Abstract
Background
There has been a campaign by the National Education on Sleeping Habits and Living Environment, to reduce the incidence of sudden infant death syndrome (SIDS). However, more than 100 infants die suddenly and unexplainably before the age of 1 year in Korea. Long QT syndrome (LQTS), an inheritable cardiac disease, has been reported to likely be associated with up to 14% of SIDS cases. However, genetic studies of the association between SIDS and LQTS have not yet been conducted in Korea.
Methods
We conducted genetic analysis using genomic DNA extracted from paraffin-embedded tissue blocks from 200 SIDS cases autopsied between 2005 and 2013. We analyzed the following genetic mutations associated with LQTS, KCNQ1, SCN5A, KCNE1, KCNE2, KCNJ2, and CAV3.
Results
Of the 200 SIDS cases, 58% involved male infants (116 male and 84 female infants, respectively), the mean age was 140 days (median, 107 days; range, 24–270 days), and they were all of Asian-Korean ethnicity. SIDS IA category criteria comprised 45 cases (22.5%) while the rest were SIDS IB. Fifteen infants (7.5%) had R1193Q in SCN5A, of doubtful pathogenicity, and no pathogenic LQTS variants were observed.
Conclusion
This genetic investigation of LQTS in SIDS showed a low diagnostic yield. These findings suggest that LQTS molecular autopsy could be cautiously conducted in selected cases with family involvement to improve the available genetic counseling information. Meanwhile, a national SIDS registry should be established to document and evaluate the genetic risk of SIDS in Korea.
Keywords: Sudden Infant Death Syndrome, Long QT Syndrome, Molecular Autopsy, Genetic
Graphical Abstract
INTRODUCTION
Sudden unexplained infant deaths (SUIDs) are defined as the death of an infant less than 1-year-old in which investigation, autopsy, medical history review, and appropriate laboratory testing fail to identify a specific cause of death, which includes cases that meet the definition of sudden infant death syndrome (SIDS).1 We previously reviewed 355 cases of SIDS in Korea, focusing on the sleep environment.2 Subsequently, the campaign for safe sleep environments including “back to sleep” was widely launched but more than 100 infants still die suddenly and unexplainably before the age of 1 year in Korea.
Long QT syndrome (LQTS) is a group of inheritable primary electric diseases of the heart. The disease cluster was first noted in a family where several children with congenital hearing loss exhibited QT prolongation in electrocardiography (ECG) examinations, and experienced recurrent syncope and sudden cardiac death, with an autosomal recessive inheritance (Jervelle and Lange-Nielsen [JLN] syndrome).3 LQTS is now understood to be a cardiac channelopathy involving ventricular repolarization delay due to a prolonged duration of the myocardial action potential. Postmortem genetic testing or molecular autopsy has revealed a strong association between LQTS variation and SIDS.4,5 It has been reported that 5.2%–14.0% of SIDS cases may be linked to LQTS,6,7 which affects the cardiac conduction system. Therefore, postmortem genetic testing has recently been recommended as a routine procedure in the autopsy of SIDS cases.6,8 Unfortunately, the molecular diagnosis of postmortem examinations is not yet a routine practice in Korea. In this study, we retrospectively reviewed 200 SIDS cases from a genetic viewpoint to determine the number of cases in Korea that may have been associated with LQTS variation and accumulate information to support the value of the availability of postmortem genetic testing for SIDS in Korea.
METHODS
We designed a retrospective study to test for the presence of genetic risk factors involving LQTS in SIDS in Korea. The cases analyzed were retrieved from a nationwide pool of infant deaths recorded between January 2005 and December 2013. After a thorough review of the police investigative and autopsy reports as well as a histological re-examination, 200 cases of SIDS IA and IB were selected according to Krous et al.9 The SIDS IA group was defined based on the following characteristics: cases aged 21 to 270 days at the time of death with a normal clinical history, term delivery, normal growth and development, no familial history of sudden unexplained deaths, and no suspicious scenes. A comprehensive postmortem investigation including toxicological, microbiological, vitreous chemistry, or metabolic screening studies with death scene investigation by police authorities was conducted. Category IB consisted of infant deaths that met the general definition as well as all the criteria for category IA except for this investigation. Infants of the SIDS category II, which includes deaths outside the SIDS I age range (21 days to 9 months) and cases where accidental asphyxia, were excluded in this study.9 The obtained data included the position in which the deceased was found.
The genomic DNA from 200 SIDS patients was extracted from the organ (the heart or liver) tissue paraffin-embedded blocks using the QIAamp DNA FFPE tissue kit (QIAGEN, Hilden, Germany). Owing to expected low DNA yields from the paraffin-embedded tissue samples from each patient, all DNA quantities were determined using a NANODROP™ LITE spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Table 1 shows the 85 DNA variants from 6 genes (KCNQ1, SCN5A, KCNE1, KCNE2, KCNJ2, and CAV3), ascertained from the literature using PubMed (www.ncbi.nlm.nih.gov) based on their reported involvement in SIDS,10,11,12,13,14,15,16,17,18,19,20 which were investigated using a real-time polymerase chain reaction (PCR) single nucleotide polymorphism (SNP) assay.
Table 1. LQT-related genes investigated in this study.
| Genes | Protein change | Mutation point | Frequency |
|---|---|---|---|
| CAV3 | T78M | c233t | 0 |
| V14L | g40c | 0 | |
| T78K | c233a | 0 | |
| KCNH2 | G604S | g1825a | 0 |
| G628S | g1882a | 0 | |
| R1047L | g3140t | 0 | |
| KCNQ1 | P117L | c350t | 0 |
| E146K | g436a | 0 | |
| KCNE1 | G25V | g74t | 0 |
| G60D | g179a | 0 | |
| SCN5A | A997D | c2990a | 0 |
| A997S | g2989t | 0 | |
| A997T | g2989a | 0 | |
| C1004R | t3010c | 0 | |
| D1041N | g3121a | 0 | |
| E1015K | g3043a | 0 | |
| E1784K | g5350a | 0 | |
| F1293S | t3878c | 0 | |
| F1473S | t4418c | 0 | |
| F1522Y | t4565a | 0 | |
| F919L | c2757a | 0 | |
| G514C | g1540t | 0 | |
| G615E | g1844a | 0 | |
| G709V | g2126t | 0 | |
| G833R | g2497a | 0 | |
| G969C | g2905t | 0 | |
| H558R | a1673g | 36 | |
| I1005T | t3014c | 0 | |
| I1835T | t5504c | 0 | |
| I759F | a2275t | 0 | |
| K1018E | a3052g | 0 | |
| L1308F | c3922t | 0 | |
| L461F | g1383t | 0 | |
| L567Q | t1700a | 0 | |
| L995F | c2983t | 0 | |
| N1325S | a3974g | 0 | |
| N1774D | c5302a | 0 | |
| N291H | a871c | 0 | |
| N406K | c1218a | 0 | |
| P1002S | c3004t | 0 | |
| P1008S | c3022t | 0 | |
| P1011L | c3032t | 0 | |
| P1011S | c3031t | 0 | |
| P1021S | c3061t | 0 | |
| P1090L | c3269t | 9 | |
| P648L | c1943t | 0 | |
| Q1000L | a2999t | 0 | |
| R1023C | c3067t | 0 | |
| R1023H | g3068a | 0 | |
| R1023P | g3068c | 0 | |
| R1193Q | g3578a | 15 | |
| R1826H | g5474a | 0 | |
| R222Q | g665a | 0 | |
| R367H | g1100a | 0 | |
| R620H | g1859a | 0 | |
| R680H | g2039a | 0 | |
| R689C | c2065t | 0 | |
| R689H | g2066a | 0 | |
| R811H | g2432a | 0 | |
| R975W | t2923c | 0 | |
| R986Q | g2957a | 0 | |
| S1103Y | c3308a | 0 | |
| S1218I | g3653t | 0 | |
| S1787N | g5360a | 0 | |
| S216L | c647t | 0 | |
| S524Y | c1571a | 0 | |
| T1007I | c3020t | 0 | |
| T1016M | c3047t | 0 | |
| T1304m | g5050a | 0 | |
| V1951L | g5870a | 0 | |
| V232I | g694a | 0 | |
| V411M | g1231a | 0 | |
| Y1494N | t4480a | 0 | |
| KCNJ2 | C54F | g161t | 0 |
| R67Q | g200a | 0 | |
| D71N | g211a | 0 | |
| T75A | a223g | 0 | |
| D78Y | g232t | 0 | |
| R82Q | g245a | 0 | |
| C101R | t301c | 0 | |
| G144S | g430a | 0 | |
| G146S | g436a | 0 | |
| T192A | a574g | 0 | |
| G215D | g644a | 0 | |
| R218Q | g653a | 0 |
LQT = long QT.
The SNP genotyping with real-time PCR used a pair of primers and a specific dye-labeled probe for each allele: allele 1 (normal) was labeled with FAM, and the other allele 2 (mutation) was labeled with HEX. During amplification, the generation of FAM, HEX, or both types of fluorescence indicate an allele 1 homozygote, allele 2 homozygote, and a heterozygote, respectively. The quencher dye at the 3ʹ end of each probe was BHQ®-1 (black hole quencher® 1). Some primers and probes were designed firsthand whereas others were purchased from Applied Biosystems (AB) Taqman SNP assays mto human SM (Life Technologies, Carlsbad, CA, USA).
Real-time PCR was performed with 20 ng gDNA using the iQ probe SuperMix and CFX Connect™ real-time PCR (Bio-Rad, Hercules, MA, USA) according to the manufacturer's protocol. The PCR conditions were as follows: an initial incubation at 95°C for 5 minutes, followed by 50 cycles of denaturation at 95°C for 15 seconds and annealing and extension at 60°C–68°C (temperatures depended on primers) for 30 seconds. Multiplex genotyping strategies were used to confirm the results with TaqMan. The PCR for sequence analysis was performed using the Prime STAR™ HS (premix, TAKARA, Shiga, Japan) according to the manufacturer's protocol.
Ethics statement
The study was confirmed as a research activity qualifies as non-human participant by the Institutional Review Board of Seoul National University Hospital (No. 2018-001). All data were analyzed anonymously. The requirement for informed consent was waived by the board.
RESULTS
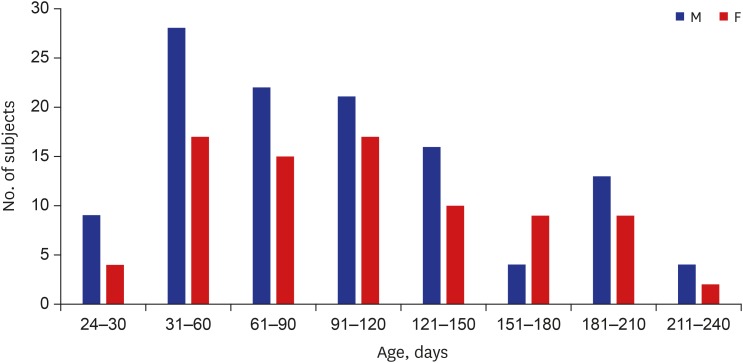
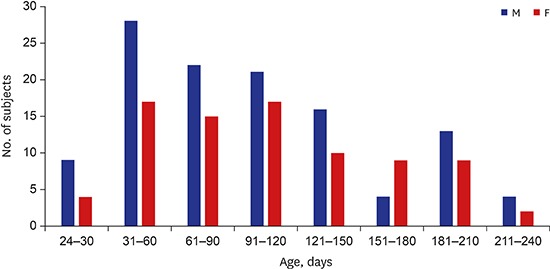
The results showed that 58% of the 200 SIDS cases were male infants (116 male and 84 female infants, respectively). In addition, the mean age was 140 days (median, 107 days; range, 24 to 270 days) and all the infants were of Asian ethnicity (Fig. 1). SIDS IA category criteria were fulfilled in 45 cases (22.5%) whereas the rest were determined to be SIDS IB (Table 2). All infants died in their sleep.
Fig. 1. Age and sex distribution of SIDS subjects.
SIDS = sudden infant death syndrome, M = male, F = female.
Table 2. Demographics, history, investigations, cardiac testing in 15 cases, which showed a genetic variant (R1193Q in SCN5A).
| ID No. | Sex | Age, day | Position found at death scene | SIDS category |
|---|---|---|---|---|
| 10 | M | 30 | Supine | IA |
| 14 | M | 31 | Supine | IB |
| 21 | M | 45 | Supine | IA |
| 28 | M | 53 | Supine | IA |
| 38 | M | 60 | Supine | IB |
| 59 | M | 65 | Supine | IB |
| 68 | M | 77 | Side | IB |
| 70 | M | 81 | Prone | IB |
| 99 | M | 103 | Supine | IA |
| 103 | M | 112 | Supine | IB |
| 105 | M | 118 | Prone | IB |
| 134 | F | 122 | Supine | IB |
| 137 | F | 143 | Supine | IB |
| 161 | M | 162 | Side | IB |
| 164 | F | 174 | Supine | IA |
SIDS = sudden infant death syndrome, M = male, F = female.
Table 1 shows the variants of LQTS-related genes detected in SIDS in this study. A previously reported SNP,12,21,22 H558R in exon 12 and P1090L in exon 18 of the SCN5A gene was found in 36 and 9 (18.0% and 4.5%) SIDS cases, respectively. No variants indicating possible pathogenicity were found in this study.
Fifteen cases (7.5%) had R1193Q in SCN5A (Table 1), which has been reported in association with LQTS and Brugada syndrome (BS),22,23 but a recent study showed it to be a common polymorphism in Asians.12 The prevalence of the R1193Q mutation showed no statistical significance in relation to sex, SIDS category, and position of the infant at the death scene.
DISCUSSION
Although a molecular autopsy is not included as a standard protocol in SIDS cases, postmortem genetic testing is increasingly being recommended, especially focusing on arrhythmia syndrome.24,25 Tester and Ackerman26 estimated that approximately 10% of all SIDS cases were actually caused by cardiac channelopathies resulting in LQTS. A previous large population-based study also reported that pathogenic variants associated with KCNH2 and SCN5A were found in 9.5% of subjects.18
From the start of this study, we cautiously selected category IA and IB SIDS cases and excluded those in category II to exclude the mechanical asphyxia and assess the precise relationship between SIDS and LQTS. We hypothesized that approximately 5%–10% of the pathogenic variants would be identified in this SIDS cohort. However, pathogenic variants associated with KCNQ1, SCN5A, KCNE1, KCNE2, KCNJ2, and CAV3 were not observed. In particular, detection of the R1193Q mutation in 15 cases was confusing. Previously, the R1193Q variant was reported in association with LQTS and BS.21,22 This variant was present in 0.3%, 8%, and 12% of Caucasians, Asians,12 and the Han Chinese,22 respectively.
Subsequently, the variant R1193Q was considered a common polymorphism in Asian populations.27 However, the influence of the R1193Q mutation in BS and LQTS currently remains unclear. The genetic risk might be polygenic and, thus, the R1193Q mutation in the SCN5A gene could influence variants of other genes, not previously reported.28
Routine analysis of LQTS-related genes in postmortem examination of SIDS cases has been recommended since channelopathies may be caused by pathogenic variants in genes associated with structural heart disease.29 However, a recent review reported that the overall diagnostic yield of gene variants in SIDS cases was substantially lower than that in the Exome Aggregation Consortium (ExAC) 14% vs. 41%, respectively.30 No significant differences were found between SIDS and ExAC yields for any genes. The New Zealand study also showed that no significant pathologic variants were found in the non-selected series of SIDS.28 In our study, the diagnostic rate of the pathological variants related to LQTS was zero, subsequently could raise the question as to whether a routine molecular autopsy associated with LQTS would be necessary for all cases of SIDS.
The New Zealand study also showed few positive variants in SIDS study, which suggests that postmortem genetic testing in SIDS should be conducted in cases with a familial clinical history of sudden death or cardiac arrhythmia and the absence of risk factors such as a bed-sharing. Rare mutations associated with inherited cardiac diseases including LQTS, BS, and catecholaminergic polymorphic ventricular tachycardia (CPVT) could still explain more than 14% of SIDS cases.30 However, a molecular autopsy with large cohorts using next-generation sequencing (NGS) is still necessary.
Forensic pathologists have established archives of formalin-fixed, paraffin-embedded (FFPE) tissue samples. Although molecular analysis using FFPE tissue samples has shown comparable quality,31 the cost of genetic testing has increased compared to the test using DNA extracted from blood. Presently, there is an urgent need to establish a national tissue (blood) bank for SIDS in Korea. Moreover, to elucidate the genetic risk associated with SIDS, simultaneous genetic testing of parents and siblings with a familial history of sudden death, syncope, and clinically proven arrhythmia would be reasonable, which is also effective for genetic counseling.
However, in Korea, the decision to perform a medico-legal autopsy is made by public prosecutors, and the autopsy is typically performed if warranted, such as in suspicious deaths associated with a likely crime. Therefore, numerous cases of SUIDs were not pathologically investigated and no genetic counseling to prevent the sudden deaths of siblings has been established in Korea. Forensic molecular autopsy and valuable genetic counseling by clinicians should be performed to prevent SIDS.
This study has some potential limitations that need to be considered. First, FFPE could cause DNA fragmentation, resulting in low DNA yields, which could increase the risk of allelic locus dropout.32 In this study, the DNA extraction and TaqMan SNP assays were performed in triplicate for all the point mutations, which were confirmed using sequence analysis. Second, recent studies used large gene panels investigated using NGS,33,34 which would likely be a more promising method to discover the etiology of SIDS than other methods. However, there was no blood preservation for SIDS cases and, therefore, we had to analyze small panels of LQTS genes. Currently, we are preserving blood samples of SIDS cases to perform prospective studies using NGS to target all channelopathy-associated genes including those related to LQTS, Brugada, CPVT, and structural cardiac genes.
Despite these limitations, this study has many strengths. For instance, to the best of our knowledge, this is the first report of genetic analysis in Korean SIDS. Most previous studies were conducted in Western countries.10,11,12,13,14,15,16,17,18,19,20 Since other factors such as racial and environmental factors might be associated with SIDS, nationwide studies would have crucial implications. Second, this is the first study to focus on LQTS in SIDS in Korea. The collection of medico-legal autopsy data that provides the criteria for molecular autopsies in SIDS with clinical information including familial history for cardiac events is especially challenging in qualitative research in SIDS.
In conclusion, only 15 of the 85 DNA variants tested from 6 genes involved in the LQTS exhibited R1193Q in SCN5A with doubtful pathogenicity, and no pathogenic variants were observed. Considering the diagnostic yield in this study was close to zero, these findings suggest that molecular autopsy should be cautiously conducted in select cases with a familial clinical history to improve the quality and availability of genetic counseling for the families of victims. Furthermore, other factors affecting the sudden death of vulnerable infants should be investigated through comprehensive autopsy examinations since the development of SIDS is complex and multifactorial.
Moreover, it is necessary to establish a national young sudden death registry and investigative program including genetic counseling to explore the genetic background of inherited cardiac conditions including LQTS, BS, and CPVT as well as discover and assess genetic risk factors for SIDS in Korea.
ACKNOWLEDGMENTS
We thank all members of the National Forensic Service for their assistance in providing the data for this study.
Footnotes
Funding: This work was supported by National Forensic Service (2015-forensic medicine-05), Ministry of the Interior and Safety, and National Research Fund (800-20160361), Ministry of the Education, Republic of Korea.
Disclosure: The authors have no potential conflicts of interest to disclose.
Author Contributions: Conceptualization: Yoo SH. Data curation: Yang KM, Lee BW, Choi BH, Yoo SH. Investigation: Son MJ, Kim MK. Writing - original draft: Song MJ, Yoo SH. Writing - review & editing: Yoo SH.
References
- 1.Task Force on Sudden Infant Death Syndrome. Moon RY. SIDS and other sleep-related infant deaths: expansion of recommendations for a safe infant sleeping environment. Pediatrics. 2011;128(5):1030–1039. doi: 10.1542/peds.2011-2284. [DOI] [PubMed] [Google Scholar]
- 2.Yoo SH, Kim AJ, Kang SM, Lee HY, Seo JS, Kwon TJ, et al. Sudden infant death syndrome in Korea: a retrospective analysis of autopsy-diagnosed cases. J Korean Med Sci. 2013;28(3):438–442. doi: 10.3346/jkms.2013.28.3.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jervell A, Lange-Nielsen F. Congenital deaf-mutism, functional heart disease with prolongation of the Q-T interval and sudden death. Am Heart J. 1957;54(1):59–68. doi: 10.1016/0002-8703(57)90079-0. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz PJ, Priori SG, Dumaine R, Napolitano C, Antzelevitch C, Stramba-Badiale M, et al. A molecular link between the sudden infant death syndrome and the long-QT syndrome. N Engl J Med. 2000;343(4):262–267. doi: 10.1056/NEJM200007273430405. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz PJ, Priori SG, Bloise R, Napolitano C, Ronchetti E, Piccinini A, et al. Molecular diagnosis in a child with sudden infant death syndrome. Lancet. 2001;358(9290):1342–1343. doi: 10.1016/S0140-6736(01)06450-9. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Shah KR, Um SY, Eng LS, Zhou B, Lin Y, et al. Cardiac channelopathy testing in 274 ethnically diverse sudden unexplained deaths. Forensic Sci Int. 2014;237:90–99. doi: 10.1016/j.forsciint.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Ackerman MJ, Siu BL, Sturner WQ, Tester DJ, Valdivia CR, Makielski JC, et al. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA. 2001;286(18):2264–2269. doi: 10.1001/jama.286.18.2264. [DOI] [PubMed] [Google Scholar]
- 8.Semsarian C, Ingles J, Wilde AA. Sudden cardiac death in the young: the molecular autopsy and a practical approach to surviving relatives. Eur Heart J. 2015;36(21):1290–1296. doi: 10.1093/eurheartj/ehv063. [DOI] [PubMed] [Google Scholar]
- 9.Krous HF, Beckwith JB, Byard RW, Rognum TO, Bajanowski T, Corey T, et al. Sudden infant death syndrome and unclassified sudden infant deaths: a definitional and diagnostic approach. Pediatrics. 2004;114(1):234–238. doi: 10.1542/peds.114.1.234. [DOI] [PubMed] [Google Scholar]
- 10.Tester DJ, Ackerman MJ. Genetics of long QT syndrome. Methodist DeBakey Cardiovasc J. 2014;10(1):29–33. doi: 10.14797/mdcj-10-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun A, Xu L, Wang S, Wang K, Huang W, Wang Y, et al. SCN5A R1193Q polymorphism associated with progressive cardiac conduction defects and long QT syndrome in a Chinese family. J Med Genet. 2008;45(2):127–128. doi: 10.1136/jmg.2007.056333. [DOI] [PubMed] [Google Scholar]
- 12.Ackerman MJ, Splawski I, Makielski JC, Tester DJ, Will ML, Timothy KW, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1(5):600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Mank-Seymour AR, Richmond JL, Wood LS, Reynolds JM, Fan YT, Warnes GR, et al. Association of torsades de pointes with novel and known single nucleotide polymorphisms in long QT syndrome genes. Am Heart J. 2006;152(6):1116–1122. doi: 10.1016/j.ahj.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Kwon HW, Lee SY, Kwon BS, Kim GB, Bae EJ, Kim WH, et al. Long QT syndrome and dilated cardiomyopathy with SCN5A p.R1193Q polymorphism: cardioverter-defibrillator implantation at 27 months. Pacing Clin Electrophysiol. 2012;35(8):e243–e246. doi: 10.1111/j.1540-8159.2012.03409.x. [DOI] [PubMed] [Google Scholar]
- 15.Eckhardt LL, Farley AL, Rodriguez E, Ruwaldt K, Hammill D, Tester DJ, et al. KCNJ2 mutations in arrhythmia patients referred for LQT testing: a mutation T305A with novel effect on rectification properties. Heart Rhythm. 2007;4(3):323–329. doi: 10.1016/j.hrthm.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olesen MS, Bentzen BH, Nielsen JB, Steffensen AB, David JP, Jabbari J, et al. Mutations in the potassium channel subunit KCNE1 are associated with early-onset familial atrial fibrillation. BMC Med Genet. 2012;13(1):24. doi: 10.1186/1471-2350-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapa S, Tester DJ, Salisbury BA, Harris-Kerr C, Pungliya MS, Alders M, et al. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120(18):1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnestad M, Crotti L, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, et al. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115(3):361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 19.Delannoy E, Sacher F, Maury P, Mabo P, Mansourati J, Magnin I, et al. Cardiac characteristics and long-term outcome in Andersen-Tawil syndrome patients related to KCNJ2 mutation. Europace. 2013;15(12):1805–1811. doi: 10.1093/europace/eut160. [DOI] [PubMed] [Google Scholar]
- 20.Oshima Y, Yamamoto T, Ishikawa T, Mishima H, Matsusue A, Umehara T, et al. Postmortem genetic analysis of sudden unexpected death in infancy: neonatal genetic screening may enable the prevention of sudden infant death. J Hum Genet. 2017;62(11):989–995. doi: 10.1038/jhg.2017.79. [DOI] [PubMed] [Google Scholar]
- 21.Hwang HW, Chen JJ, Lin YJ, Shieh RC, Lee MT, Hung SI, et al. R1193Q of SCN5A, a Brugada and long QT mutation, is a common polymorphism in Han Chinese. J Med Genet. 2005;42(2):e7. doi: 10.1136/jmg.2004.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Zhao J, Barrane FZ, Champagne J, Chahine M. Nav1.5/R1193Q polymorphism is associated with both long QT and Brugada syndromes. Can J Cardiol. 2006;22(4):309–313. doi: 10.1016/s0828-282x(06)70915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan BH, Valdivia CR, Rok BA, Ye B, Ruwaldt KM, Tester DJ, et al. Common human SCN5A polymorphisms have altered electrophysiology when expressed in Q1077 splice variants. Heart Rhythm. 2005;2(7):741–747. doi: 10.1016/j.hrthm.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 24.Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10(12):1932–1963. doi: 10.1016/j.hrthm.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8(8):1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 26.Tester DJ, Ackerman MJ. The molecular autopsy: should the evaluation continue after the funeral? Pediatr Cardiol. 2012;33(3):461–470. doi: 10.1007/s00246-012-0160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsusue A, Yuasa I, Umetsu K, Nakayashiki N, Dewa K, Nishimukai H, et al. The global distribution of the p.R1193Q polymorphism in the SCN5A gene. Leg Med (Tokyo) 2016;19:72–76. doi: 10.1016/j.legalmed.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Glengarry JM, Crawford J, Morrow PL, Stables SR, Love DR, Skinner JR. Long QT molecular autopsy in sudden infant death syndrome. Arch Dis Child. 2014;99(7):635–640. doi: 10.1136/archdischild-2013-305331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaver EC, Versluijs GM, Wilders R. Cardiac ion channel mutations in the sudden infant death syndrome. Int J Cardiol. 2011;152(2):162–170. doi: 10.1016/j.ijcard.2010.12.051. [DOI] [PubMed] [Google Scholar]
- 30.Baruteau AE, Tester DJ, Kapplinger JD, Ackerman MJ, Behr ER. Sudden infant death syndrome and inherited cardiac conditions. Nat Rev Cardiol. 2017;14(12):715–726. doi: 10.1038/nrcardio.2017.129. [DOI] [PubMed] [Google Scholar]
- 31.Van Niekerk C, Van Deventer BS, du Toit-Prinsloo L. Long QT syndrome and sudden unexpected infant death. J Clin Pathol. 2017;70(9):808–813. doi: 10.1136/jclinpath-2016-204199. [DOI] [PubMed] [Google Scholar]
- 32.Klopfleisch R, Weiss AT, Gruber AD. Excavation of a buried treasure--DNA, mRNA, miRNA and protein analysis in formalin fixed, paraffin embedded tissues. Histol Histopathol. 2011;26(6):797–810. doi: 10.14670/HH-26.797. [DOI] [PubMed] [Google Scholar]
- 33.Neubauer J, Lecca MR, Russo G, Bartsch C, Medeiros-Domingo A, Berger W, et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur J Hum Genet. 2017;25(4):404–409. doi: 10.1038/ejhg.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertz CL, Christiansen SL, Larsen MK, Dahl M, Ferrero-Miliani L, Weeke PE, et al. Genetic investigations of sudden unexpected deaths in infancy using next-generation sequencing of 100 genes associated with cardiac diseases. Eur J Hum Genet. 2016;24(6):817–822. doi: 10.1038/ejhg.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]