Abstract
Purpose
The aim of this study was to analyze and report pathogenic variants in the ABCA4 gene in Brazilian patients with a clinical diagnosis of Stargardt disease.
Methods
This retrospective study evaluated variants in the ABCA4 gene in Brazilian patients with Stargardt disease. The patients’ visual acuity and age of symptom onset were obtained from previous medical records. The patients were classified according to the autofluorescence patterns.
Results
Fifty patients aged between 10 and 65 years from 44 families were included in the study. Among these cases, the mean age of symptom onset was 14 years (range, 5–40 years). ABCA4 gene sequencing was conclusive in 40 patients (80%), negative in two patients (4%), and inconclusive in eight patients (16%). Four families carried homozygous pathogenic variants. Segregation analysis results were available for 23 families. One novel variant was found: p.Ala2084Pro. The most frequent pathogenic variant in this group was p.Arg602Trp (12/100 alleles). Based on the phenotypic characteristics assessed with fundus autofluorescence imaging, 12 patients were classified as having type I phenotype, 16 as having type II, and 18 patients as having type III. The cases classified as type III phenotype included patients who were homozygous for the p.Asn96Asp and p.Arg2030* variants. One patient with a type I phenotype carried the homozygous intronic variant c.3862+1G>A.
Conclusions
Next-generation sequencing was effective for the molecular diagnosis of genetic diseases and specifically allowed a conclusive diagnosis in 80% (40/50) of the patients. As the ABCA4 gene does not show a preferential region for pathogenic variants, the diagnosis of Stargardt disease depends on broader analysis of the gene. The most common pathogenic variants in the ABCA4 gene described in the literature were also found in these Brazilian patients. Although some genotype–phenotype correlations were found, more studies regarding the progression of Stargardt disease will help increase our understanding of the pathogenicity of these gene variants.
Introduction
Stargardt disease (STGD, OMIM 248200) is an autosomal recessive retinal degenerative disease caused by defects in the ABCA4 gene (OMIM 601691). The disease causes functional and structural retinal damage, and the main characteristic is central vision impairment. Stargardt disease exhibits great clinical variability and may diffusely alter the retina in the most severe cases. Since the first descriptions of this disease, diagnosis has been based on phenotypic characteristics [1,2]. Because the gene responsible for this disease has been identified, clinical diagnosis of these patients may be confirmed genetically with gene sequencing [3].
Decreased central vision is the initial symptom of Stargardt disease; it classically occurs between the first and second decades of life and progressively worsens with age [1,2]. This progressive decline in bilateral central vision is related to retinal atrophy of the macular region due to loss of the external segments of photoreceptors and retinal pigment epithelium (RPE) cells [4,5]. Lipofuscin deposits in the RPE and photoreceptors (called flecks) are key markers for a diagnosis of Stargardt disease. In addition, these clinical signs, which are clearly identified with a fundus autofluorescence examination, determine the phenotypic classification of patients with this disease [6]. For example, areas of RPE atrophy are hypoautofluorescent on retinal autofluorescence examination, whereas the hyperautofluorescence points identified in this examination are related to lipofuscin accumulation in RPE cells [6]. Lipofuscin accumulation in the RPE layer masks choroidal fluorescence on fluorescein angiography, an effect is known as the dark or silent choroid sign [7].
The ABCA4 gene is exclusively expressed in photoreceptors, cones and rods [8,9]. The loss or decreased function of the protein encoded by the ABCA4 gene causes deposition of the main component of lipofuscin [N-retinylidene-N-retinylethanolamine (A2-E)] in the RPE and the formation of retinal flecks. This accumulation slows adaptation to darkness [10,11]. The presence of lipofuscin in RPE cells causes oxidative damage to these cells, which become apoptotic. Moreover, loss of the RPE layer leads to secondary photoreceptor death, resulting in reduced vision [11-13].
ABCA4 is a polymorphic gene with low conservation due to the high frequency of allelic variation in the gene. It presents many benign variants, with a frequency higher than 1% in the population, as are hundreds of pathogenic variants [14]. The majority of variants are missense mutations, followed by protein-truncating variants [15]. Complex alleles and intronic variants have also been described for the ABCA4 gene [14-17]. The presence of pathogenic variants of this gene is the most common cause of retinal diseases among children and young adults and is primarily related to autosomal recessive Stargardt disease [14,18]. Involvement of ABCA4 has also been reported in cases of cone–rod dystrophy and autosomal recessive retinitis pigmentosa [19,20].
The phenotypic characteristics of each case demonstrate particularities corresponding to the combination of pathogenic variants in the ABCA4 gene in compound heterozygous and homozygous individuals. The phenotypic characteristics of certain variants, including p.Gly1961Glu and the complex allele p.[Leu541Pro; Ala1038Val], have been established [21,22]. Overall, the clinical outcome depends on the severity of the combined alleles [15].
Little is known to date about the pathogenic variants in the Brazilian population with Stargardt disease. Thus, the aim of this study was to analyze and describe pathogenic variants in the ABCA4 gene in Brazilian patients with a clinical diagnosis of Stargardt disease.
Methods
This retrospective study evaluated variants in the ABCA4 gene in Brazilian patients with a Stargardt disease diagnosis. Cases were selected based on a review of 254 genetic tests for retinal dystrophy performed between January 2009 and January 2017 at the Federal University of São Paulo (UNIFESP) and the Ocular Genetics Institute (IGO), São Paulo, Brazil. All patients signed informed consent. The study was approved by the Research Ethics Committee of UNIFESP (Protocol No. 6159), Brazil, and was conducted in accordance with the ethical standards of the 1964 Declaration of Helsinki and the Association for Research in Vision and Ophthalmology (ARVO) statement on human subjects, and its subsequent amendments.
From a pool of 254 retinal dystrophy tests, patients with Stargardt for whom the ABCA4 gene was sequenced using next-generation sequencing (NGS) were included in the study. Patients with a clinical diagnosis of Stargardt disease and conclusive molecular testing regarding other Stargardt genes were excluded. Among the study group, there were patients with clinically Stargardt with a conclusive ABCA4 test, an inconclusive test with one variant, and patients with no variant.
The Ion PGMTM System (Life Technologies, Carlsbad, CA) and MiSeq by Illumina (Illumina, San Diego, CA) platforms were used for the molecular diagnosis of the cases. The ABCA4 gene was included in gene panels related to Stargardt disease or macular dystrophies. Only coding regions and immediately flanking intron sequences were examined. Changes in the promoter region, deep intronic regions, or other non-coding regions of the gene were not evaluated. Among the patients, only those with an initial clinical diagnosis of Stargardt disease were selected for inclusion in this study.
Segregation results available in medical records were analyzed and included in the study. Sanger sequencing was used to identify in relatives pathogenic variants found in probands.
Gene sequencing data collected from medical records were compared with variants available in the following databases: Human Gene Mutation Database (HGMD) [23], ClinVar [24], 1000 Genomes Project [25], Exome Aggregation Consortium (ExAC) [26], and Leiden Open Variation Database (LOVD) [27]. The following programs were used to predict the pathogenicity of new variants: CADD (deleterious threshold: 20) [28], PolyPhen-2 [29], MutationTaster [30], and Sorting Intolerant from Tolerant (SIFT) [31]. The nucleotide numbering is based on the reference sequence NM_000350, with the A of the initiation codon (ATG) representing number 1.
Data on the patients’ visual acuity and age of symptom onset were collected from medical records. Complementary examinations, including retinography (RG), fundus autofluorescence imaging (FAF), and optical coherence tomography (OCT), were reassessed to confirm the clinical diagnosis of Stargardt disease.
The patients were classified according to signs found in the autofluorescence test (Heidelberg Retina Angiograph, HRA 2, Heidelberg Engineering, Heidelberg, Germany), as described by Fujinami et al. [6]. This classification separates patients into the following three types: type I (an area of foveal hypoautofluorescence surrounded by a retina with a homogeneous appearance, with or without foci of high or low autofluorescent signals), type II (an area of foveal hypofluorescence surrounded by an area with a heterogeneous appearance and hyperautofluorescent and hypoautofluorescent points that extend to the temporal arcades, giving the retina a reticulate appearance), and type III (extensive areas of hypoautofluorescence in the posterior pole and a heterogeneous appearance of the remaining retinal areas, with hyperautofluorescent and hypoautofluorescent points) [6].
Results
A total of 50 patients from 44 families (including four sibling pairs and two mother-daughter pairs) aged between 10 and 65 years were included in the study. Three families had a history of consanguinity (siblings 4 and 6 and patients 33 and 40). Table 1, Table 2, Table 3, Table 4 shows the molecular analysis results and clinical characteristics of the Stargardt disease cases, including the age of symptom onset (range, 5–40 years; mean, 14 years) and visual acuity, which ranged from 20/40 to counting fingers at 20 cm during the examination.
Table 1. Fundus autofluorescence type I patients. Pathogenic variants in the ABCA4 gene and clinical data.
| Pt | S |
Pathogenic variants |
Visual acuity |
Age of onset/ data collect/ disease duration (years) | |||
|---|---|---|---|---|---|---|---|
|
Allele 1 |
Allele 2 |
||||||
| Nucleotide change | Protein change | Nucleotide change | Protein change | RE LE | |||
| 1• |
F |
c.4926C>G |
p.Ser1642Arg |
c.5882G>A |
p.Gly1961Glu |
20/200
20/200 |
12/30/18 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
|
|
||||
| 9 |
M |
c.6112C>T |
p.Arg2038Trp |
c.6320G>A |
p.Arg2107His |
20/200
20/150 |
11/35/24 |
| 14 |
M |
not detected |
|
not detected |
|
FC 2m
20/40 |
30/42/12 |
| 31• |
M |
c.1622T>C |
p.Leu541Pro |
c.3386G>T |
p.Arg1129Leu |
N/A
N/A |
N/A/23/ N/A |
| c.4328G>A |
p.Arg1443His |
|
|
||||
| 49• |
M |
c.5282C>G |
p.Pro1761Arg |
c.2345G>A |
p.Trp782* |
20/400
20/400 |
15/25/10 |
| c.6316C>T |
p.Arg2106Cys |
|
|
||||
| 43• |
F |
c.5882G>A |
p.Gly1961Glu |
c.66G>T |
p.Lys22Asn |
20/60
20/60 |
19/20/1 |
| 51• |
M |
c.1622T>C |
p.Leu541Pro |
c.5882G>A |
p.Gly1961Glu |
20/30
20/30 |
10/11/1 |
| c.3113C>T |
p.Ala1038Val |
|
|
||||
| 28 |
M |
c.70C>T |
p.Arg24Cys |
c.1804C>T |
p.Arg602Trp |
20/50
20/60 |
16/26/10 |
| 33§• |
M |
c.3862+1G>A† |
p.? |
c.3862+1G>A† |
p.? |
20/200
20/200 |
10/10/0 |
| 35• |
M |
c.6079C>T |
p.Leu2027Phe |
c.6250G>C |
p.Ala2084Pro |
20/25
20/25 |
20/20/0 |
| 48• |
F |
c.5882 G>A |
p.Gly1961Glu |
c.1364T>A |
p.Leu455Gln |
20/60
20/150 |
32/32/0 |
| c.3113 C>T |
p.Ala1038Val |
|
|
||||
| 36• | F | c.1804C>T | p.Arg602Trp | not detected | 20/25 20/25 | 7/28/21 | |
Pt, Patient; S, Sex; N/A, not available; FC, Finger counting. Novel variant. • Segregation analysis performed †Homozygous variants ‡c.5044_del15bp, nonframeshift deletion (c.5044_5058delGTTGCCATCTGCGTG). § Consanguinity Families: Siblings: 41(Table 4) and 49. Mother 44 (Table 3) and daughter 43. Mother 45 (Table 3) and daughter 36.
Table 2. Fundus autofluorescence type II patients. Pathogenic variants in the ABCA4 gene and clinical data.
| Pt | S |
Pathogenic variants |
Visual acuity |
Age of onset/ data collect/ disease duration (years) | |||
|---|---|---|---|---|---|---|---|
|
Allele 1 |
Allele 2 |
||||||
| Nucleotide change | Protein change | Nucleotide change | Protein change | RE LE | |||
| 3• |
F |
c.4926C>G |
p.Ser1642Arg |
c.2791G>A |
p.Val931Met |
20/150
20/150 |
14/37/23 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
|
|
||||
| 5 |
F |
c.3898C>T |
p.Arg1300* |
c.1804C>T |
p.Arg602Trp |
20/200
20/200 |
7/17/10 |
| 8 |
F |
not detected |
|
not detected |
|
20/400
20/400 |
32/44/12 |
| 11 |
M |
c.6079C>T |
p.Leu2027Phe |
not detected |
|
FC1m
20/200 |
12/24/12 |
| 12• |
M |
c.4328G>A |
p.Leu541Pro |
c.2743G>A |
p.Asp915Asn |
20/200
20/200 |
8/13/5 |
| c.1622T>C |
p.Arg1443His |
|
|
||||
| 16 |
M |
c.4720G>T |
p.Glu1574* |
not detected |
|
20/150
20/200 |
6/38/32 |
| 18 |
M |
c.634C>T |
p.Arg212Cys |
c.1804C>T |
p.Arg602Trp |
20/400
20/400 |
6/31/25 |
| 19 |
M |
c.634C>T |
p.Arg212Cys |
c.1804C>T |
p.Arg602Trp |
FC1.5m
FC1.5m |
11/26/15 |
| 21 |
M |
c.3386G>T |
p.Arg1129Leu |
not detected |
|
20/150
20/150 |
16/32/16 |
| 22• |
M |
c.4926C>G |
p.Ser1642Arg |
c.4340A>T |
p.Glu1447Val |
20/200
FC2m |
8/10/2 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
|
|
||||
| 29 |
F |
c.223T>G |
p.Cys75Gly |
c.6088C>T |
p.Arg2030* |
N/A
N/A |
8/16/8 |
| 32 |
M |
c.455G>A |
p.Arg152Gln |
not detected |
|
20/30
FC10cm |
36/39/3 |
| 46 |
F |
c.5461–10T>C |
p.[Thr1821Valfs*13, Thr1821Aspfs*6] |
c.1804C>T |
p.Arg602Trp |
20/200
20/200 |
9/10/1 |
| 47• |
F |
c.1622T>C |
p.Leu541Pro |
c.286A>G |
p.Asn96Asp |
20/400
20/400 |
7/26/19 |
| c.4328G>A |
p.Arg1443His |
|
|
||||
| 50• |
M |
c.1804C>T |
p.Arg602Trp |
c.5381C>A |
p.Ala1794Asp |
20/400
20/400 |
15/31/16 |
| 26 | F | c.5714+5G>A | p.? | not detected | 20/80 20/80 | 9/14/5 | |
Pt, Patient; S, Sex; N/A, not available; FC, Finger counting.• Segregation analysis performed ‡c.5044_del15bp, nonframeshift deletion (c.5044_5058delGTTGCCATCTGCGTG).Families: Siblings: 18 and 19.
Table 3. Fundus autofluorescence type III patients. Pathogenic variants in the ABCA4 gene and clinical data.
| Pt | S |
Pathogenic variants |
Visual acuity |
Age of onset/ data collect/ disease duration (years) | |||
|---|---|---|---|---|---|---|---|
|
Allele 1 |
Allele 2 |
||||||
| Nucleotide change | Protein change | Nucleotide change | Protein change | RE LE | |||
| 4§ |
F |
c.286A>G |
p.Asn96Asp† |
c.286A>G |
p.Asn96Asp† |
20/400
20/400 |
23/39/16 |
| 6§ |
M |
c.286A>G |
p.Asn96Asp† |
c.286A>G |
p.Asn96Asp† |
FC2m
FC2m |
14/47/33 |
| 7 |
M |
c.3862+1G>A |
p.? |
not detected |
|
FC20cm
FC20cm |
16/63/47 |
| 10• |
F |
c.4926C>G |
p.Ser1642Arg |
c.3056C>T |
p.Thr1019Met |
FC2m
FC1m |
5/26/21 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
|
|
||||
| 13• |
M |
c.4926C>G |
p.Ser1642Arg |
c.1804C>T |
p.Arg602Trp |
FC50cm
FC2m |
8/26/18 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
|
|
||||
| 15 |
F |
c.5714+5G>A |
p.? |
c.6005+1G>T |
p.? |
FC20cm
FC10cm |
12/40/28 |
| 17• |
F |
c.1622T>C |
p.Leu541Pro |
c.3329–2A>T |
p.? |
HM
FC10cm |
7/59/52 |
| c.3113C>T |
p.Ala1038Val |
|
|
||||
| 20• |
M |
c.1622T>C |
p.Leu541Pro |
c.3329–2A>T |
p.? |
HM
HM |
8/65/57 |
| c.3113C>T |
p.Ala1038Val |
|
|
||||
| 23• |
F |
c.1622T>C |
p.Leu541Pro |
c.1804C>T |
p.Arg602Trp |
FC1.5m
FC1.5m |
7/31/24 |
| c.4328G>A |
p.Arg1443His |
|
|
||||
| 24 |
M |
c.4457C>T |
p.Pro1486Leu |
c.1804C>T |
p.Arg602Trp |
FC1.5m
FC1.5m |
15/46/31 |
| 44• |
F |
c.4926C>G |
p.Ser1642Arg |
c.66G>T |
p.Lys22Asn |
N/A
N/A |
N/A |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
|
|
||||
| 30• |
F |
c.6088C>T |
p.Arg2030* † |
c.6088C>T |
p.Arg2030* † |
FC1.5m
20/400 |
7/38/31 |
| 37 |
F |
c.4003_4004delCC |
p.Pro1335Argfs*86 |
c.658C>T |
p.Arg220Cys |
20/30
20/30 |
28/31/3 |
| 39 |
F |
c.1648G>A |
p.Gly550Arg |
not detected |
|
FC1.5m
FC1.5m |
8/36/28 |
| 42• |
M |
c.2894 A>G |
p.Asn965Ser |
c.5381C>A |
p.Ala1794Asp |
20/400
20/400 |
17/38/21 |
| 25 |
M |
c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
c.1804C>T |
p.Arg602Trp |
20/400
20/400 |
7/26/19 |
| 34• |
M |
c.4926C>G |
p.Ser1642Arg |
c.1622T>C |
p.Leu541Pro |
N/A
N/A |
7/33/26 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del‡ |
c.4328G>A |
p.Arg1443His |
||||
| 45• | F | c.1804C>T | p.Arg602Trp | c.3386G>T | p.Arg1129Leu | 20/400 20/400 | 18/61/43 |
Pt, Patient; S, Sex; N/A, not available; HM, Hand movement; FC, Finger counting.• Segregation analysis performed †Homozygous variants. ‡c.5044_del15bp, nonframeshift deletion (c.5044_5058delGTTGCCATCTGCGTG). § Consanguinity Families: Siblings:4 and 6; 17 and 20. Mother 44 and daughter 43 (Table 1). Mother 45 and daughter 36 (Table 1).
Table 4. Pathogenic variants in the ABCA4 gene and clinical data of patients with no fundus autofluorescence classification.
| Pt | S |
Pathogenic variants |
Visual acuity |
Age of onset/ data collect/ disease duration (years) | |||
|---|---|---|---|---|---|---|---|
|
Allele 1 |
Allele 2 |
||||||
| Nucleotide change | Protein change | Nucleotide change | Protein change | RE LE | |||
| 40§• |
M |
c.1622T>C |
p.Leu541Pro† |
c.1622T>C |
p.Leu541Pro† |
N/A
N/A |
N/A/35/N/A |
| c.4328G>A |
p.Arg1443His† |
c.4328G>A |
p.Arg1443His† |
||||
| 41• |
F |
c.5282C>G |
p.Pro1761Arg |
c.2345G>A |
p.Trp782* |
20/200
20/200 |
N/A/27/N/A |
| c.6316C>T |
p.Arg2106Cys |
|
|
||||
| 27Ω |
F |
c.868C>T |
p.Arg290Trp |
c.5882G>A |
p.Gly1961Glu |
20/400
20/400 |
40/61/21 |
| c.2690C>T |
p.Thr897Ile |
|
|
||||
| 38• | M | c.32T>C | p. Leu11Pro | c.3113C>T | p. Ala1038Val | 20/400 20/400 | 20/21/1 |
Pt, Patient; S, Sex; N/A, not available • Segregation analysis performed †Homozygous variants. § Consanguinity Families: Siblings: 41 and 49 (Table 1). ΩPatient 27 present 3 patogenic variants, but without segregation analisys it was impossible to determinate which of them are combined in cis.
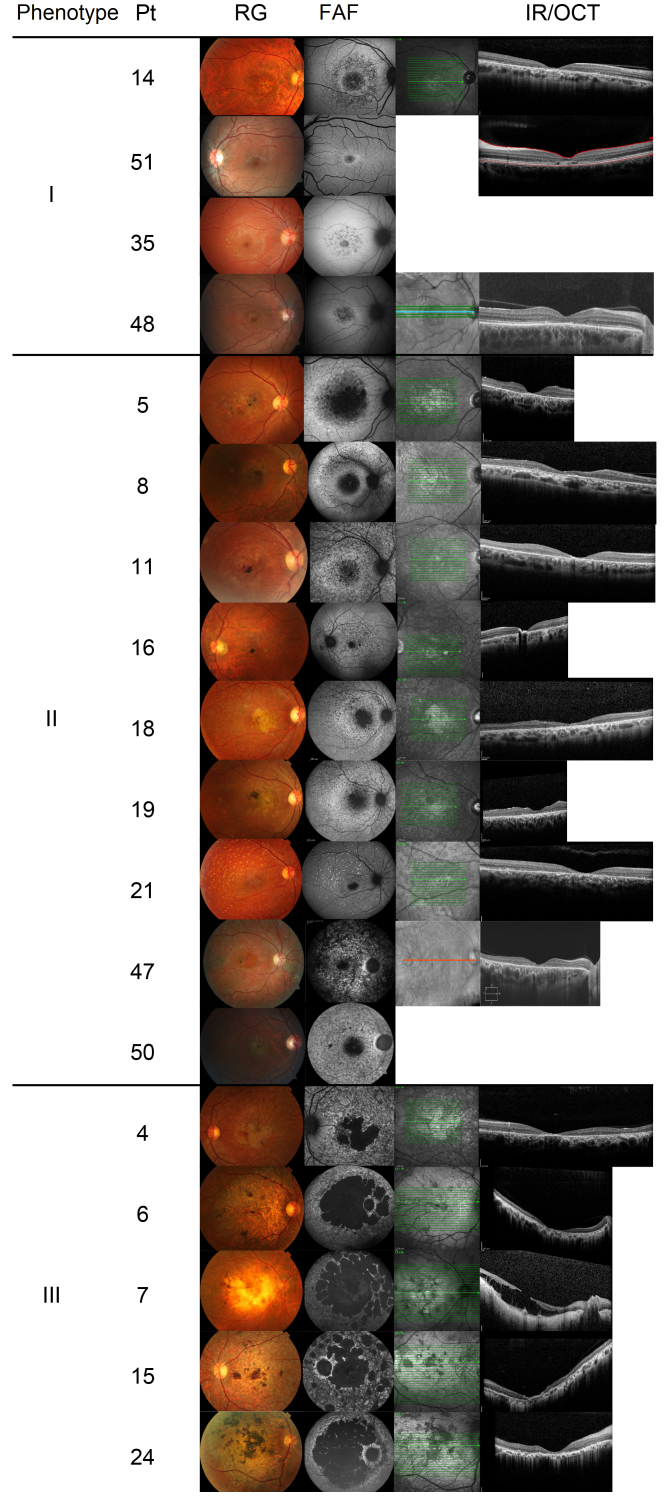
All patients included showed changes in pigmentation or macular atrophy on retinography examination. Retinal flecks were found in almost all cases. OCT revealed focal areas of photoreceptor layer discontinuity corresponding to macular atrophy (Figure 1). The patients were classified into three types of phenotypic characteristics according to the fundus autofluorescence examination results (Table 1, Table 2, Table 3 and Figure 2) [6]. Twelve patients were classified as type I (Table 1), 16 patients as type II (Table 2), and 18 patients as type III (Table 3). Records of the fundus autofluorescence examination were lacking for four patients (Table 4), who, therefore, were not classified.
Figure 1.
Type I FAF phenotype: patients 14, 51, 35, and 48. Type II FAF phenotype: patients 5, 8, 11, 16, 18, 19, 21, 47, and 50. Type III FAF phenotype: patients 4, 6, 7, 15, and 24. Negative cases at molecular testing: patients 14 and 8. Inconclusive cases at molecular testing: patients 11, 16, 21, and 7. Patient, Pt; retinography, RG; fundus autofluorescence, FAF; infrared imaging, IR; optical coherence tomography, OCT.
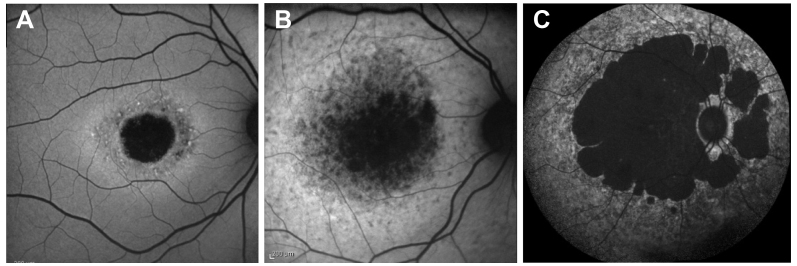
Figure 2.
Fundus autofluorescence pictures of Stargardt cases. A: Patient 9 with the type I phenotype. B: Patient 5 with the type II phenotype. C: Patient 6 with the type III phenotype.
The ABCA4 gene sequencing results were conclusive for 40 patients (80%) but negative for patients 8 and 14 (4%). An additional eight patients (7, 11, 16, 21, 26, 32, 36, and 39) carried only one pathogenic variant in the ABCA4 gene, rendering their molecular test inconclusive (16%). Patients 8, 14, 7, 11, 16, 21, and 39 were evaluated using a small Stargardt disease panel that included the ABCA4 (ID: 24, OMIM: 601691), ELOVL4 (ID: 6785, OMIM: 605512), and PROM1 (ID: 8842, OMIM: 604365) genes, although no pathogenic variants in those other genes were found. Patients 26 and 32 were assessed for the ABCA4, ELOVL4, PROM1, BEST1 (ID: 7439, OMIM: 607854), and PRPH2 (ID: 5961, OMIM: 179605) genes and patient 36 for the ABCA4, ELOVL4, PROM1, BEST1, PRPH2, CDH3 (ID: 1001, OMIM: 114021), EFEMP1 (ID: 2202, OMIM: 601548), IMPG1 (ID: 3617, OMIM: 602870), IMPG2 (ID:50939, OMIM: 607056) and TIMP3 (ID: 7078, OMIM: 188826) genes. No pathogenic variants were found in the other genes.
In the group of 40 conclusive patients, five (12.5%) had homozygous pathogenic variants in the ABCA4 gene, with 35 being compound heterozygous variants (85.5%); segregation analysis data were available for 26 patients (65%) [32]. Segregation analysis confirmed complex alleles in 20 patients (50% of 40 patients), all of whom showed three pathogenic variants; confirmation that the third variation was in trans rendered these cases conclusive for molecular diagnosis. Although patient 27 showed three pathogenic variants, segregation analysis data were not available for this patient.
The following six complex alleles were found: p.[Leu541Pro; Arg1443His]; p.[Leu541Pro; Ala1038Val]; p.[Ser1642Arg; Val1682_Val1686del]; p.[Pro1761Arg; Arg2106Cys] and p.[Ala1038Val; Gly1961Glu] p.[Ala1038Val; Met669Ile]. All of these alleles were confirmed with segregation analysis to be in cis.
One novel variant, p.Ala2084Pro (Table 5), not present in genetic variant (HGMD, ClinVar and LOVD) or population (1000 Genomes and ExAC) databases was found. Pathogenicity prediction software (CADD, SIFT, MutationTaster, and PolyPhen-2) classified the variant p.Ala2084Pro as disease-causing. It was found in one allele of patient 35 and was considered pathogenic to conclude the genotype test. In addition, three variants that had previously been described without pathogenic classification or with conflicting pathogenicity classifications (p.Met669Ile, p.Arg152Gln, and p.Thr897Ile) were analyzed, as outlined in Table 5 [33-36].
Table 5. Novel variant and variants with uncertain significance.
| Patient | 35 | 38 | 32 | 27 |
|---|---|---|---|---|
|
Nucleotide change |
c.6250G>C |
c.2007G>C |
c.455G>A |
c.2690C>T |
|
Protein change |
p.Ala2084Pro |
p.Met669Ile |
p.Arg152Gln |
p.Thr897Ile |
|
dbSNP |
N/A |
N/A |
rs62646862 |
rs61749440 |
|
ClinVar |
N/A |
N/A |
Conflicting |
Conflicting |
|
CADD score/pathogenicity |
34/
Deleterious |
23.9/
Deleterious |
21.5/
Deleterious |
23.6/
Deleterious |
|
MutationTaster score/pathogenicity |
1/
Disease-causing |
1/
Disease-causing |
0.775/
Disease-causing |
0.989/
Disease-causing |
|
SIFT score/pathogenicity |
0.019/
Deleterious |
0.189/
Tolerated |
0.124/
Tolerated |
0.188/
Tolerated |
|
PolyPhen-2 HumVar score/pathogenicity |
1.000/
Damage |
0.413/
Benign |
0.015/
Benign |
0.994/
Damage |
|
1000 genomes |
N/A |
N/A |
0,00159744 |
0,00159744 |
|
ExAC |
N/A |
N/A |
0,0024 |
0,0011 |
| References | Novel variant | 35 | 33 and 34 | 32 |
N/A, Not available
Among the four families (patients 4 and 6 (siblings), 30, 33, and 40) with homozygous variants (Table 1, Table 3, Table 4) only patient 30 denied a history of consanguinity. The most frequent variant in this population was p.Arg602Trp (12/100 alleles). It was present in two cases with phenotype type I, five with type II, and five with type III.
Other frequent variants among patients with Stargardt disease, such as p.Gly1961Glu, p.[Leu541Pro; Ala1038Val], and c.5461–10T>C, were also found in the study population [15,37]. The variants identified in this study are distributed throughout the ABCA4 gene, including intronic and exonic regions (Table 6, Table 7), and no preferential region for the occurrence of the variants was observed.
Table 6. Distribution of pathogenic variants in the ABCA4 gene.
| Complex alleles | |||
|---|---|---|---|
| Nucleotide Change | Protein Change | Location | Allele† |
| c.4926C>G |
p.Ser1642Arg |
35 |
7 |
| c.5044_5058del15‡ |
p.Val1682_Val1686del |
36 |
|
| c.1622T>C |
p.Leu541Pro |
12 |
7 |
| c.4328G>A |
p.Arg1443His |
29 |
|
| c.5282C>G |
p.Pro1761Arg |
37 |
2 |
| c.6316C>T |
p.Arg2106Cys |
46 |
|
| c.5882 G>A |
p.Gly1961Glu |
42 |
1 |
| c.3113 C>T |
p.Ala1038Val |
21 |
|
| c.1622T>C |
p.Leu541Pro |
12 |
3 |
| c.3113C>T |
p.Ala1038Val |
21 |
|
| c.3113C>T | p.Ala1038Val | 21 | 1 |
† Number of alleles that each complex allele has apear.
Table 7. Distribution of pathogenic variants in the ABCA4 gene.
| Single alleles | |||
|---|---|---|---|
| Nucleotide Change | Protein Change | Location | Allele† |
| c.32T>C |
p.Leu11Pro |
1 |
1 |
| c.66G>T |
p.Lys22Asn |
1 |
2 |
| c.70C>T |
p.Arg24Cys |
2 |
1 |
| c.223T>G |
p.Cys75Gly |
3 |
1 |
| c.286A>G |
p.Asn96Asp |
3 |
5 |
| c.455G>A |
p.Arg152Gln |
5 |
1 |
| c.634C>T |
p.Arg212Cys |
6 |
2 |
| c.658C>T |
p.Arg220Cys |
6 |
1 |
| c.1364 T>A |
p.Leu455Gln |
11 |
1 |
| c.1648G>A |
p.Gly550Arg |
12 |
1 |
| c.1804C>T |
p.Arg602Trp |
13 |
12 |
| c.2345G>A |
p.Trp782* |
15 |
2 |
| c.2743G>A |
p.Asp915Asn |
19 |
1 |
| c.2791G>A |
p.Val931Met |
19 |
1 |
| c.2894 A>G |
p.Asn965Ser |
19 |
1 |
| c.3056C>T |
p.Thr1019Met |
21 |
1 |
| c.3329–2A>T |
p.? |
Intron 22 |
2 |
| c.3386G>T |
p.Arg1129Leu |
23 |
3 |
| c.3862+1G>A |
p.? |
Intron 26 |
3 |
| c.3898C>T |
p.Arg1300* |
27 |
1 |
| c.4003_4004delCC |
p.Pro1335Argfs*86 |
27 |
1 |
| c.4340A>T |
p.Glu1447Val |
29 |
1 |
| c.4457C>T |
p.Pro1486Leu |
30 |
1 |
| c.4720G>T |
p.Glu1574* |
33 |
1 |
| c.5044_5058del15 |
p.Val1682_Val1686del |
36 |
1 |
| c.5381C>A |
p.Ala1794Asp |
38 |
2 |
| c.5461–10T>C |
p.[Thr1821Valfs*13, Thr1821Aspfs*6] |
Intron 38 |
1 |
| c.5714+5G>A |
p.? |
Intron 40 |
2 |
| c.6005+1G>T |
p.? |
Intron 43 |
1 |
| c.6079C>T |
p.Leu2027Phe |
44 |
2 |
| c.6088C>T |
p.Arg2030* |
44 |
3 |
| c.6112C>T |
p.Arg2038Trp |
44 |
1 |
|
c.6250G>C |
p.Ala2084Pro |
45 |
1 |
| c.6320G>A | p.Arg2107His | 46 | 1 |
Novel vatiant.† Number of alleles that each variant has apear.Patient 27 was excluded of this table due to the absence of segregation analysis. FAF, Fundus autofluorescence; Pt, Patient; N/A, not available; HM, Hand movement; FC, Finger counting. Novel variant. †Homozygous variants. ‡c.5044_del15bp, nonframeshift deletion (c.5044_5058delGTTGCCATCTGCGTG). Families: Siblings: 4 and 6; 17 and 20; 18 and 19; 41 and 49. Mother 44 and daughter 43. Mother 45 and daughter 36. § Consanguinity •Patient 27 presented with 3 pathogenic variants, but without segregation analysis it was impossible to determinate which of them are combined in cis.
Discussion
The ABCA4 gene is highly polymorphic, and there are a large number of benign variants in addition to the more than 900 pathogenic variants that have been described to date [14,15,17]. The allelic heterogeneity and distribution of pathogenic variants along the 50 exons and introns of this gene demonstrate the complexity of the encoded protein and its role in Stargardt disease (Table 6, Table 7) [38]. Moreover, the large number of variants complicates predictions of disease-causing mutations [39]. Another characteristic of the ABCA4 gene is the presence of pathogenic variants in cis (Table 6) [32,40]. For this reason, segregation analysis is crucial for clarifying the family origin of each pathogenic variant in compound heterozygotes and for reaching a conclusive molecular diagnosis.
Phenotypic severity is directly linked to the protein damage caused by a pathogenic variant [12,15,22], and clinical heterogeneity can be explained by different combinations of pathogenic variants in the ABCA4 gene [40,41]. Therefore, knowledge of variant pathogenicity may affect disease prognosis and genetic counseling in each case [15]. Some studies have addressed the presence of homozygous mutations in the ABCA4 gene to assess the effect of a pathogenic variant alone [6,15].
Among the four families with homozygous pathogenic variants in this study, three could be classified based on their phenotypic characteristics. Patients 4 and 6, who were siblings, carried the homozygous variant p.Asn96Asp and were classified as having the type III phenotype 16 and 33 years, respectively, after symptom onset. Despite denying family consanguinity, patient 30 carried the homozygous pathogenic variant p.Arg2030* and was also classified with the type III phenotype at 38 years of age. These clinical characteristics suggest that those changes are associated with a worse disease prognosis. Conversely, the phenotype of patient 33, with the homozygous intronic variant c.3862+1G>A, at diagnosis at 10 years of age was type I. The pathogenic variant c.3862+1G>A causes a splicing defect, and this intronic variant has been found in cases of cone–rod degeneration and retinitis pigmentosa in addition to cases of Stargardt disease [42,43]. Follow-up of the phenotypic progression of patient 33 will indicate the prognosis for this variant.
The pathogenic variant most commonly found in this group was p.Arg602Trp, which was present in 12 patients (12 alleles). Wiszniewski et al. described the missense p.Arg602Trp as being responsible for mislocalization of the ABCA4 protein, resulting in the absence of physiologic protein function [21]. These authors further suggested that this missense variant is a severe variant due to the early age of symptom onset when associated with another mislocalization or nonsense variant [21]. Symptoms in patient 5, who presented p.Arg1300* and p.Arg602Trp, started at age 7, with phenotype II showing after 10 years (Table 2). However, the results suggest that the severity of the disease does not depend exclusively on the pathogenic effect of p.Arg602Trp. Patients with this variant exhibited all three phenotypes, demonstrating that the combination of pathogenic variant effects in both alleles is important for determining the phenotype (or the Stargardt disease severity). This dependence is illustrated in the following cases. Patient 46 presented the first symptoms at age 9, and after only 1 year, her phenotype was type II (Table 2). She carried p.Arg602Trp and c.5461–10T>C variants, and the latter is described as a severe pathogenic variant [37] because it leads to abnormal mRNA and ABCA4 protein truncation. In addition, patients homozygous for this variant have a severe phenotype. In contrast, patient 28 presented two missense variants (p.Arg24Cys and p.Arg602Trp). His first symptoms were at age 16, and after 10 years, his phenotype was type I (Table 1). Although the age of onset of the disease may be related to its severity, the natural history of the disease may better reflect the pathogenicity of variants.
The variant p.Gly1961Glu is associated with milder disease and causes late onset of visual symptoms, with a phenotype that does not evolve to type III [44]. This characteristic was also observed in patients 1, 43, 48, and 51, who carried this variant and showed symptoms after 10 years of age. All patients were classified as type I based on the fundus autofluorescence examinations, and patient 1 maintained these phenotypic characteristics after 18 years of the disease (Table 1). The phenotypic characteristics of these four patients corroborate the hypothesis that variant p.Gly1961Glu causes milder disease.
Variants c.5714+5G>A and c.6005+1G>T were present in patient 15. According to Burke et al., the latter is likely to completely abolish splicing and to cause a frameshift that results in severe protein dysfunction [45]. Conversely, variant c.5714+5G>A is associated with a mild pathogenic effect and a later median age of onset [15,46,47]; nonetheless, Cremers et al. proposed that this variant has a moderately severe effect [19]. Patient 15 was classified as the type III phenotype at 28 years after symptom onset (Table 3), suggesting a severe effect of those variants.
Analysis of the relationship between the genetic variants and the phenotype of each patient showed that sibling patients had the same mutations and phenotypic classifications. Conversely, the two mother-and-daughter families showed an autosomal dominant inheritance pattern, which suggests that another gene is responsible for their Stargardt disease, even though molecular testing confirmed Stargardt disease linked to the ABCA4 gene. Because of different mutation combinations, the mother and daughter exhibited different phenotypes. The high frequency of variants in ABCA4 may have led to the identification of three different pathogenic variants in the same family.
This study reports pathogenic variants in the ABCA4 gene in Brazilian patients with a clinical diagnosis of Stargardt disease. Thus far, little is known about the genetic characteristics of the Brazilian population with retinal dystrophy. The presence of variants previously described and with a founder effect known for ethnic group-specific populations [48] confirms the miscegenation of Brazilians, including p.Arg1129Leu of Spanish origin [42,49] and p.[Leu541Pro; Ala1038Val] and p.Gly1961Glu of German origin [47,50].
Six complex alleles were found in this study (Table 6). These complex alleles were present in 20 patients distributed among all three phenotypes (Table 1, Table 2, Table 3) and no phenotype–genotype correlation was observed. Although the complex allele p.[Leu541Pro; Ala1038Val] has reportedly high frequency, the allele was present in only three patients in this study [50]. Conversely, the variant p.Leu541Pro formed a complex allele with p.Arg1443His in patients 12, 23, 31, 34, 40, and 47 [6]. This high frequency of complex alleles (20 of 40 conclusive cases) and the possibility of different variant combinations in cis emphasize the importance of segregation analysis for a correct molecular diagnosis [32].
NGS has been found to be effective for the molecular diagnosis of patients with Stargardt disease [51] and was used for molecular diagnosis in all cases reported in this study. These patients were assessed using a gene panel for Stargardt disease or macular dystrophies. In this study, the rate of conclusive molecular diagnosis was 80% (40/50), higher than the rate reported in other studies with patients with Stargardt disease using NGS (ranging from 50% to 60%) [51-54].
The negative and inconclusive cases may be explained by mutations in non-sequenced regions, such as intronic, untranslated, and regulatory regions, and/or a lack of knowledge of the pathogenicity of new variants in the ABCA4 gene. In addition, other genes not analyzed or not yet related to the disease might cause these phenotypes [14]. New studies are evaluating other regions of the ABCA4 gene that were previously considered irrelevant for protein formation, including deep-intronic regions [52,53], in an effort to validate variants as disease causing and to improve molecular diagnosis [14,55,56]. Genes other than ABCA4 have been evaluated to clarify inconclusive cases [35,57-59].
The importance of a conclusive molecular diagnosis is directly related to new treatments for retinal dystrophies [15]. Identifying pathogenic variants responsible for disease enables the inclusion of patients in clinical trials and renders them eligible for future commercial therapies [14,60].
CONCLUSION
NGS is effective for the molecular diagnosis of genetic diseases, and this approach specifically allowed a conclusive diagnosis in 80% (40/50) of the patients. The ABCA4 gene does not show a preferential region for pathogenic variants; thus, the diagnosis of Stargardt disease depends on broader analysis of the gene. The most common pathogenic variants in ABCA4 described in the literature were also found in the Brazilian patients in this study. Some genotype–phenotype correlations were found, but more studies on the progression of Stargardt disease will help us understand the pathogenicity of variants in genes.
Acknowledgments
This research was supported by research funds from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; Process No. 2012/50454–5). The authors are grateful for fellowship support from CAPES (FLM, MVS and KAC), CNPq (RFS) and FAPESP (RPM). The authors sincerely appreciate the help provided by R. Allikmets during the analysis of the results. DECLARATION OF INTERESTS: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article. Despite the similarity in names, Molecular Vision Laboratory is not related to the journal, Molecular Vision. The Molecular Vision Laboratory, Inc., is a medical diagnostic laboratory and subsidiary of Centrillion Technology Holdings Ltd. There are no financial or intellectual relationships to be disclosed.
References
- 1.Stargardt K. Ueber familiare, progressive Degeneration in der Makulagegend des Auges. Albrecht von Graefes Arch Klin Exp Ophthal. 1909;71:534–49. [Google Scholar]
- 2.Fishman GA. Fundus flavimaculatus. A clinical classification. Arch Ophthalmol. 1976;94:2061–7. doi: 10.1001/archopht.1976.03910040721003. [DOI] [PubMed] [Google Scholar]
- 3.Allikmets R. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;17:122. doi: 10.1038/ng0997-122a. [DOI] [PubMed] [Google Scholar]
- 4.Gomes NL, Greenstein VC, Carlson JN, Tsang SH, Smith RT, Carr RE, Hood DC, Chang S. A comparison of fundus autofluorescence and retinal structure in patients with Stargardt disease. Invest Ophthalmol Vis Sci. 2009;50:3953–9. doi: 10.1167/iovs.08-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke TR, Tsang SH. Allelic and phenotypic heterogeneity in ABCA4 mutations. Ophthalmic Genet. 2011;32:165–74. doi: 10.3109/13816810.2011.565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujinami K, Sergouniotis PI, Davidson AE, Mackay DS, Tsunoda K, Tsubota K, Robson AG, Holder GE, Moore AT, Michaelides M, Webster AR. The Clinical Effect of Homozygous ABCA4 Alleles in 18 Patients. Ophthalmology. 2013;120:2324–31. doi: 10.1016/j.ophtha.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Passerini I, Sodi A, Giambene B, Mariottini A, Menchini U, Torricelli F. Novel mutations in of the ABCR gene in Italian patients with Stargardt disease. Eye (Lond) 2010;24:158–64. doi: 10.1038/eye.2009.35. [DOI] [PubMed] [Google Scholar]
- 8.Sun H, Nathans J. Stargardt’s ABCR is localized to the disc membrane of retinal rod outer segments. Nat Genet. 1997;17:15–6. doi: 10.1038/ng0997-15. [DOI] [PubMed] [Google Scholar]
- 9.Molday LL, Rabin AR, Molday RS. ABCR expression in foveal cone photoreceptors and its role in Stargardt macular dystrophy. Nat Genet. 2000;25:257–8. doi: 10.1038/77004. [DOI] [PubMed] [Google Scholar]
- 10.Cideciyan AV, Aleman TS, Swider M, Schwartz SB, Steinberg JD, Brucker AJ, Maguire AM, Bennett J, Stone EM, Jacobson SG. Mutations in ABCA4 result in accumulation of lipofuscin before slowing of the retinoid cycle: a reappraisal of the human disease sequence. Hum Mol Genet. 2004;13:525–34. doi: 10.1093/hmg/ddh048. [DOI] [PubMed] [Google Scholar]
- 11.Weng J, Mata NL, Azarian SM, Tzekov RT, Birch DG, Travis GH. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 12.Koenekoop RK. The gene for Stargardt disease, ABCA4, is a major retinal gene: a mini-review. Ophthalmic Genet. 2003;24:75–80. doi: 10.1076/opge.24.2.75.13996. [DOI] [PubMed] [Google Scholar]
- 13.Molday RS, Zhong M, Quazi F. The role of the photoreceptor ABC transporter ABCA4 in lipid transport and Stargardt macular degeneration. Biochim Biophys Acta. 2009;1791:573–83. doi: 10.1016/j.bbalip.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun TA, Mullins RF, Wagner AH, Andorf JL, Johnston RM, Bakall BB, Deluca AP, Fishman GA, Lam BL, Weleber RG, Cideciyan AV, Jacobson SG, Sheffield VC, Tucker BA, Stone EM. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22:5136–45. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis SS, Bax NM, Zernant J, Allikmets R, Fritsche LG, den Dunnen JT, Ajmal M, Hoyng CB, Cremers FP. In Silico Functional Meta-Analysis of 5,962 ABCA4 Variants in 3,928 Retinal Dystrophy Cases. Hum Mutat. 2017;38:400–8. doi: 10.1002/humu.23165. [DOI] [PubMed] [Google Scholar]
- 16.Michaelides M, Chen LL, Brantley MA, Jr, Andorf JL, Isaak EM, Jenkins SA, Holder GE, Bird AC, Stone EM, Webster AR. ABCA4 mutations and discordant ABCA4 alleles in patients and siblings with bull’s-eye maculopathy. Br J Ophthalmol. 2007;91:1650–5. doi: 10.1136/bjo.2007.118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanna P, Strauss RW, Fujinami K, Michaelides M. Stargardt disease: clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2017;101:25–30. doi: 10.1136/bjophthalmol-2016-308823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan J, Gerber S, Larget-Piet D, Rozet JM, Dollfus H, Dufier JL, Odent S, Postel-Vinay A, Janin N, Briard ML, Frézal J, Munnich A. A gene for Stargardt’s disease (fundus flavimaculatus) maps to the short arm of chromosome 1. Nat Genet. 1993;5:308–11. doi: 10.1038/ng1193-308. [DOI] [PubMed] [Google Scholar]
- 19.Cremers FP, van de Pol DJ, van Driel M, den Hollander AI, van Haren FJ, Knoers NV, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, Pinckers AJ, Deutman AF, Hoyng CB. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum Mol Genet. 1998;7:355–62. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 20.Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000;67:960–6. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiszniewski W, Zaremba CM, Yatsenko AN, Jamrich M, Wensel TG, Lewis RA, Lupski JR. ABCA4 mutations causing mislocalization are found frequently in patients with severe retinal dystrophies. Hum Mol Genet. 2005;14:2769–78. doi: 10.1093/hmg/ddi310. [DOI] [PubMed] [Google Scholar]
- 22.Lee W, Schuerch K, Zernant J, Collison FT, Bearelly S, Fishman GA, Tsang SH, Sparrow JR, Allikmets R. Genotypic spectrum and phenotype correlations of ABCA4-associated disease in patients of south Asian descent. Eur J Hum Genet. 2017;25:735–43. doi: 10.1038/ejhg.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 28.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. Vol 11. United States2014. p. 361–2. [DOI] [PubMed] [Google Scholar]
- 31.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 32.Salles MV, Motta FL, Dias da Silva E, Varela P, Costa KA, Filippelli-Silva R, Martin RP, Chiang JP, Pesquero JB, Sallum JMF. Novel Complex ABCA4 Alleles in Brazilian Patients With Stargardt Disease: Genotype-Phenotype Correlation. Invest Ophthalmol Vis Sci. 2017;58:5723–30. doi: 10.1167/iovs.17-22398. [DOI] [PubMed] [Google Scholar]
- 33.Simonelli F, Testa F, de Crecchio G, Rinaldi E, Hutchinson A, Atkinson A, Dean M, D’Urso M, Allikmets R. New ABCR mutations and clinical phenotype in Italian patients with Stargardt disease. Invest Ophthalmol Vis Sci. 2000;41:892–7. [PubMed] [Google Scholar]
- 34.Schulz HL, Grassmann F, Kellner U, Spital G, Ruther K, Jagle H, Hufendiek K, Rating P, Huchzermeyer C, Baier MJ, Weber BH, Stohr H. Mutation Spectrum of the ABCA4 Gene in 335 Stargardt Disease Patients From a Multicenter German Cohort-Impact of Selected Deep Intronic Variants and Common SNPs. Invest Ophthalmol Vis Sci. 2017;58:394–403. doi: 10.1167/iovs.16-19936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Downes SM, Packham E, Cranston T, Clouston P, Seller A, Nemeth AH. Detection rate of pathogenic mutations in ABCA4 using direct sequencing: clinical and research implications. Arch Ophthalmol. 2012;130:1486–90. doi: 10.1001/archophthalmol.2012.1697. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Zhang VW, Feng Y, Tian X, Li FY, Truong C, Wang G, Chiang PW, Lewis RA, Wong LJ. Dependable and efficient clinical utility of target capture-based deep sequencing in molecular diagnosis of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014;55:6213–23. doi: 10.1167/iovs.14-14936. [DOI] [PubMed] [Google Scholar]
- 37.Sangermano R, Bax NM, Bauwens M, van den Born LI, De Baere E, Garanto A, Collin RW, Goercharn-Ramlal AS, den Engelsman-van Dijk AH, Rohrschneider K, Hoyng CB, Cremers FP, Albert S. Photoreceptor Progenitor mRNA Analysis Reveals Exon Skipping Resulting from the ABCA4 c.5461–10T→C Mutation in Stargardt Disease. Ophthalmology. 2016;123:1375–85. doi: 10.1016/j.ophtha.2016.01.053. [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Tsybovsky Y, Kolesnikov AV, Rozanowska M, Swider M, Schwartz SB, Stone EM, Palczewska G, Maeda A, Kefalov VJ, Jacobson SG, Cideciyan AV, Palczewski K. Protein misfolding and the pathogenesis of ABCA4-associated retinal degenerations. Hum Mol Genet. 2015;24:3220–37. doi: 10.1093/hmg/ddv073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allikmets R. Simple and complex ABCR: genetic predisposition to retinal disease. Am J Hum Genet. 2000;67:793–9. doi: 10.1086/303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis RA, Shroyer NF, Singh N, Allikmets R, Hutchinson A, Li Y, Lupski JR, Leppert M, Dean M. Genotype/Phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet. 1999;64:422–34. doi: 10.1086/302251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maia-Lopes S, Aguirre-Lamban J, Castelo-Branco M, Riveiro-Alvarez R, Ayuso C, Silva ED. ABCA4 mutations in Portuguese Stargardt patients: identification of new mutations and their phenotypic analysis. Mol Vis. 2009;15:584–91. [PMC free article] [PubMed] [Google Scholar]
- 42.Riveiro-Alvarez R, Lopez-Martinez MA, Zernant J, Aguirre-Lamban J, Cantalapiedra D, Avila-Fernandez A, Gimenez A, Lopez-Molina MI, Garcia-Sandoval B, Blanco-Kelly F, Corton M, Tatu S, Fernandez-San Jose P, Trujillo-Tiebas MJ, Ramos C, Allikmets R, Ayuso C. Outcome of ABCA4 disease-associated alleles in autosomal recessive retinal dystrophies: retrospective analysis in 420 Spanish families. Ophthalmology. 2013;120:2332–7. doi: 10.1016/j.ophtha.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briggs CE, Rucinski D, Rosenfeld PJ, Hirose T, Berson EL, Dryja TP. Mutations in ABCR (ABCA4) in patients with Stargardt macular degeneration or cone-rod degeneration. Invest Ophthalmol Vis Sci. 2001;42:2229–36. [PubMed] [Google Scholar]
- 44.Zernant J, Lee W, Collison FT, Fishman GA, Sergeev YV, Schuerch K, Sparrow JR, Tsang SH, Allikmets R. Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J Med Genet. 2017;54:404–12. doi: 10.1136/jmedgenet-2017-104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burke TR, Allikmets R, Smith RT, Gouras P, Tsang SH. Loss of peripapillary sparing in non-group I Stargardt disease. Exp Eye Res. 2010;91:592–600. doi: 10.1016/j.exer.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fakin A, Robson AG, Fujinami K, Moore AT, Michaelides M, Pei-Wen Chiang J. G EH, Webster AR. Phenotype and Progression of Retinal Degeneration Associated With Nullizigosity of ABCA4. Invest Ophthalmol Vis Sci. 2016;57:4668–78. doi: 10.1167/iovs.16-19829. [DOI] [PubMed] [Google Scholar]
- 47.Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, Jurklies B, Lorenz B, Scholl HP, Apfelstedt-Sylla E, Weber BH. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet. 2000;67:800–13. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zernant J, Xie YA, Ayuso C, Riveiro-Alvarez R, Lopez-Martinez MA, Simonelli F, Testa F, Gorin MB, Strom SP, Bertelsen M, Rosenberg T, Boone PM, Yuan B, Ayyagari R, Nagy PL, Tsang SH, Gouras P, Collison FT, Lupski JR, Fishman GA, Allikmets R. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum Mol Genet. 2014;23:6797–806. doi: 10.1093/hmg/ddu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valverde D, Riveiro-Alvarez R, Bernal S, Jaakson K, Baiget M, Navarro R, Ayuso C. Microarray-based mutation analysis of the ABCA4 gene in Spanish patients with Stargardt disease: evidence of a prevalent mutated allele. Mol Vis. 2006;12:902–8. [PubMed] [Google Scholar]
- 50.Sciezynska A, Ozieblo D, Ambroziak AM, Korwin M, Szulborski K, Krawczynski M, Stawinski P, Szaflik J, Szaflik JP, Ploski R, Oldak M. Next-generation sequencing of ABCA4: High frequency of complex alleles and novel mutations in patients with retinal dystrophies from Central Europe. Exp Eye Res. 2016;145:93–9. doi: 10.1016/j.exer.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 51.Fujinami K, Zernant J, Chana RK, Wright GA, Tsunoda K, Ozawa Y, Tsubota K, Webster AR, Moore AT, Allikmets R, Michaelides M. ABCA4 gene screening by next-generation sequencing in a British cohort. Invest Ophthalmol Vis Sci. 2013;54:6662–74. doi: 10.1167/iovs.13-12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weisschuh N, Mayer AK, Strom TM, Kohl S, Glockle N, Schubach M, Andreasson S, Bernd A, Birch DG, Hamel CP, Heckenlively JR, Jacobson SG, Kamme C, Kellner U, Kunstmann E, Maffei P, Reiff CM, Rohrschneider K, Rosenberg T, Rudolph G, Vamos R, Varsanyi B, Weleber RG, Wissinger B. Mutation Detection in Patients with Retinal Dystrophies Using Targeted Next Generation Sequencing. PLoS One. 2016;11:e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang JP, Trzupek K. The current status of molecular diagnosis of inherited retinal dystrophies. Curr Opin Ophthalmol. 2015;26:346–51. doi: 10.1097/ICU.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 54.O’Sullivan J, Mullaney BG, Bhaskar SS, Dickerson JE, Hall G, O’Grady A, Webster A, Ramsden SC, Black GC. A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J Med Genet. 2012;49:322–6. doi: 10.1136/jmedgenet-2012-100847. [DOI] [PubMed] [Google Scholar]
- 55.Bax NM, Sangermano R, Roosing S, Thiadens AA, Hoefsloot LH, van den Born LI, Phan M, Klevering BJ, Westeneng-van Haaften C, Braun TA, Zonneveld-Vrieling MN, de Wijs I, Mutlu M, Stone EM, den Hollander AI, Klaver CC, Hoyng CB, Cremers FP. Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum Mutat. 2015;36:43–7. doi: 10.1002/humu.22717. [DOI] [PubMed] [Google Scholar]
- 56.Bauwens M, De Zaeytijd J, Weisschuh N, Kohl S, Meire F, Dahan K, Depasse F, De Jaegere S, De Ravel T, De Rademaeker M, Loeys B, Coppieters F, Leroy BP, De Baere E. An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in Belgian Stargardt patients. Hum Mutat. 2015;36:39–42. doi: 10.1002/humu.22716. [DOI] [PubMed] [Google Scholar]
- 57.Jonsson F, Westin IM, Osterman L, Sandgren O, Burstedt M, Holmberg M, Golovleva I. ATP-binding cassette subfamily A, member 4 intronic variants c.4773+3A>G and c.5461–10T>C cause Stargardt disease due to defective splicing. Acta Ophthalmol. 2018 doi: 10.1111/aos.13676. [DOI] [PubMed] [Google Scholar]
- 58.Imani S, Cheng J, Shasaltaneh MD, Wei C, Yang L, Fu S, Zou H, Khan MA, Zhang X, Chen H, Zhang D, Duan C, Lv H, Li Y, Chen R, Fu J. Genetic identification and molecular modeling characterization reveal a novel PROM1 mutation in Stargardt4-like macular dystrophy. Oncotarget. 2017;9:122–41. doi: 10.18632/oncotarget.22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donato L, Scimone C, Rinaldi C, Aragona P, Briuglia S, D’Ascola A, D’Angelo R, Sidoti A. Stargardt Phenotype Associated With Two ELOVL4 Promoter Variants and ELOVL4 Downregulation: New Possible Perspective to Etiopathogenesis? Invest Ophthalmol Vis Sci. 2018;59:843–57. doi: 10.1167/iovs.17-22962. [DOI] [PubMed] [Google Scholar]
- 60.Schindler EI, Nylen EL, Ko AC, Affatigato LM, Heggen AC, Wang K, Sheffield VC, Stone EM. Deducing the pathogenic contribution of recessive ABCA4 alleles in an outbred population. Hum Mol Genet. 2010;19:3693–701. doi: 10.1093/hmg/ddq284. [DOI] [PMC free article] [PubMed] [Google Scholar]