptional regulation of gene expression by lncRNAs is through regulation of mRNA stability in the cytoplasm. These examples (Figure 1) demonstrate that lncRNAs can have effects in both nuclear and cytosolic compartments that could be dictated by their subcellular localization.
Figure 1. Examples of molecular functions of lncRNAs.
A) Xist recruits PRC2 complex to the X chromosome resulting in methylation of H3K27. B) HOTAIR binds to both PRC2 complex and LSD1/CoRest/REST complex modularly to promote methylation of H3K27 and demethylation of H3K4. C) Methylation of H3K27 by PRC2 recruited by asFGFR2 affects the splicing of FGFR2 transcripts transcribed from the opposite strand. D) MALAT1 binds to SR proteins which impact its function as splicing factors in the nucleus. E) H19 is a host transcript for miR-675. F) Linc-MD1 binds to miR-133 and miR-135, affecting their impact on target mRNAs. G) 1/2-sbsRNA binds to mRNAs promoting Staufen1-mediated mRNA decay of the duplexed RNAs.
LncRNAs are increasingly being recognized as important players in the development of diabetes as well as in diabetic complications. Transcriptome studies on pancreatic β cells revealed that several lncRNAs are aberrantly expressed in Type 2 diabetes and GWAS have associated genetic variants in lncRNA loci with Type1 Diabetes (T1D) [17, 18]. With the increased recognition of lncRNAs associated with diabetes, there is heightened interest in the function of lncRNAs in diabetic complications. Here in this review, we focus on lncRNAs associated with diabetes vascular complications with a major focus on vascular smooth muscle cell, endothelial cell and monocyte/macrophage dysfunctions (For a review on lncRNAs related to vessel wall biology, we refer the reader to [36]).
LncRNAs in vascular cells
Vascular cells including endothelial cells, vascular smooth muscle cells, and pericytes, have all been shown to express lncRNAs linked to vascular diseases. (For a full review please see [37, 38]). In vascular endothelial cells, LOC100129973 lncRNA has been shown to prevent apoptosis, a process important in the development of cardiovascular disease. Specifically, LOC100129973 binds two miRNAs, miR-4707-5p and miR-4767, thus leading to the upregulation of the miRNA targets: API5 and BCL2L12 mRNAs, both important for inhibiting apoptosis [39]. Interestingly, the targeting of miR-4767 may not be unique to LOC100129973, as Linc00152, in human umbilical vein vascular endothelial cells has also been show to bind this miRNA under pro-inflammatory conditions induced by oxidized low density lipoprotein (oxLDL) [40]. These data suggest this lncRNA is important for preventing apoptosis of endothelial cells in cardiovascular disease setting. LncRNAs can also impact other processes related to endothelial cell function. For example, PUNISHER lncRNA impacts vessel maturation as well as low-density lipoprotein uptake, all important for endothelial cell function [41]. In vascular smooth muscle cells (VSMC), a number of lncRNAs have been implicated in normal processes as well as in disease states [42]. For example, in human smooth muscle cells, SENCR lncRNAs has been shown to be involved in the regulation of the important transcriptional regulator MYOCD as well as in the regulation of genes responsible for contractile phenotype[43]. Angiotensin II (Ang II) has several pathological effects in VSMCs related to hypertension and atherosclerosis. Using a combination of transcriptomic and epigenomic sequencing (RNA-seq and ChIP-seq) in rat VSMCs, it was observed that Ang II drives the upregulation of a number of novel lncRNAs, including Lnc-Ang362. This lncRNA was found to have growth promoting effects in VSMC [42]. In the diseased state, SMILR lncRNA has been observed to be induced by mitogenic stimuli in VSMCs and increased in atherosclerotic plaques from human patients, suggesting its potential role in VSMC dysfunction in cardiovascular disease [44]. Correspondingly, knockdown of SMILR leads to decreased VSMC proliferation in line with its potential role in atherosclerosis [44]. In human pericytes, HypERlnc is expressed and low expression of HypERlnc leads to decreased cell viability and proliferation and ultimately pericyte dedifferentiation [45]. Interestingly, this decrease in HypERlnc is observed in patients with heart failure and those with idiopathic pulmonary arterial hypertension, suggesting HypERlnc has a role in promoting human cardiopulmonary disease [45]. Further studies on the mechanisms of action of these vascular lncRNAs can lead to approaches to enhance or curb their effects and thereby modulate vascular disease progression.
LncRNAs in diabetic vascular complications
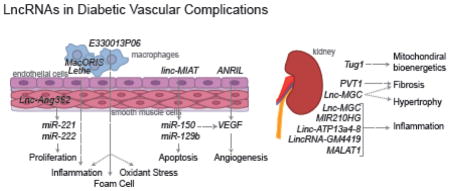
Vascular disease is a known complication of diabetes. In recent years, there have been data indicating that lncRNAs are involved including diabetes associated nephropathy, retinopathy, hypertension, and atherosclerosis [46].
Under high glucose (HG) conditions mimicking the diabetic state, a number of lncRNAs are dysregulated in vascular-related cells. In human retinal endothelial cells, ANRIL lncRNA is upregulated by HG treatment leading to the upregulation of VEGF, and major mitogen for endothelial cells involved in microvascular complications like retinopathy [47]. Concomitantly, loss of ANRIL expression through siRNA-targeted knockdown of the lncRNA attentuates HG induced upregulation of VEGF. The role of ANRIL in endothelial cells is not restricted to retinal endothelial cells, as this lncRNA has also been shown to be involved in a number of other vascular diseases including atherosclerosis and hypertension [48]. ANRIL can also promote angiogenesis through VEGF by activing the NF-κB pathway in diabetes [49]. Impacting the VEGF pathway appears to be a common role of lncRNAs in the retinas. For example, HG-stimulated retinal endothelial cells have elevated expression of lnc-MIAT lncRNA, which act as a ceRNA for miR-150 which targets VEGF [50, 51]. Lnc-MIAT can furthermore act to suppress miR-29b to promote apoptosis. Additional lncRNAs are also implicated in microvascular dysfunction in retinas in diabetic mouse models. The lncRNA MEG3 has been shown to be inhibited by HG and oxidative stress in streptozotocin (STZ)-induced diabetic mice [52]. These data suggest that MEG3 may have functions in human retinas in diabetes, but further studies are needed.
As discussed above, lncRNAs can have physiological and pathophysiological effects in VSMCs. Additionally, under diabetic conditions, angiotensin II (Ang II) is often elevated, which promotes hypertension and atherosclerosis. Notably, as indicated earlier, treatment of rat VSMCs with Ang II upregulates several novel lncRNAs, including Lnc-Ang362 [42]. This lncRNA functions as a host transcript for two miRNAs, miR-221 and miR-222, which cause proliferation of VSMC. Knockdown of Lnc-Ang362 reduces not only Lnc-Ang362, but also miR-221 and miR-222 and VSMC proliferation, suggesting that Ang II upregulation of Lnc-Ang362 drives cell proliferation through the upregulation of the two miRNAs. More recently it was found that key Ang II induced lncRNAs can overlap Ang II regulated enhancers [53]. CRISPR-Cas9 editing of the lncRNA overlapping enhancer regions could downregulate the expression of Ang II regulated nearby genes in VSMC [53], demonstrating functional crosstalk between the two epigenetic layers. Thus, the dysregulation of Lnc-Ang362 as well as other as yet un-investigated lncRNAs can play significant roles in Ang II effects in VSMC, including hypertension and atherosclerosis.
Inflammatory and immune cells also play a role in diabetic vascular complications because enhanced inflammation with monocyte/macrophage infiltration into target organs is a common feature of most complications. Monocytes infiltrate the vasculature, and subsequently differentiate into macrophages which take up lipid to form foam cells and promote hyper-inflammatory state which drives the development of atherosclerosis. Many monocyte/macrophage lncRNAs are shown to have pro- or anti-inflammatory functions in monocyte/macrophages that are not discussed here, but much less is known under diabetic conditions. Transcriptomic profiling (RNA-Seq) of bone marrow macrophages derived from obese, insulin resistant type 2 diabetic mouse model (Leprdb/db), showed elevated expression of lncRNAs, including E330013P06 relative to control non-diabetic Leprdb/+ mice [54]. This lncRNA was also found to be upregulated in macrophages from mice fed a high fat diet, as well as in monocytes obtained from human type 2 diabetic subjects, the latter demonstrating conservation in humans. Overexpression of this lncRNA in cultured macrophages enhances the response of macrophages to inflammatory signals and also promotes foam cell formation. These data suggest that lncRNAs are also involved in the aberrant response of macrophages to the hyperglycemic environment and promote the inflammatory state often associated with diabetes in the vascular system. More recently, another lncRNA named Lethe, has been reported to have a direct impact on inflammatory pathways in the mouse genome. Lethe binds to NF-κB, a critical transcription factor regulating inflammatory genes [55]. Furthermore, under HG conditions, Lethe expression is decreased which coincides with increase reactive oxygen species (ROS) [56]. Overexpression of Lethe reduces ROS production, though the mechanism or direct impact of this in vivo is unclear. Interestingly, the human genome does not appear to have an ortholog of Lethe, but it is possible another lncRNA functions to bind NF-κB in a similar fashion. Recently, deep sequencing of human macrophages identified many lncRNAs that are associated with cardiometabolic diseases [57]. Among those, almost a hundred lncRNAs are differentially expressed during macrophage activation, suggesting they have roles in regulating inflammatory functions of macrophages. Interestingly, the authors found RP11-472N13.3, an annotated lncRNA, has top trait association and contains single nucleotide polymorphisms (SNPs) associated with central obesity. Knockdown of this lncRNA resulted in increase of IFN-γ induced JAK2 and STAT1 phosphorylation in THP-1 human macrophages and hence was named MacORIS (macrophage-enriched obesity-associated lncRNA serving as a repressor of IFN-γ signaling) [57]. Microarray profiling of whole blood samples from subjects newly diagnosed with type 2 diabetes identified several dysregulated lncRNAs with potential roles in inflammation and insulin resistance [58] Together, these studies suggest that targeting monocyte-macrophage-specific lncRNAs could be a therapeutic option in treating obesity, atherosclerosis and other related coronary artery diseases.
One of the most common microvascular complications of diabetes is diabetic nephropathy which manifests as increased fibrosis in most renal cells, loss of kidney function and ultimately renal failure. Although miRNAs have been clearly shown to play a role in diabetic nephropathy [59], the role of lncRNAs has emerged more recently. LncRNAs in renal cells have been shown to be involved in fibrosis, the accumulation of extracellular matrix (ECM). In the human kidney, PVT1 is a lncRNA that is upregulated in renal mesangial cells treated with HG indicating its potential involvement in diabetic nephropathy [60]. Indeed, loss of PVT1 expression results in reduced ECM accumulation showing that its upregulation may promote fibrosis [60]. Molecularly, PVT1 is the host gene to miR-1207, which functions independently to regulate a number of important ECM-related genes including transforming growth factor-beta1 (TGF-β1), plasminogen activator inhibitor-1 (PAI-1) and fibronectin (FN1) [60]. Furthermore, the PVT1 locus has long been implicated in end-stage renal disease from genome-wide association studies [61]. In a more recent study, it was found that a key lncRNA, Lnc-MGC, also promotes fibrosis as well as growth in mesangial cells related to diabetic nephropathy pathogenesis. This lncRNA, which is regulated by endoplasmic reticulum (ER) stress-related transcription factor, CHOP, is upregulated by HG and TGF-β1 in glomerular mouse and human mesangial cells [62]. Lnc-MGC is a host transcript to a mega cluster of over 40 miRNAs (termed miR-379 cluster). Expression of Lnc-MGC leads to an increase of this cluster of miRNAs which are derived from Lnc-MGC, most of which are upregulated in mouse models of diabetic renal disease [62]. Interestingly, CHOP knockout mice show a decrease in the expression of both Lnc-MGC as well as key miRNAs within the cluster. Furthermore, knockdown of Lnc-MGC utilizing a modified antisense oligonucleotide (GapmeR) reduces expression of the miRNAs, prevents accumulation of ECM in the glomeruli, and ultimately decreases the renal glomerular hypertrophy associated with early stages of diabetic nephropathy demonstrating the translational potential of targeting renal lncRNAs [62]. Analysis of renal transcriptomes reveal that additional lncRNAs, including Erbb4-IR, are regulated by Smad3, a transcription factor regulating key fibrotic genes and an effector of TGF-β1 [63]. Using a non-diabetic mouse model, it was recently found that TGF-β mediates renal fibrosis through the Smad3-Erbb4-IR lncRNA axis and targeting this lncRNA could be one approach to reduce renal fibrosis [64]. A recent study shows diabetic ANRIL knockout (ANRIL KO) mice have reduced levels of renal extracellular matrix proteins and urinary albumin relative to diabetic wild type mice, suggesting ANRIL also regulates key renal and vascular functions related to diabetic nephropathy[65]. It is likely there are many more renal lncRNAs that modulate genes and factors associated with fibrosis in the diabetic kidney. Additionally, lncRNAs are differentially expressed under hypoxic and inflammatory states in human proximal tubule epithelial cells [66]. The lncRNAs MIR210HG and linc-ATP13A4-8 are highly induced by hypoxia treatment and linc-KIAA1737-2 is induced by cytokine treatment. Thus lncRNAs identified here in renal epithelial and mesangial cells are relevant to human kidney disease and are poised for future human translational studies.
In the glomerulus, diabetes also results in podocyte effacement, apoptosis and dysfunction. In these cells, a number of lncRNAs have been described. Tug1 (taurine-upregulated 1) modulates the expression of peroxisome proliferator-activated receptor γ (PPARγ) coactivator α (PGC-1α). This interaction impacts the PGC-1α targets that are important for mitochondrial biogenesis. Under diabetic conditions, Tug1 is downregulated and leads to dysregulation of mitochondrial bioenergetics and overexpression of Tug1 is sufficient to reverse the impacts on the mitochondria as well as improve diabetic nephropathy in diabetic mice [67].
Other lncRNAs in the kidney can promote inflammation, a process also associated with the development of nephropathy. In mesangial cells, LincRNA-GM4419 has been shown to directly interact with the p50 subunit of NF-κB, regulating its activity and promoting inflammation. In mice, reduction of LincRNA-GM4419 decreases inflammation in mesangial cells [68]. MALAT1 is upregulated by HG in endothelial cells and in renal tissues from diabetic mice [69]. The upregulation of this lncRNA is accompanied by increase in serum amyloid antigen 3 (SAA3), and increase in inflammatory genes including TNFα and IL-6. These inflammatory gene changes were reduced by knockdown of MALAT1 expression [69]. These data suggest that MALAT1 is a key regulator of inflammation in endothelial cells under diabetic conditions.
Other non-coding RNAs of note
Enhancers are non-coding genomic elements that enhance the target gene expression [70]. Recent studies have shown that enhancer regions possess binding sites for transcription factors and have the ability to be transcribed, producing what are called enhancer RNAs (eRNAs) with important functional roles [71, 72]. SNPs with disease or trait association have been detected in these enhancers [73]. As noted earlier, in VSMCs, Ang II promotes dynamic alterations to enhancers, some of which overlap Ang II regulated lncRNAs, suggesting that such potential eRNAs can also be involved in Ang II-driven pathologies [53]. However, more work is needed to validate the eRNA characteristics of these lncRNAs.
A relatively new group of non-coding RNAs are tRNA fragments (tRFs). These RNAs has been implicated to play a role in the inheritance of metabolic changes from paternal diet in mouse models. Specifically, sperm and male reproductive tract express tRFs, which are affected by paternal diet [74, 75]. As such, offspring metabolism, mostly through gene expression, is affected by the dysregulation of tRFs in their father’s germ cells. Additionally, sperm from obese men have altered transcriptome profiles and DNA methylation levels, suggesting that obesity and diabetes in humans can also impact offspring health and risk of disease via changes in non-coding RNAs [76]. How tRFs may be involved in diabetes and associated complications are not currently known but may be an interesting avenue of investigation.
PERSPECTIVE
LncRNAs have garnered much interest in recent years due to their growing numbers of cellular functions under normal and diseased states. In the field of diabetic complications, lncRNAs are increasingly being implicated in the physiology and pathology of vascular disease related cells. As we begin to profile more vascular tissues from diabetic animal models and human patients, we will without a doubt uncover many more lncRNAs involved in the diseased state. Interestingly, many lncRNA loci have been shown to contain SNPs found in GWAS studies of cardiometabolic traits [77]. These data along with other GWAS studies have implicated lncRNA loci as important molecules related to disease.
As we learn more about the optimum approaches to interfere with the expression of lncRNAs in vivo, therapeutic approaches to target these lncRNAs can be developed to reduce cardiovascular disease. This may be an effective strategy as lncRNA expression has been shown to be highly cell-type specific. In mouse models, we have utilized GapmeR antisense oligonucleotides to specifically target an lncRNA expressed in the kidney [62] to slow down the progression of diabetic nephropathy. Strategies such as this can be utilized against lncRNAs in vascular cells to reduce diabetic complications, which are mainly responsible for the mortality and morbidity of diabetes. As lncRNAs can be localized in the nucleus or cytoplasm, the targeting approach has to be accordingly designed. Other challenges include the relatively low expression of lncRNAs compared to coding mRNAs, as well as poor conservation amongst species. More emphasis on the study of lncRNAs that are expressed in human normal and diseased cells can help in exploiting them as biomarkers of early detection, or targets for the prevention of human diabetic vascular and inflammatory complications. Clearly, this is an exciting emerging field that is likely to see much growth in the upcoming years.
Table 1.
LncRNAs in Diabetic vascular complications
| LncRNA | Biological Function | Mechanism | References |
|---|---|---|---|
| ANRIL | Atherosclerosis, Hypertension, Diabetes, Retinopathy, Nephropathy | Regulation of CDKN2A/B | [48] |
| MIAT | Retinal microvascular dysfunction | Sponging miR-150-5p | [51] |
| MALAT1 | Inflammation in diabetes | Upregulation of IL-6 and TNFα | [69] |
| MEG3 | Microvascular dysfunction | PI3K/Akt signal activation | [52] |
| Lnc-Ang362 | Ang II induced VSMC proliferation | Expression of miR-221 and miR-222 | [42] |
| E330013P06 | Macrophage inflammation in Diabetes | Upregulation of IL-6 and Ptgs2 | [54] |
| Lethe | Regulation of macrophage ROS production | Modulating Nox2 gene expression | [56] |
| Pvt1 | ECM accumulation & diabetic nephropathy | Regulation of Fn1 and Col4a1 expression | [60] |
| Lnc-MGC | Renal fibrosis and early diabetic nephropathy | Regulation of EDEM3 expression | [62] |
| Tug1 | Mitochondrial energetics of podocytes in diabetes | Interaction with PGC-1α | [67] |
Acknowledgments
The authors gratefully acknowledge funding from the National Institutes of Health, R01 DK 065073, R01 HL106089, R01 DK081705 (to RN) and K01 DK104993 (to AL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nathan DM. Diabetes: Advances in Diagnosis and Treatment. JAMA. 2015;314(10):1052–1062. doi: 10.1001/jama.2015.9536. [DOI] [PubMed] [Google Scholar]
- 2.Nathan DM. Long-term complications of diabetes mellitus. N Engl J Med. 1993;328(23):1676–1685. doi: 10.1056/NEJM199306103282306. [DOI] [PubMed] [Google Scholar]
- 3.Rask-Madsen C, King GL. Vascular Complications of Diabetes: Mechanisms of Injury and Protective Factors. Cell Metabolism. 2013;17(1):20–33. doi: 10.1016/j.cmet.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes JM, Cooper ME. Mechanisms of Diabetic Complications. Physiol Rev. 2013;93(1):137–188. doi: 10.1152/physrev.00045.2011. [DOI] [PubMed] [Google Scholar]
- 5.Shah MS, Brownlee M. Molecular and Cellular Mechanisms of Cardiovascular Disorders in Diabetes. Circ Res. 2016;118(11):1808–1829. doi: 10.1161/CIRCRESAHA.116.306923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37(Database issue):D122–126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 9.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn RA, Chang HY. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Cell Stem Cell. 2014;14(6):752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends in cell biology. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Dechamethakun S, Muramatsu M. Long noncoding RNA variations in cardiometabolic diseases. Journal of human genetics. 2017;62(1):97–104. doi: 10.1038/jhg.2016.70. [DOI] [PubMed] [Google Scholar]
- 13.McPherson R, Pertsemlidis A, Kavaslar N, Stewart AFR, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. Faseb J. 2011;25(2):444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 17.Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakic N, Garcia-Hurtado J, Rodriguez-Segui S, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirza AH, Kaur S, Brorsson CA, Pociot F. Effects of GWAS-associated genetic variants on lncRNAs within IBD and T1D candidate loci. PLoS One. 2014;9(8):e105723. doi: 10.1371/journal.pone.0105723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao R, Wang LJ, Wang HB, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 22.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 23.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 24.Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 25.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21(12):2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu S, Yuan GC, Shao Z. The PRC2-binding long non-coding RNAs in human and mouse genomes are associated with predictive sequence features. Sci Rep. 2017;7:41669. doi: 10.1038/srep41669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22(5):370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong CG, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470(7333):284. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–U245. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miano JM, Long X. The short and long of noncoding sequences in the control of vascular cell phenotypes. Cell Mol Life Sci. 2015;72(18):3457–3488. doi: 10.1007/s00018-015-1936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchida S, Dimmeler S. Long Noncoding RNAs in Cardiovascular Diseases. Circ Res. 2015;116(4):737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 38.Jian L, Jian D, Chen Q, Zhang L. Long Noncoding RNAs in Atherosclerosis. J Atheroscler Thromb. 2016;23(4):376–384. doi: 10.5551/jat.33167. [DOI] [PubMed] [Google Scholar]
- 39.Lu W, Huang SY, Su L, Zhao BX, Miao JY. Long Noncoding RNA LOC100129973 Suppresses Apoptosis by Targeting miR-4707-5p and miR-4767 in Vascular Endothelial Cells. Sci Rep-Uk. 2016:6. doi: 10.1038/srep21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teng W, Qiu CG, He ZH, Wang GL, Xue YL, Hui XZ. Linc00152 suppresses apoptosis and promotes migration by sponging miR-4767 in vascular endothelial cells. Oncotarget. 2017;8(49):85014–85023. doi: 10.18632/oncotarget.18777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131(14):1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel Long Noncoding RNAs Are Regulated by Angiotensin II in Vascular Smooth Muscle Cells. Circ Res. 2013;113(3):266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arteriosclerosis, thrombosis, and vascular biology. 2014;34(6):1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation. 2016;133(21):2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bischoff FC, Werner A, John D, Boeckel JN, Melissari MT, Grote P, Glaser SF, Demolli S, Uchida S, Michalik KM, et al. Identification and Functional Characterization of Hypoxia-Induced Endoplasmic Reticulum Stress Regulating lncRNA (HypERlnc) in Pericytes. Circ Res. 2017;121(4):368. doi: 10.1161/CIRCRESAHA.116.310531. [DOI] [PubMed] [Google Scholar]
- 46.Leung A, Natarajan R. Long Noncoding RNAs in Diabetes and Diabetic Complications. Antioxidants & redox signaling. 2017 doi: 10.1089/ars.2017.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas AA, Feng BA, Chakrabarti S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Invest Ophth Vis Sci. 2017;58(1):470–480. doi: 10.1167/iovs.16-20569. [DOI] [PubMed] [Google Scholar]
- 48.Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: Molecular Mechanisms and Implications in Human Health. Int J Mol Sci. 2013;14(1):1278–1292. doi: 10.3390/ijms14011278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B, Wang D, Ji TF, Shi L, Yu JL. Overexpression of lncRNA ANRIL up-regulates VEGF expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating NF-kappa B signaling pathway in a rat model. Oncotarget. 2017;8(10):17347–17359. doi: 10.18632/oncotarget.14468. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Awata T, Yamashita H, Kurihara S, Morita-Ohkubo T, Miyashita Y, Katayama S, Mori K, Yoneya S, Kohda M, Okazaki Y, et al. A Genome-Wide Association Study for Diabetic Retinopathy in a Japanese Population: Potential Association with a Long Intergenic Non-Coding RNA (vol 9, e111715, 2014) Plos One. 2015;10(4) doi: 10.1371/journal.pone.0111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan BA, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT Regulates Microvascular Dysfunction by Functioning as a Competing Endogenous RNA. Circ Res. 2015;116(7):1143. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 52.Qiu GZ, Tian W, Fu HT, Li CP, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Bioph Res Co. 2016;471(1):135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 53.Das S, Senapati P, Chen Z, Reddy MA, Ganguly R, Lanting L, Mandi V, Bansal A, Leung A, Zhang S, et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat Commun. 2017:8. doi: 10.1038/s41467-017-01629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy MA, Chen Z, Park JT, Wang M, Lanting L, Zhang Q, Bhatt K, Leung A, Wu XW, Putta S, et al. Regulation of Inflammatory Phenotype in Macrophages by a Diabetes-Induced Long Noncoding RNA. Diabetes. 2014;63(12):4249–4261. doi: 10.2337/db14-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rapicavoli NA, Qu K, Zhang JJ, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013:2. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed]
- 56.Zgheib C, Hodges MM, Hu JY, Liechty KW, Xu JW. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. Plos One. 2017;12(5) doi: 10.1371/journal.pone.0177453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Xue C, Wang Y, Shi J, Zhang X, Li W, Nunez S, Foulkes AS, Lin J, Hinkle CC, et al. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated With Cardiometabolic Diseases. Journal of the American Heart Association. 2017;6(11) doi: 10.1161/JAHA.117.007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Chang X, Zhang P, Fan L, Zhou T, Sun K. Aberrant Expression of Long Non-Coding RNAs in Newly Diagnosed Type 2 Diabetes Indicates Potential Roles in Chronic Inflammation and Insulin Resistance. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017;43(6):2367–2378. doi: 10.1159/000484388. [DOI] [PubMed] [Google Scholar]
- 59.Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann Ny Acad Sci. 2015;1353:72–88. doi: 10.1111/nyas.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One. 2011;6(4):e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanson RL, Craig DW, Millis MP, Yeatts KA, Kobes S, Pearson JV, Lee AM, Knowler WC, Nelson RG, Wolford JK. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genonte-wide single nucleotide polymorphism association study. Diabetes. 2007;56(4):975–983. doi: 10.2337/db06-1072. [DOI] [PubMed] [Google Scholar]
- 62.Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L, Deshpande S, Jia Y, Lai JY, O’Connor CL, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Q, Chung AC, Huang XR, Dong Y, Yu X, Lan HY. Identification of novel long noncoding RNAs associated with TGF-beta/Smad3-mediated renal inflammation and fibrosis by RNA sequencing. The American journal of pathology. 2014;184(2):409–417. doi: 10.1016/j.ajpath.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 64.Feng M, Tang PM, Huang XR, Sun SF, You YK, Xiao J, Lv LL, Xu AP, Lan HY. TGF-beta Mediates Renal Fibrosis via the Smad3-Erbb4-IR Long Noncoding RNA Axis. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas AA, Feng B, Chakrabarti S. ANRIL regulates production of extracellular matrix proteins and vasoactive factors in Diabetic Complications. American journal of physiology Endocrinology and metabolism. 2017 doi: 10.1152/ajpendo.00268.2017. ajpendo 00268 02017. [DOI] [PubMed] [Google Scholar]
- 66.Lin J, Zhang X, Xue CY, Zhang HR, Shashaty MGS, Gosai SJ, Meyer N, Grazioli A, Hinkle C, Caughey J, et al. The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am J Physiol-Renal. 2015;309(11):F901–F913. doi: 10.1152/ajprenal.00290.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, Green NH, Chang BH, Overbeek PA, Danesh FR. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. The Journal of clinical investigation. 2016;126(11):4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi H, Peng R, Zhang LY, Sun Y, Peng HM, Liu HD, Yu LJ, Li AL, Zhang YJ, Jiang WH, et al. LincRNA-Gm4419 knockdown ameliorates NF-kappaB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell death & disease. 2017;8(2):e2583. doi: 10.1038/cddis.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Puthanveetil P, Chen SL, Feng BA, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19(6):1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 71.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Q, Yan MH, Cao ZH, Li X, Zhang YF, Shi JC, Feng GH, Peng HY, Zhang XD, Zhang Y, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 75.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun FY, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, Mortensen B, Appel EVR, Jorgensen N, Kristiansen VB, et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metabolism. 2016;23(2):369–378. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Ballantyne RL, Zhang X, Nunez S, Xue C, Zhao W, Reed E, Salaheen D, Foulkes AS, Li M, Reilly MP. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Human molecular genetics. 2016;25(14):3125–3141. doi: 10.1093/hmg/ddw154. [DOI] [PMC free article] [PubMed] [Google Scholar]