Abstract
From a liquid biopsy, cell-free DNA (cfDNA) can provide information regarding basal tumoral genetic patterns and changes upon treatment. In a prospective cohort of 30 diffuse large B-cell lymphomas (DLBCL), we determined the clinical relevance of cfDNA using targeted next-generation sequencing and its correlation with PET scan imaging at the time of diagnosis and during treatment. Using a dedicated DLBCL panel, mutations were identified at baseline for 19 cfDNAs and profiles were consistent with expected DLBCL patterns. Tumor burden-related clinical and PET scan features (LDH, IPI, and metabolic tumor volume) were significantly correlated with the quantity of tumoral cfDNA. Among the four patients presenting additional mutations in their cfDNAs, three had high metabolic tumor volumes, suggesting that cfDNA more accurately reflects tumor heterogeneity than tissues biopsy itself. Mid-treatment, four patients still had basal mutations in their cfDNAs, including three in partial response according to their Deauville scores. Our study highlights the major interests in liquid biopsy, in particular in the context of bulky tumors where cfDNA allows capturing the entire tumoral mutation profile. Therefore, cfDNA analysis in DLBCL represents a complementary approach to PET scan imaging.
Introduction
The concept of liquid biopsy has been well known for several years mainly among solid tumors, for which it has been possible to demonstrate the presence of circulating tumor cells, circulating tumor DNA (ctDNA) contained in plasma cell-free circulating DNA (cfDNA), and circulating micro-RNAs1–4. In healthy subjects, cfDNA arises from the apoptosis of nucleated cells and is found at low levels in the blood, demonstrating that detection of ctDNA requires highly sensitive technologies, such as those currently available with next-generation sequencing (NGS) approaches or digital PCR. In cancer patients, it is known that plasma ctDNA is mainly released by apoptotic tumor cells, but may also be released by necrotic tumor cells or actively secreted by tumor cells5, as illustrated by the large range of DNA fragment sizes that are detected (from 150 base pairs to several kilobases).
The value of liquid biopsies was recently highlighted in a few series of diffuse large B-cell lymphoma (DLBCL) patients but also in primary central nervous system lymphoma (PCNSL) cases in whom high-throughput sequencing of a panel of target genes was performed, demonstrating the successful detection of somatic variants in both the tumors and the plasma with similar mutational profiles6–8.
Combined immunochemotherapy and targeted therapies have considerably changed the management of lymphomas over the past decade. Nevertheless, response to treatment is often heterogeneous, and while some patients will remain relapse-free after therapy, others experience early disease progression and may develop chemo-refractoriness due to the acquisition of tumor resistance mechanisms and the clonal evolution of tumor cells. Therefore, ctDNA analyses theoretically offer information regarding potentially actionable mutations, clonal evolution, treatment response, and genetic mechanisms of resistance.
Similar to ctDNA detection, 18F-fluoro-2-dexoxy-D-glucose (FDG) positron-emission tomography/computed tomography (PET/CT) is a sensitive, non-invasive tool for DLBCL staging, and has been shown to predict therapeutic response and outcomes in this setting. Interim PET (iPET) performed after two cycles of treatment or at mid-treatment has been considered a strong predictor of outcomes using response criteria, such as the Deauville 5-point scale (5-PS) or the reduction in maximum SUV (ΔSUVmax) method9. CtDNA detection and PET/CT imaging have been simultaneously reported in two studies using immunoglobulin high-throughput sequencing. These works demonstrated that detection of molecular disease in the plasma may precede PET/CT detection of relapse and improve specificity with similar sensitivity compared with PET/CT10–12.
On behalf the LYSA (Lymphoma Study Association), we recently designed and validated a set of 34 genes (termed “Lymphopanel”) selected according to the literature and a whole-exome sequencing study of relapsed/refractory DLBCL patients. The Lymphopanel was informative for 96% of 215 patients enrolled in LYSA trials, highlighting the molecular heterogeneity of subtypes and identifying somatic mutations with therapeutic and prognostic impacts13. Its relevance and accuracy for ctDNA was not evaluated.
The primary aim of the current study was to evaluate our Lymphopanel as a liquid biopsy tool in a prospective cohort of DLBCL. The secondary purposes were to determine its modification upon treatment and to correlate baseline and dynamic ctDNA characteristics with PET scan imaging metrics obtained at baseline and during follow-up.
Subjects and methods
Study design and patients
Thirty DLBCL or primary mediastinal B-cell lymphoma (PMBL) patients were enrolled prospectively. Biological material at the time of diagnosis, including DNA and RNA from biopsies, blood and bone marrow, was collected before any treatment, as well as clinical features (bone marrow biopsy results, stage according to the Ann Arbor classification, and international prognostic index (IPI) calculation) were recorded. Patients were followed after rituximab-cyclophosphamide-doxorubicin-vincristine-prednisone (R-CHOP) or R-CHOP-like chemotherapies, and blood samples (for cfDNA extraction) were collected at mid-treatment, end of treatment, and 1 year after end of treatment. An 18FDG-PET-TDM was performed at the time of diagnosis and during the follow-up (mid-treatment and end of treatment, according to treatment strategies).
Patients provided written informed consent in accordance with the Declaration of Helsinki, and the institutional review board approved the protocol (Registration clinical.gov number: NCT02339805).
Sample collection and processing
Tumor genomic DNA (gDNA) was isolated from fresh or formalin-fixed paraffin-embedded (FFPE) diagnostic tissue biopsies. Bone marrow samples were collected at the time of diagnosis. From fresh tissue and bone marrow, gDNA was extracted using proteinase K followed by salt and ethanol precipitation and was stored at −20 °C in 10 mM Tris-Cl and 1 mM EDTA (pH 8) buffer. From FFPE tissue, gDNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Courtaboeuf, France), according to the manufacturer’s instructions.
CtDNA monitoring was completed on serial plasma samples. For each patient, plasma samples were collected at baseline and interim monitoring was performed on samples obtained at mid-treatment, at the end of treatment, 6 months after the end of treatment, and in case of relapse (Supplementary Fig. S1). Blood was collected in EDTA tubes that were centrifuged for 10 min at 3000 rpm within 3 h of collection. Plasma was aliquoted into 1 mL in microtubes and stored at −80 °C until extraction. After thawing, plasma aliquots (from 1 to 3 mL) were centrifuged for 5 min at 13,000 rpm. Then, cfDNA was extracted from the supernatant using the QIAamp circulating nucleic acid kit (Qiagen) and quantified using QuBit High Sensitivity dsDNA (Thermo Fisher Scientific, Illkirch, France).
GCB/ABC phenotyping
For tumoral samples, cell of origin (COO) molecular classification was obtained using the Hans algorithm14. Immunohistochemistry staining was performed for CD10, BCL6, MUM1, MYC, BCL2, FOXP1, and IgM as described previously15,16.
Sequencing and copy number variations (CNV) detection
At the time of diagnosis, gDNA and cfDNA were sequenced with the entire Lymphopanel. Ion Torrent Personal Genome Machine (Thermo Fisher Scientific) Sequencing was performed using our Lymphopanel as previously described. Briefly, the Lymphopanel was designed to identify mutations in 34 genes important for lymphomagenesis grouped into 8 specific pathways. This design covers 87,703 bases and generates 872 amplicons. Variant analysis was performed using an in-house generated bioinformatics pipeline that was previously described6,13,17. CNV detection was performed using ONCOCNV software as previously described17,18.
Sanger sequencing was also performed to validate a portion of the mutations found in tumoral gDNA using the BigDye® Terminator v3.1 Cycle Sequencing Kit and an ABI PRISM 3130 analyzer (Applied Biosystems, Forster City, CA, USA).
During follow-up, to increase the sensitivity and for cost-effectiveness of the sequencing, only mutated amplicons detected at baseline were sequenced and used to detect minimal residual disease in cfDNA.
The tumoral cfDNA (ctDNA) concentrations were expressed in haploid genome equivalents per mL of plasma (hGE/mL) and calculated by multiplying the mean variant allelic frequency (VAF) by the concentration of cfDNA (pg/mL of plasma) and dividing by 3.3, as previously described in the publication by Scherer et al12.
PET scan analysis
All patients underwent [18F]fluorodeoxyglucose-PET/CT (FDG-PET/CT) before the onset of chemotherapy in the nuclear medicine department, according to standard procedures. The following parameters were determined on the baseline scan: (1) SUVmax, the highest SUVmax measured in the tumor sites and (2) total metabolic tumor volume (MTV), which was obtained by summing the metabolic volumes of all the nodal and extranodal lesions, a volume of interest was set around each lesion (node or organ involvement) as previously described19. The bone marrow involvement was only included in the volume measurement if there was focal uptake. The spleen was considered as involved if there was focal uptake or diffuse uptake higher than 150% of the liver background. Total lesion glycolysis (TLG) was also calculated as the sum of the product of the metabolic volume of each local tumor based on its SUVmean (TLG = ΣMTV × SUVmean). For segmentation, the 41% SUVmax threshold method was used19,20. iPET was planned in all treated patients at mid-treatment, allowing us to assess early response using the 5-point Deauville score (5-DS) and the DeltaSUV (ΔSUVmax) calculation as previously reported21. PET was also performed at the end of treatment and the response was assessed according to the current recommendations (Supplementary Fig. S1)22.
Statistical analysis
All statistical analyses were performed using R software version 3.1.223. Progression-free survival (PFS) was evaluated from the date of enrollment to the date of disease progression, relapse or death from any cause. Overall survival (OS) was evaluated from the date of enrollment to the date of death from any cause. Log-rank tests were used to assess differences in the OS and PFS rates calculated by Kaplan–Meier estimates. Statistical differences between parameters in box plots were determined using the Wilcoxon test. P-values < 0.05 were considered statistically significant.
Results
Patient population description
A prospective series of 30 consecutive untreated DLBCL or PMBL patients were enrolled and followed after R-CHOP or R-CHOP-like chemotherapies. Patients’ clinical features are listed in Table 1. One patient died before beginning any chemotherapy. From mid-treatment, 17 patients achieved complete response (Deauville score ≤ 3), 2 were stable (absence of metabolic response), and 10 showed a partial response (Deauville score > 3). At the end of treatment, 1 more patient had died, 6 patients presented a partial response, 4 a progression, and 18 a complete response. At the end of the trial, 1 patient decided to stop follow-up, 20 patients showed a complete response, 3 relapses were observed including 1 with premature death, 2 progressive patients had died, and the 2 others still presented a progression (Supplementary Fig. S2, Supplementary Table S2).
Table 1.
Patients characteristics
| Characteristics | Patients (%) |
|---|---|
| Men/women | 17/13 |
| Median age | 67 [20–93] |
| IPI score | |
| 0–1 | 8 (27) |
| 2–3 | 14 (46) |
| 4–5 | 8 (27) |
| LDH > normal value | 12 (40) |
| Stade III–IV | 21 (70) |
| Bone marrow involvement | 0/25 (0) |
| COO classification | |
| Hans (Non-GCB/GCB/NA) | 9 (34)/17 (66)/5 |
| MYC + expression | 7/25 (28) |
| BCL2 + expression | 16/24 (66) |
| Dual expression | 5/21 (24) |
| First-line treatment | |
| RCHOP | 16 (55) |
| RCHOP-like regimen (RACVBP/R miniCHOP) | 13 (45) |
| Treatment response | |
| CR | 20 |
| PR | 5 |
| PD | 1 |
| TEP base line features | |
| SUV max | 22.61 [4.66–43.03] |
| Metabolic tumor volume (MTV) | 399.28 [0.29–2846] |
| Total lesion glycolysis (TLG) | 4697.50 [1.17–31149] |
| Delta SUV at interim PET | |
| >70% | 22 |
| <70% | 7 |
| Deauville score at interim PET | |
| 1–3 | 17 |
| 4–5 | 12 |
| Deauville score at final PET | |
| 1–3 | 14 |
| 4–5 | 10 |
Plasma genotyping at baseline
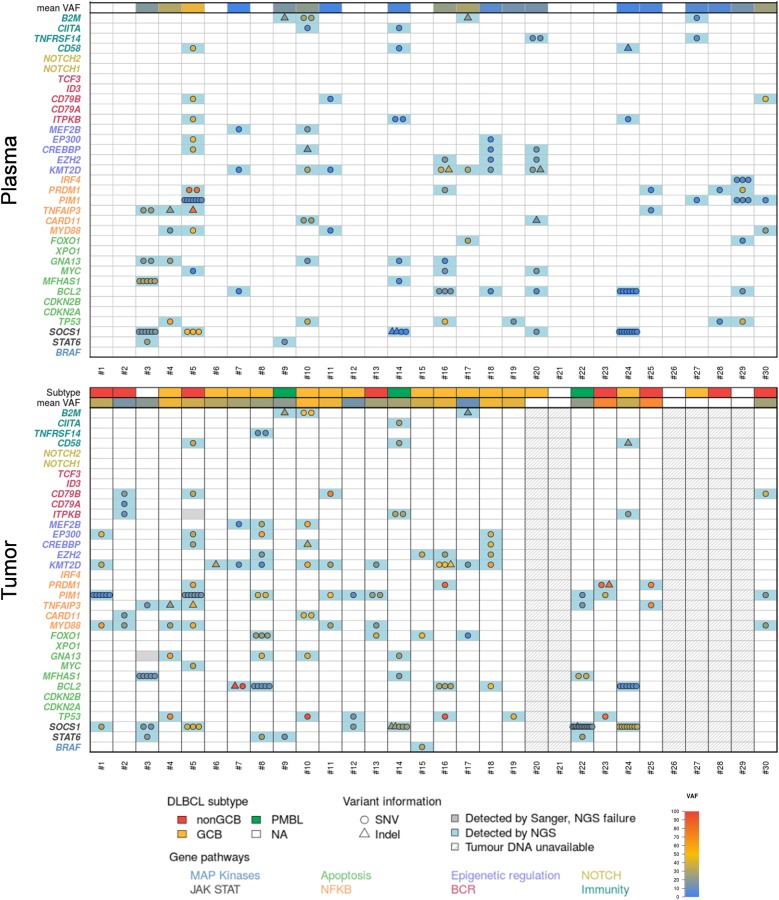
To determine the clinical relevance of liquid biopsy at the time of diagnosis, we first assessed the basal DLBCL genetic patterns with cfDNA by targeted NGS. CfDNA was available from the 30 patients. The mean plasma DNA concentration was 74 ng/mL [14.6–433], and sequencing was performed with the entire panel with a mean depth of 3750×. cfDNA mutations were identified in 19/30 patients (63%). The mutation profiles were consistent with patterns usually observed in DLBCL and previously reported by analyzing the tumor tissues13,17 (Fig. 1).
Fig. 1. Basal genetic patterns in plasma cfDNA and matched tumoral DNA.
Representation of the prevalence and molecular spectrum of somatic mutations (SNVs and Indels) identified in cfDNA (upper panel) and paired tumoral DNA (lower panel) at the time of diagnosis. The 34 genes in the Lymphopanel are grouped by pathway
KMT2D (MLL2) and PIM1 were the most mutated genes, with somatic mutations in 36.7% of cases (n = 11/30), followed by SOCS1 and TP53 in 26.7% (n = 8/30); MYD88 and BCL2 in 23.3% (n = 7/30); GNA13 and PRDM1 in 20% (n = 6/30); EZH2, FOXO1, and TNFAIP3 in 16.7% (n = 5/30); ITPKB, CD79B, B2M, STAT6, EP300, and CREBBP in 13.3% (n = 4/30); CARD11, MFHAS1, TNFRSF14, MEF2B, CD58, and MYC in 10% (n = 3/30); CIITA in 6.7% (n = 2/30); and IRF4, CD79A, and BRAF in 3.3% (n = 1/30). The mutation distribution was associated with the COO subtype, such as MYD88 (particularly L265P), PIM1, and CD79B in ABC DLBCL; EZH2, BCL2, and GNA13 in GCB DLBCL or STAT6, SOCS1, and MFHAS1 in PMBL. Multiple mutations were identified in genes such as IRF4, SOCS1, PIM1, and BLC2, suggesting an AID process as previously described13.
The cfDNA VAF was very variable with a mean of 18.2% [0.8–87.4]. The mean concentration of ctDNA was 4604 hGE/mL [0–39,151]. A high concentration of cfDNA signaled the presence of circulating DNA from tumoral origins, as no plasma samples with cfDNA concentrations above the average were negative for mutations (p = 5.6 × 10−4, Supplementary Fig. S3).
Tumor genotyping at baseline and concordance with plasma genotyping
When available, matched gDNA from tissue biopsies was sequenced with the Lymphopanel and compared to the cfDNA results. Enough material was obtained for 25 patients, including 11 frozen and 14 FFPE biopsies.
The sequencing results were interpretable for 24 cases and not interpretable for 1 case (#28) (Fig. 1). Mutations were found in the 24 interpretable cases; for the 6 remaining cases without workable tumoral materials, an informative mutational pattern was successfully obtained in 4 matched cfDNA. Finally, the mutations at baseline were detected either in gDNA or ctDNA in 28/30 cases, giving an informativity rate of 93%.
In five cases, additional mutations were only detected in cfDNA compared to gDNA (#3, #5, #10, and #16). Conversely, some mutations found in gDNA were not found in cfDNA, as it has been previously described6,7. In these cases, the mutations did not necessarily have the lowest VAF in the gDNA, showing that the mutations were not subclonal. For paired samples, mutations were detected in cfDNA up to a VAF of 4.6% in gDNA (patient #7). The mean VAF was significantly higher in tumor DNA (33%) compared to cfDNA (p = 5.6 × 10−7), but in some cases the VAF in cfDNA was nearly similar to the VAF in gDNA, reflecting the abundance of circulating DNA from tumoral origins.
After sequencing the tumors, we confirmed the results in cfDNA by performing a new sequencing method targeting only the identified mutations to increase sensitivity (Supplementary Table).
CNV analysis at baseline
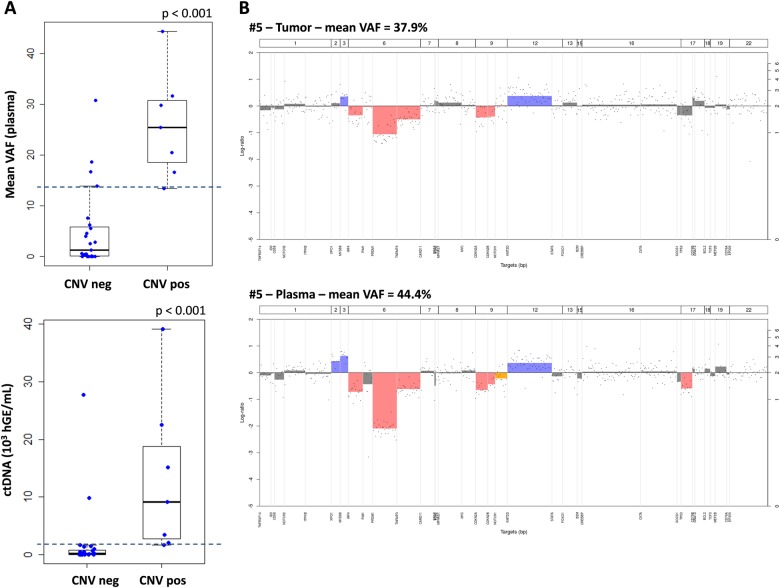
CNV analysis was performed in both plasma and tumor samples using the entire panel. As expected in DLBCL cases, we frequently found deletions at the 6q and 9p loci (33% and 37%, respectively) as well as TP53 deletions (13%) in the DLBCL cases. The gains and/or losses identified were similar in the paired samples from a patient. Discordances were principally due to the quantity of tumoral DNA contained in the sample from the tissue biopsy or plasma (according to mean VAF, Supplementary Fig. S4).
In cfDNAs, CNVs were detectable when tumoral circulating DNA was abundant; according to the mean VAF from the seven positive patients, this was when the tumoral circulating DNA ranged from 13 to 45% (p < 0.001, Fig. 2, Fig. S4). Above this threshold, patients negative for CNVs in cfDNA were also negative in tumoral samples, except for one patient (#19). For this particular case, the mean VAF was 14%, but with only one mutation, while patient #29 had a mean VAF of 13.4% calculated from 10 mutations with frequencies between 6 and 40%. Therefore, with a threshold of 13.4% for mean VAF in the cfDNA, CNVs were detectable in 91% of the cases (10/11). If we expressed these results based on the quantity of ctDNA using a threshold of 1630 hGE/mL for ctDNA, all the positive plasmas were above this value, highlighting the importance of both the VAF and cfDNA concentration at the time of diagnosis.
Fig. 2. CNV analysis in plasma and tumor.
a Boxplot representation of mean VAF (upper panel) or ctDNA amount (lower panel) according to the detection of the CNVs in the plasma samples (neg = negative and pos = positive). The dashed blue lines show the thresholds of detection for CNVs in cfDNA, corresponding to 13.4% for the mean VAF and 1630 hGE/mL for ctDNA. b Example of CNVs from patient #5 in tumor (upper panel) and plasma (lower panel) showing the concordance of the two results when cfDNA of tumoral origin is abundant
Correlation between cfDNA and clinical or TEP baseline features
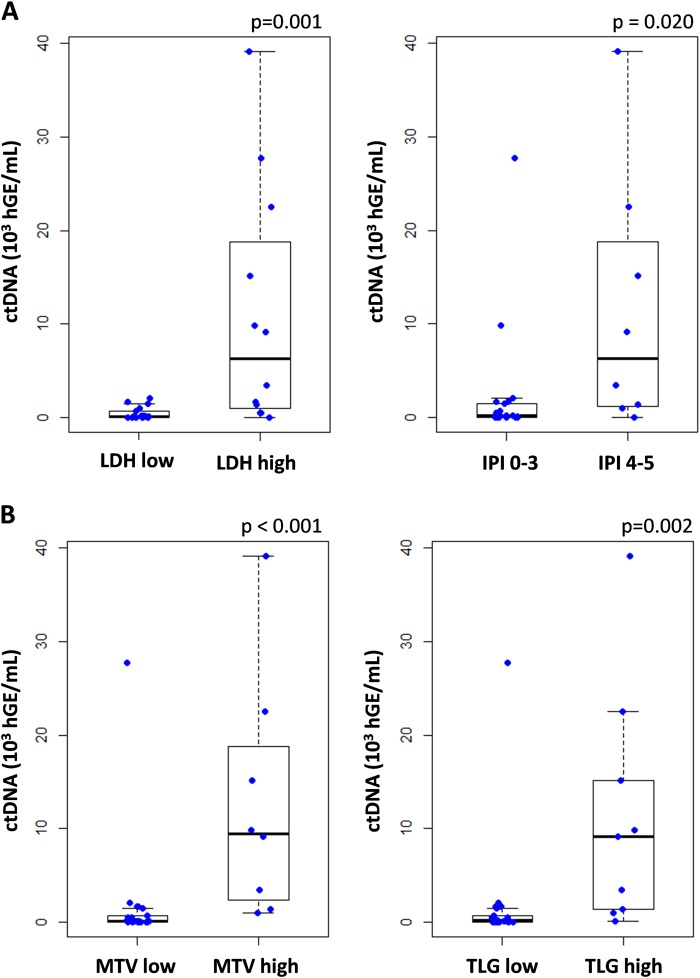
We first compared the presence of ctDNA with standard clinical indices. CfDNA mutations were observed in all patients but one with elevated serum lactate dehydrogenase (LDH), but the presence of mutations was not correlated to the disease stage. The mean VAF in cfDNA, as well as cfDNA concentration, were significantly correlated with LDH and the IPI. The quantity of ctDNA was more significantly correlated with these two features (p = 0.001 and p = 0.02, respectively, Fig. 3a), once again highlighting the importance of both the VAF and cfDNA concentration at the time of diagnosis.
Fig. 3. Correlations between tumoral cfDNA amounts and clinical/PET baseline features.
CtDNA amount is significantly correlated with the clinical indices LDH (threshold = 480 UI/L) and IPI (a), and with the PET features MTV and TLG (threshold = mean) at the time of diagnosis (b)
PET scan imaging (18F-FDG PET/CT) data were collected for all the patients at baseline. The mean MTV was 743.69 cm3 (0.29–2846.62) and the mean TLG was 8341.96 (1.17–31149.00). At the time of diagnosis, both MTV and TLG correlated strongly with the ctDNA concentration (p < 0.001 and p = 0.002, respectively, Fig. 3b), and more significantly than the concentration or VAF alone, as for clinical features. Patients with an MTV below the median (399.28 cm3) had a superior OS and PFS compared with those with a high MTV (p = 0.016 and p = 0.009, respectively, Fig. S5).
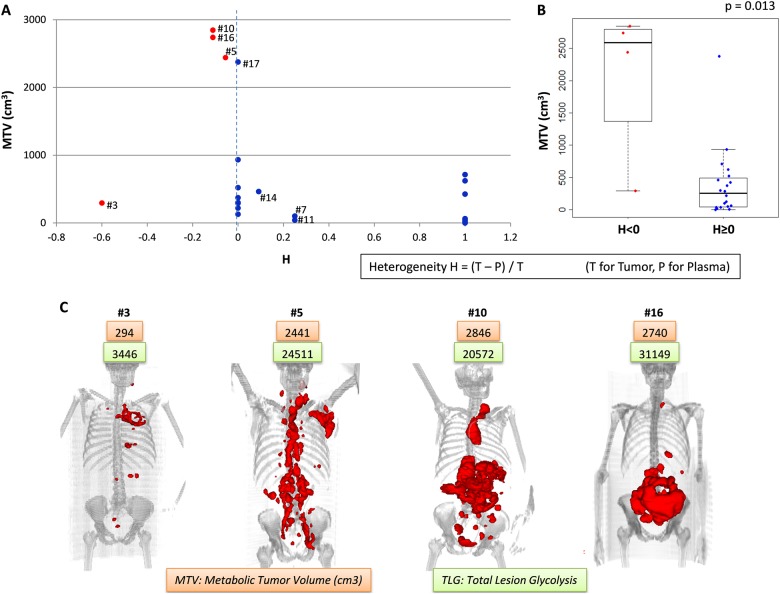
We calculated a heterogeneity coefficient H representing the number of mutations found in the cfDNA compared to gDNA (H = (Number of mutations in gDNA−Number of mutations in cfDNA)/Number of mutations in gDNA). This coefficient was significantly correlated with the MTV (p = 0.013, Fig. 4b). Three in four patients (#5, #10, and #16) presenting additional mutations in the cfDNA, compared to gDNA (h < 0), had a high MTV above 2000 cm3 (Fig. 4a). The 3D view of the PET images showed that two of these patients had many tumoral masses (#5 and #10), as well as a fourth patient who had a low MTV despite additional mutations in its cfDNA (#3, Fig. 4c). These data suggest that cfDNA mutation analysis more accurately reflects the spatial tumor heterogeneity than tissue biopsy analysis. Conversely, the three patients (#7, #11, and #14) with more mutations in tumor DNA (0 < h < 1) had lower MTV values (<500 cm3). Interestingly, patient #17 presented the same mutations in its cfDNA and tumor DNA (h = 0) and an important MTV (2375 cm3), but the mutation VAFs were high in the plasma sample compared with the tumor sample. All the patients with negative plasma (h = 1) had MTVs less than 800 cm3.
Fig. 4. Tumoral heterogeneity.
a Metabolic tumor volume according to the coefficient of heterogeneity (H = (Number of mutations in tumor DNA−Number of mutations in cfDNA)/Number of mutations in tumor DNA). The red dots represent the patients with more mutations detected in the cfDNA than in the tumoral DNA (H < 0). b Boxplot representation of the MTV values according to the presence (red dots, H < 0) or absence (blue dots, H ≥ 0) of supplementary mutations in the cfDNA. c 3D view of PET scan images for the four patients presenting supplementary mutations in the cfDNA, with MTV in orange boxes and TLG in green boxes
CfDNA monitoring and correlation with iPET and final analysis
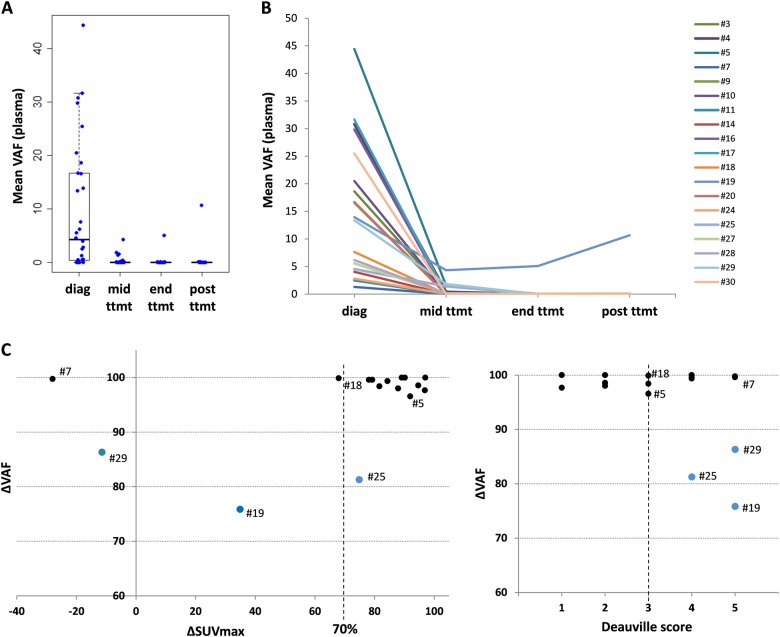
Longitudinal analysis of the plasma samples collected at mid-treatment and at the end of treatment or at relapse/progression was performed and correlated with the PET scan characteristics (ΔSUVmax and Deauville score).
Upon treatment with R-CHOP/R-CHOP-like regimens, a rapid clearance of cfDNA mutations was observed in the 16 baseline positive cases available at mid-treatment (Fig. 5 and Supplementary Table S1). In four cases (#5, #19, #25, and #29), the basal DLBCL mutations did not completely disappear at mid-treatment. Among these four patients, three had a delta VAF (ΔVAF) less than 90% and were in partial response according to their Deauville Scores (#19, #25, and #29), including two with a ΔSUVmax <70% (#19 and #29). Patient (#25) had a ΔSUVmax of 75%, though the ΔVAF was 81% and ctDNA was still detectable (VAF = 1.4%). Moreover, patient #29 finally showed progression and died before the end of treatment. Conversely, for patient #7, the mutations disappeared from the cfDNA at mid-treatment (ΔVAF near 100%) despite the absence of metabolic response (Deauville score of 5) and a ΔSUVmax <70% (−28).
Fig. 5. Longitudinal assessment of mutation abundance in plasma cfDNA upon R-CHOP treatment according to interim PET scan.
a Distribution of the cfDNA mean VAFs of the patients at the different times of follow-up (diagnosis, mid-treatment, end of treatment, and 6 months post-treatment). b Evolution of the cfDNA mean VAFs for each patient throughout treatment. c ΔVAF values in plasma according to the ΔSUVmax (left) or Deauville score (right) between diagnosis and mid-treatment. The vertical dashed lines represent the cut-off ΔSUVmax of 70% (left) or Deauville score of 3 (right), and the blue dots represent patients with ΔVAF <90%
Unfortunately, at the end of treatment, some important plasma samples were missing (only 20 available). Patient #10, who presented progressive disease, had no plasma samples either at the end of treatment or after. Patient #25 had a partial response and circulating DNA of tumoral origin at mid-treatment, but no plasma samples were collected for further follow-up.
Patients’ examples
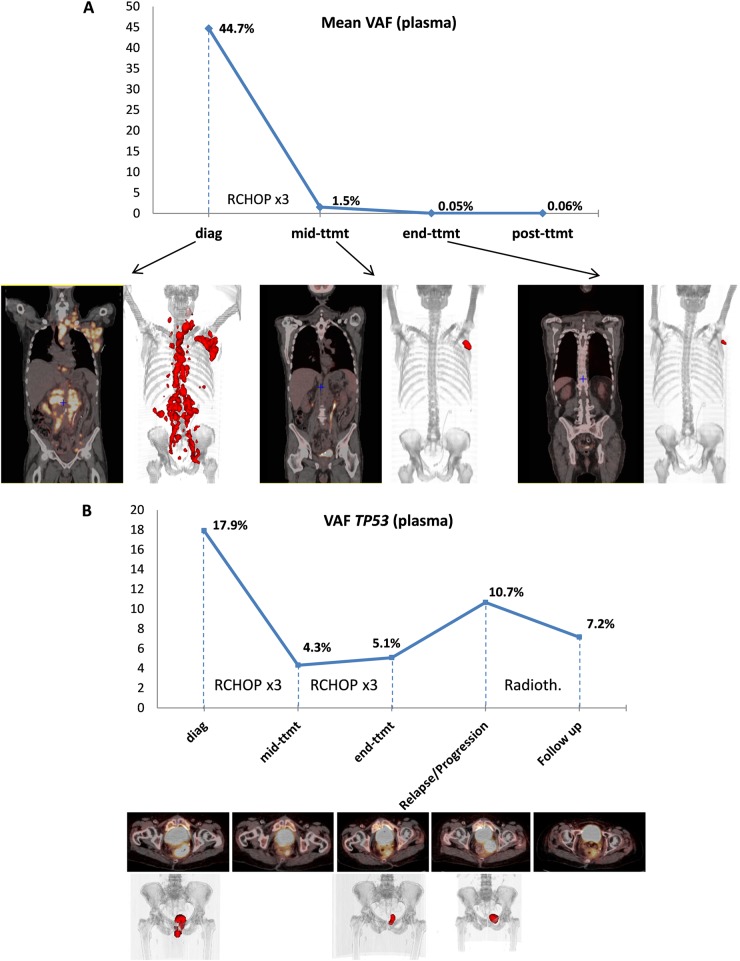
As an example, two emblematic patients were more extensively described (Fig. 6).
Fig. 6. Examples of non-invasive real-time monitoring of the DLBCL clonal evolution in plasma cfDNA.
Mean VAF and PET scan images at different times during follow-up for patient #5 (a) and patient #19 (b)
Patient #5 had one of the highest MTVs in its baseline PET scan (2441 cm3), with an important dissemination of the tumoral mass showed by the 3D view of the PET images (Figs. 4c, 6a). The plasma sample was very rich in tumoral cfDNA according to the important VAF values, which had a mean of 44.4% (ctDNA amount of 22,508 hGE/mL), and the CNV analysis showed the same gains and deletions in the plasma and tumor (Fig. 2b). More mutations were observed in the cfDNA compared with tumoral DNA (Fig. 4 and Supplementary Table S1), indicating spatial tumor heterogeneity that was probably correlated with the dissemination observed in the PET images. At mid-treatment, this patient showed a complete response with a Deauville score of 3 and a ΔSUVmax of 92%. The ΔVAF was 97%, but we still detected tumoral DNA circulating in the plasma with a mean mutation VAF of 1.5%. At the end of treatment and 6 months post treatment, this patient had yet a complete response and no more tumoral circulating DNA was found. Despite an unfavorable somatic mutation profile, including MYD88 mutations and a CDKN2A deletion, the patient is still considered in CR after 2 years of follow-up.
Patient #19 had a TP53 mutation detected both in its cfDNA (VAF = 14%) and tumoral gDNA (Supplementary Table S1). The plasma sample was informative despite the low MTV (128 cm3). This mutation was still present in the cfDNA at mid-treatment, but with a decreased VAF (4.3%, ΔVAF = 75%), though the patient was partially responding to treatment with a ΔSUVmax of 35% and a Deauville score of 5. Of note, even if we observed a decrease in the TP53 mutation frequency, the ΔVAF of 75% was the lowest ΔVAF in the cohort. At the end of treatment, the patient was still in partial response according to the PET scan images and still harbored tumoral cfDNA, as shown by a persistent detectable TP53 mutation with a VAF of 5% (Fig. 6b). Four months after the end of treatment, the patient had progressive disease and the frequency of the TP53 mutation increased to 10.7%. Five months later, treatment by radiotherapy led to an improvement according to PET scan data, though the two following PET scans showed progression and a stable persistent hypermetabolism that was not specific. Finally, we analyzed an extra plasma sample (nearly 2 years after the end of treatment) and observed that the same TP53 mutation was still present at a VAF of 7.16%, supporting the better sensitivity of the liquid biopsy.
Discussion
In this prospective study we assessed basal DLBCL genetic patterns, their modification upon treatment using plasma cfDNA analysis, and their correlation with PET scan imaging by NGS. In complement with previously published results obtained from tumor biopsies from patients enrolled in LYSA clinical trials, we demonstrated here that the 34-gene Lymphopanel (87,703 bp) is also a reliable genotyping tool to characterize tumoral cfDNA in plasma13,17. CfDNA mutations were identified in 19/30 patients (63%) cases, a similar rate to that reported by Rossi et al.7 in the training cohort (2/30, 66%). In the validation cohort, 17/20 (85%) DLBCL patients harbored cfDNA mutations, a higher rate that can be explained by another distinct technology (CAPP-Seq) and a larger gene panel (59 genes, 207,299 bp). However, despite the quantitative and qualitative differences of the interrogated genes, CAPP-Seq and the Lymphopanel-Ampliseq-based technologies allowed the recovery of at least one clonal mutation in 92.6 and 96% of DLBCL patients, respectively, as documented by in silico validation or cohort sequencing7,13. This suggests that the discrepancy observed between the two studies is most likely related to the higher sensitivity of the CAPP-Seq method. Nevertheless, we identified mutations in cfDNA up to a VAF of 4.6% in gDNA, while in the Rossi et al. study the cut-off for detection was 20%. Of note, we also recently tested our Lymphopanel-Ampliseq approach in PCNSL and showed that it represents a reliable genotyping tool in a setting where tumor cfDNA amounts are supposed to be low and inconstant8. For these reasons, we are currently developing new approaches of sequencing, as an alternative of CAPP-Seq, to date not widely implemented in routine, using our lymphopanel with molecular barecoding and dedicated bio informatic tools in order to improve sensitivity.
The 34-gene Lymphopanel has been extensively performed in more than 300 DLBCL cases17 and was initially design to discriminate the three main molecular subtypes, namely, the GCB, ABC, and PMBL subtypes and to provide markers for personalized medicine and targeted treatment. The results obtained by cfDNA genotyping are consistent with the patterns usually observed in the different subtypes and confirm the clinical relevance of the panel to detect targetable variants such as EZH2, CD79B, or MYD88 hotspot mutations. Of note, three among the four PMBL cases were informative in both tumor and plasma and displayed a typical genotype, suggesting that cfDNA is likely a promising tool for the management of PMBL.
Importantly, we were also able to detect CNVs that accurately and specifically matched with gDNA in some cases (Fig. 2), including the unfavorable prognostic deletion of TP53 or CDKN2A. However, in our hands and with the AmpliSeq Technology, CNVs were detectable in cfDNA in a limited number of cases, with a minimal mean VAF of 13% or 1630 hGE/mL ctDNA.
Factors that influence the amount of tumor cfDNA are partially known. Here, we confirmed previous works showing that baseline cfDNA amounts strongly correlate with tumor burden indicators including LDH level and the IPI score but also MTV or TLG12. Moreover, the patients presenting additional mutations in cfDNA, compared to gDNA, had a high MTV, suggesting that cfDNA mutations more accurately reflect tumor heterogeneity than gDNA analysis, which is limited by the unique biopsy site. Regarding the well-established prognostic value of the MTV in DLBCL, cfDNA amounts may explain the strong prognostic values recently reported in DLBCL12,24. In addition to the combined analysis of the MTV and GCB/ABC status or BCL2/MYC expression25, it appears relevant to obtain both cfDNA and MTV at baseline to risk stratify newly diagnosed DLBCL.
We performed longitudinal analysis of plasma samples collected at mid-treatment, at the end of treatment, or at relapse/progression and correlated the results with PET scan characteristics. A rapid decrease in the cfDNA mutation VAF was observed in most cases. Rapid clearance of ctDNA after two cycles of R-CHOP, higher than 2 log, has been recently reported as a strong prognostic marker24. From mid-treatment, to increase the sequencing sensitivity and for cost-effectiveness reasons, only mutations detected at baseline were monitored during the follow-up. Some patients with partial remission at mid-treatment, according to their Deauville score and ΔSUVmax (<70%), still had basal DLBCL mutations circulating in the plasma. Conversely, despite the partial response, some patients had no ctDNA in the plasma. Thus, the targeted approach is questionable because it precludes any detection of new (or subclonal) variants emerging during treatment, as it has been reported by Rossi et al7. Regardless of the liquid biopsy value, follow-up with the entire panel is recommended. These data also suggest that interim iPET and cfDNA can both be used to define early response during DLBCL treatment9. Their respective and complementary relevance remains to be determined in a larger cohort.
Interestingly, one elderly patient (#19) in partial remission had progressive disease 6 months after the end of treatment and still harbored tumoral cfDNA, as shown by a persistent detectable pathogenic TP53 mutation. However, the tumor volume still remains stable and, importantly, the mutation was also detected in the bone marrow without any evidence of invasion (according to PET scan and bone marrow sample analysis). Persistent variants should be analyzed with caution, especially those targeting TP53 in elderly patients because it has been demonstrated that blood cells from more than 2% of individuals (5–6% of people older than 70 years) contain mutations that may represent premalignant events that cause clonal hematopoietic expansion26,27. Age-associated, low-frequency TP53 mutations were also found in 100% of peripheral blood samples from 15 women with and without ovarian cancer (none with hematological disorder)28.
To conclude, our prospective study demonstrates that cfDNA genotyping of DLBCL is an accurate genotyping tool and represents a real-time and non-invasive approach for follow-up. It highlights the major interest of liquid biopsy in the context of bulky tumors, where cfDNAs are more representative than gDNA for capturing the entire tumoral mutation profile. Therefore, cfDNA analysis represents a complementary approach to PET scan imaging at baseline and during follow-up for the management of DLBCL.
Electronic supplementary material
Supplementary Table S1: Mutations and VAF in tumors and plasmas at diagnosis and during follow-up
Acknowledgements
This research was supported by grants from the Cancéropole Nord-Ouest, the Ligue Contre le Cancer, and Henri Becquerel Center.
Authors’ contributions
F.J., E.B., and P.J.V. designed the study, interpreted data, and wrote the manuscript. E.B., C.M., V.M., P.R., and P.B. performed the molecular studies. S.B., P.V., L.F.P., and H.T. contributed to the manuscript revisions. H.T., S.B., A.L.M., E.L., N.C., S.L., P.L., A.S., H.L., P.E., S.V., J.L., and L.F.P. provided study material and clinical data. J.M.P. performed the pathological revision. P.J.V. contributed to the bioinformatics analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41408-018-0111-6).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowes LE, et al. Circulating tumor cells (CTC) and cell-free DNA (cfDNA) workshop 2016: scientific opportunities and logistics for cancer clinical trial incorporation. Int. J. Mol. Sci. 2016;17:E1505. doi: 10.3390/ijms17091505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alizadeh AA, et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci. Rep. 2015;5:7610. doi: 10.1038/srep07610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anker P, Lyautey J, Lederrey C, Stroun M. Circulating nucleic acids in plasma or serum. Clin. Chim. Acta. 2001;313:143–146. doi: 10.1016/S0009-8981(01)00666-0. [DOI] [PubMed] [Google Scholar]
- 6.Bohers E, et al. Somatic mutations of cell-free circulating DNA detected by next-generation sequencing reflect the genetic changes in both germinal center B-cell-like and activated B-cell-like diffuse large B-cell lymphomas at the time of diagnosis. Haematologica. 2015;100:e280–e284. doi: 10.3324/haematol.2015.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi D, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017;129:1947–1957. doi: 10.1182/blood-2016-05-719641. [DOI] [PubMed] [Google Scholar]
- 8.Fontanilles M, et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget. 2017;8:48157–48168. doi: 10.18632/oncotarget.18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Gouill S, Casasnovas RO. Interim PET-driven strategy in de novo diffuse large B-cell lymphoma: do we trust the driver? Blood. 2017;129:3059–3070. doi: 10.1182/blood-2016-05-672196. [DOI] [PubMed] [Google Scholar]
- 10.Kurtz DM, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood. 2015;125:3679–3687. doi: 10.1182/blood-2015-03-635169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roschewski M, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16:541–549. doi: 10.1016/S1470-2045(15)70106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherer F, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016;8:364ra155. doi: 10.1126/scitranslmed.aai8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois S, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin. Cancer Res. 2016;22:2919–2928. doi: 10.1158/1078-0432.CCR-15-2305. [DOI] [PubMed] [Google Scholar]
- 14.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 15.Molina TJ, et al. Young patients with non-germinal center B-cell-like diffuse large B-cell lymphoma benefit from intensified chemotherapy with ACVBP plus rituximab compared with CHOP plus rituximab: analysis of data from the Groupe d’Etudes des Lymphomes de l’Adulte/lymphoma study association phase III trial LNH 03-2B. J. Clin. Oncol. 2014;32:3996–4003. doi: 10.1200/JCO.2013.54.9493. [DOI] [PubMed] [Google Scholar]
- 16.Jardin F, et al. Immunoglobulin heavy chain/light chain pair measurement is associated with survival in diffuse large B-cell lymphoma. Leuk. Lymphoma. 2013;54:1898–1907. doi: 10.3109/10428194.2013.767456. [DOI] [PubMed] [Google Scholar]
- 17.Dubois S, et al. Biological and clinical relevance of associated genomic alterations in MYD88 L265P and non-L265P-mutated diffuse large B-cell lymphoma: analysis of 361 cases. Clin. Cancer Res. 2017;23:2232–2244. doi: 10.1158/1078-0432.CCR-16-1922. [DOI] [PubMed] [Google Scholar]
- 18.Boeva V, et al. Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics. 2014;30:3443–3450. doi: 10.1093/bioinformatics/btu436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meignan M, et al. Metabolic tumour volumes measured at staging in lymphoma: methodological evaluation on phantom experiments and patients. Eur. J. Nucl. Med. Mol. Imaging. 2014;41:1113–1122. doi: 10.1007/s00259-014-2705-y. [DOI] [PubMed] [Google Scholar]
- 20.Boellaard R, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur. J. Nucl. Med. Mol. Imaging. 2015;42:328–354. doi: 10.1007/s00259-014-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itti E, et al. An international confirmatory study of the prognostic value of early PET/CT in diffuse large B-cell lymphoma: comparison between Deauville criteria and ΔSUVmax. Eur. J. Nucl. Med. Mol. Imaging. 2013;40:1312–1320. doi: 10.1007/s00259-013-2435-6. [DOI] [PubMed] [Google Scholar]
- 22.Barrington, S. F. et al. Role of imaging in the staging and response assessment of lymphoma: consensus of the International Conference on Malignant Lymphomas Imaging Working Group. J. Clin. Oncol.32, 3048–3058 (2014). [DOI] [PMC free article] [PubMed]
- 23.R: A language and environment for statistical computing. R Foundation for Statistical Computing, R Development Core Team. Vienna, Austria (2008). http://www.r-project.org/.
- 24.Kurtz DM, et al. Circulating tumor DNA is a reliable measure of tumor burden at diagnosis of diffuse large B cell lymphoma: an international reproducibility study. Blood. 2017;130:310–310. doi: 10.1182/blood-2016-12-758599. [DOI] [PubMed] [Google Scholar]
- 25.Cottereau AS, et al. Molecular profile and FDG-PET/CT total metabolic tumor volume improve risk classification at diagnosis for patients with diffuse large B-cell lymphoma. Clin. Cancer Res. 2016;22:3801–3809. doi: 10.1158/1078-0432.CCR-15-2825. [DOI] [PubMed] [Google Scholar]
- 26.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krimmel JD, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc. Natl Acad. Sci. USA. 2016;113:6005–6010. doi: 10.1073/pnas.1601311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Mutations and VAF in tumors and plasmas at diagnosis and during follow-up