Abstract
In this study, we provide the first description of Cystoisospora infection in Asian small-clawed otters (Aonyx cinereus). In July 2017, three juvenile otters recently imported from the Republic of Indonesia showed severe diarrhea and were diagnosed with coccidial infection; two of them eventually died. Fecal examination revealed the presence of numerous oocysts. Sporulated oocysts showed typical Cystoisospora features, measuring 24.6 ± 1.6 (22.0–27.0) × 21.8 ± 1.4 (19.0–25.0) μm, with an oocyst length/width ratio of 1.1 ± 0.1 (1.0–1.3). Each sporocyst contained four sporozoites in a head-to-tail arrangement. The Stieda body was absent, and the sporocyst residuum was present. These morphological characteristics differentiated this species from the other valid Cystoisospora species described from mustelids. Molecular analysis was conducted at two loci: the nuclear 18S ribosomal RNA and mitochondrial cytochrome c oxidase subunit I genes. The 18S sequence showed high similarity with canine Cystoisispora ohioensis (1-bp difference, 1422/1423 [99.9%]). At the cytochrome c oxidase subunit I gene locus, the sequence from otters was identical to that of feline Cystoisospora rivolta (847/847 [100%]). Phylogenetic analyses using concatenated data demonstrated that Cystoisospora sp. from otters and C. rivolta grouped together in the same Cystoisospora clade. Based on these data, we concluded that Cystoisospora sp. detected from otters appeared to be highly similar to C. rivolta.
Keywords: Cystoisospora, Isospora, Otter, Phylogenetic analysis, Coccidianl infection
Graphical abstract
Highlights
-
•
We characterized Cystoisospora infection in three Asian small-clawed otters.
-
•
Two otters showed anorexia, green diarrhea, and vomiting.
-
•
Fecal examination revealed numerous oocysts with typical Cystoisospora features.
-
•
Sporulated oocysts were morphologically differentiated from those of other Cystoisospora spp. from mustelids.
-
•
Molecular and morphological features were highly similar to feline C. rivolta.
1. Introduction
Coccidia of the genus Cystoisospora are common intestinal protozoan parasites found in mammals. Traditionally, this genus was assigned to the genus Isospora, which classified into the family Eimeriidae. Morphological and molecular characterization has differentiated the genus Isospora into two groups of parasites, i.e., the genus Isospora of the family Eimeriidae and the genus Cystoisospora of the family Sarcocystidae (Barta et al., 2005). Isospora species are found in birds and reptiles, and their sporocysts possess a Stieda body, whereas Cystoisospora oocysts are found in carnivores, primates, and swine and possess sporocysts without a Stieda body (Frenkel, 1977; Barta et al., 2005).
Otters are semiaquatic carnivorous mammals of the subfamily Lutrinae, which is part of the family Mustelidae. The subfamily Lutrinae is represented by 9 genera and 12 extant species. The Asian small-clawed otter (Aonyx cinereus, syn. Lutra cinerea) is the smallest otter species native to South and Southeast Asia. Statics by the Japanese Ministry of Health, labor and Welfare show a total of 13 captive bred individuals of Asian small-clawed otter were legally imported into Japan from 2015 to 2017 (http://www.mhlw.go.jp/, access 2018.7.2).
Reports of Cystoisospora infection in otters are rather scarce, and only a single species, Cystoisospora lutrae, has been reported in the European otter (Lutra lutra) in Spain (Torres et al., 2000). In the present study, we characterized Cystoisospora species in three juvenile Asian small-clawed otters using morphological and molecular techniques.
2. Materials and methods
2.1. Animals and specimens
Three juvenile Asian small-clawed otters (2 males and 1 female) were captive breeding littermates born on May 1st, 2017, and had been imported from the Republic of Indonesia to Japan on June 25, 2017. At the Indonesia's breeding farm, they were given milk and cat food in a cat free environment. On July 2, 2017, these three otters were diagnosed with coccidial infection based on the presence of numerous unsporulated oocysts (Fig. 1a) using a standard fecal flotation technique at Den-en-chofu Animal Hospital, Japan. Two of the three animals showed anorexia, green diarrhea, and vomiting and were administered a single dose of toltrazuril (15 mg/kg, p.o.). Although these two animals showed reduced oocyst excretion, they showed persistent diarrhea and died after 2 and 7 days, respectively.
Fig. 1.
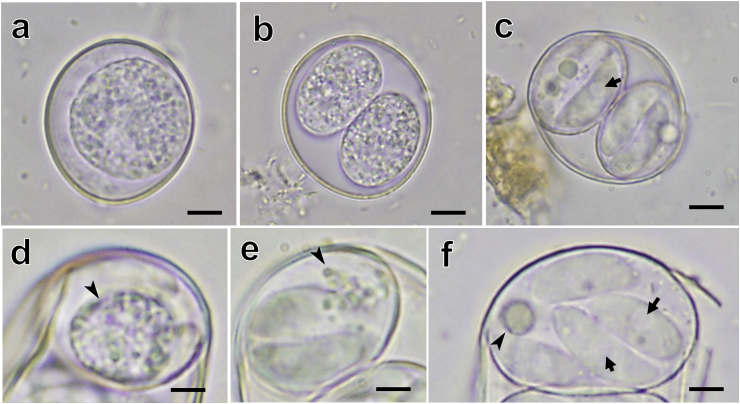
Photographs of Cystoisospora oocysts detected from Asian small-clawed otters. (a and b) Immature oocysts containing one (a) or two sporoblasts (b). (c) Mature oocysts containing two sporocysts, each with four club-shaped sporozoites. Sporozoites contained rounded retractile vacuoles (arrow). (d–f) Sporozoites containing sporocyst residuum, which was composed of numerous small granules (d and e) or a rounded granule (f).
Fecal specimens were collected from these animals, placed in separate vials with 2.0% (w/v) aqueous potassium dichromate (K2Cr2O7) solution. Oocysts sporulation was induced by incubation at room temperature for 10 days and at 4 °C for prolonged storage. For histological examination, the animal that died 7 days after diagnosis was necropsied in Nippon Veterinary and Life Science University, and the intestines, liver, lung, kidney, brain, and spleen were fixed in 10% neutral-buffered formalin. Formalin-fixed tissue specimens were embedded in paraffin, cut into 5-μm-thick sections, and stained with hematoxylin and eosin (H&E).
2.2. Microscopic analysis
Oocysts were observed with a BX41 microscope (Olympus, Japan), and photomicrographs were captured using a DP71 photomicroscope (Olympus). Line drawings were made using FireAlpaca Ver. 2.0 (http://firealpaca.com/). Measurements were made with a calibrated ocular micrometer under an oil immersion objective and are reported as means ± standard deviations with the range in parentheses.
2.3. Molecular analysis
Oocysts were purified by flotation methods from each specimens separately, and a single oocyst was isolated using a manually pulled glass micropipette under an Olympus SZX stereomicroscope (Olympus). Genomic DNA was extracted using a Power Soil DNA isolation kit (MoBio Laboratories, USA) as previously described (Tokiwa et al., 2017). Double distilled water (no fecal specimen) was included as a negative control. The resulting DNA specimens were used as templates for polymerase chain reaction (PCR) amplification.
The partial fragment of the 18S rRNA gene (18S) and the mitochondrial gene encoding cytochrome c oxidase subunit 1 (cox1) were amplified using the primer sets EF (5′-GAACTGCGAATGGCTCATT-3′)/ER (5′-CTTGCGCCTACTAGGCATTC-3′) (Kvicerová et al., 2008) and Sdae-Cox1_260F (5′-GATCTTTATGTTYTTRATGCC-3′)/Sdae-Cox1_1147R (5′-CATTACCCATAACYACACC-3′) (Ogedengbe et al., 2016), respectively. The PCR mixture contained 2.5 μL of 10× Ex Taq buffer (Takara Bio Inc., Japan), 0.2 mM dNTPs (Takara Bio, Inc.), 0.2 μM each primer, 1 U Ex Taq polymerase (Takara Bio, Inc.), and 1 μL DNA extract in a total volume of 20 μL. PCR conditions consisted of initial denaturation at 94 °C for 5 min; 35 cycles at 94 °C for 30 s, 54 °C (18S) or 50 °C (cox1) for 1 min, and 72 °C for 1 min; and a final extension step at 72 °C for 7 min (18S) or 1 min (cox1). The amplified DNA was applied to 2% agarose gels, electrophoresed, and visualized under an LED transilluminator. PCR products were sent to a sequence service (Macrogen Corp., Kyoto, Japan) and analyzed with an ABI3730xl DNA Analyzer (Thermo Fisher Scientific, USA) using the same PCR primers.
2.4. DNA sequence analyses
Sequence analyses were conducted on nucleotide sequences of the 18S and cox1 genes using MEGA version 7 software (Kumar et al., 2016). Sequence similarity was determined separately using the BLASTN program with the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/Blast.cg).
Phylogenetic trees of concatenated 18S and cox1 sequence data were generated using MEGA. Each sequence was derived from the same isolate/strain available in the GenBank/DDBJ/EMBL databases, as described by Ogedengbe et al. (2016). In total, 14 concatenated sequences, including two new sequences derived from Cystoisospora sp. detected from the otters in the present study, were aligned using MAFFT version 7 (Katoh and Standley, 2013) with the Q-I setting and ClustalW (Larkin et al., 2007), followed by manual editing. All positions with less than 95% site coverage were eliminated, and a total of 875 positions were included in the dataset. The phylogenetic tree was reconstructed by neighbor joining (NJ) and maximum likelihood (ML) methods using the Tamura-Nei model (Tamura and Nei, 1993) and General Time Reversible model with gamma distribution plus invariant sites, respectively. Bootstrap values for NJ and ML were obtained from 2000 to 1000 replicates, respectively.
3. Results
3.1. Description of Cystoisospora sp. from Asian small-clawed otters
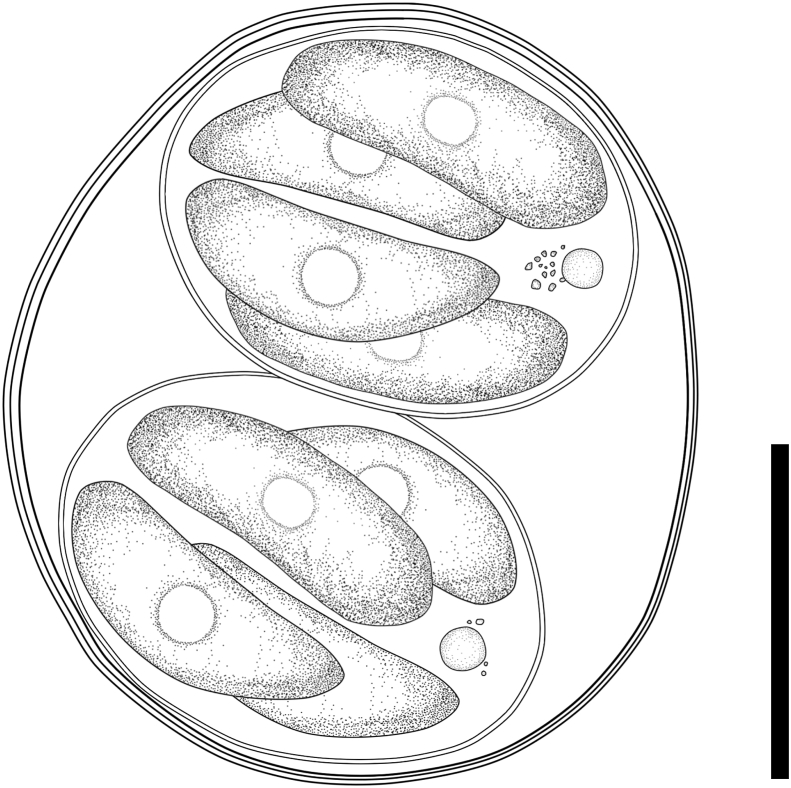
Fecal examination revealed the presence of numerous Cystoisospora oocysts. Although the accurate sporulation time could not be identified, some oocysts were mature at 70 h after sampling. The sporulated oocysts were morphologically identical in three fecal specimens. Sporulated oocysts were subspherical to spherical and measured 24.6 ± 1.6 (22.0–27.0) × 21.8 ± 1.4 (19.0–25.0) μm, with an oocyst length/width ratio of 1.1 ± 0.1 (1.0–1.3), and contained two sporocysts (Fig. 1, Fig. 2), each of which harbored four sporozoites (Fig. 1, Fig. 2). The oocyst wall was smooth and measured 0.3 μm thick, with a colorless outer layer and light yellowish-brown inner layer. The oocyst residuum, polar granule, and micropyle were absent. Sporocysts are elongate-ovoid with a smooth wall and measured 16.3 ± 1.6 (14.0–20.0) × 12.3 ± 1.0 (11.0–15.0) μm, with a sporocyst length/width ratio of 1.3 ± 0.1 (1.1–1.6) μm. Each sporocyst contained four sporozoites in a head-to-tail arrangement. Sporozoites were club-shaped, possessed a rounded refractile body at the center part, and measured 9.6 ± 0.6 (9.0–11.0) × 4.1 ± 0.3 (4.0–5.0) μm, with a length/width ratio of 2.4 ± 0.2 (2.0–2.8). The Stieda body and substieda body were absent. Sporozoites in the sporocyst residuum were initially compact masses of small granules (Fig. 1d), but gradually decreased in size and became a single rounded granule (2–5 μm in diameter) with small granules (Fig. 1, Fig. 2).
Fig. 2.
Line drawing of sporulated oocysts of Cystoisospora rivolta-like oocysts from small-clawed otters. Bar = 10 μm.
Postmortem histology analysis of the otter after toltrazuril treatment showed bacterial pneumonia and degeneration and necrosis of the epithelial cells in the mucosa of the intestine. No developmental stages of Cystoisospora sp. were observed in these legions (data not shown).
3.2. Sequence analyses of Cystoisospora sp. from Asian small-clawed otters
A 1423-bp sequence of the 18S gene obtained from the three otters was identical in each otter and was deposited in the DDBJ database under accession no. LC377842. Sequence comparisons revealed that the present sequence from otters shared the highest identity (99.9%) for C. ohioensis (1422/1423, AY618555; 1421/1423, KT1843651, KT184359, KT184366) and C. suis (1421/1423, KF854252, KF854251). Similarities of 99.8% for C. belli (1420/1423, U94787, JX025649, AB268326, DQ060659, DQ060661), 99.6% for C. canis (1417/1423, KT184368), 99.4% for C. timoni (1415/1423, AY279205, EU200732) and C. orlovi (1414/1423, AY365026), and 99.2% for C. felis (1412/1423, KT134364) and C. canis (1412/1423, KT184362, KT184363, TK184360) were also noted. Sequence identity with the near complete 18S sequence of C. rivolta (AY618554) was 81.1% (1154/1447); however, this low similarity was thought to be due to incorrect registration of the sequence because high similarities (>99%) were observed with the red yeast Rhodosporidium spp. The partial 18S sequence showed 100% identity (401/401) with that of C. rivolta (KT184367) and was identical to those of C. canis (KT184360, KT184362, KT184368), C. ohioensis (KT184351, KT184359, KT184366), C. suis (KF854252, KF854251), C. belli (U94787, JX025649, AB268326, DQ060659, DQ060661), and C. timoni (EU200792, AY279205).
An 847-bp sequence of the cox1 gene obtained from three otters was identical in each otter and deposited in the DDBJ database under accession no. LC377843. Sequence comparisons using BLAST revealed that the new sequence from otters was completely identical to that from C. rivolta (KT184383) and showed 99.8% (845/847) similarity to C. ohioensis (KT184384), 98.7% (836/847) similarity to C. canis (KT184385), and 98.4% (814/827) similarity to C. felis (KT184386). Table 1 shows the multiple alignment of cox1 sequences from 5 Cystoisospora species and the present species. A single nonsynonymous substitution occurred in between the present species and C. ohioensis (362: Ile to Leu), C. canis, and C. felis (371: Ile to Leu), and 2 nonsynonymous substitutions (362: Ile to Leu, 374: Leu to Ile) occurred in C. suis.
Table 1.
Alignment of cox1 sequences of Cystoisospora spp.
| Species | GenBank/DDBJ/EMBL accession Nos. | Nucleotide variable site |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 32 | 40 | 112 | 115 | 148 | 175 | 179 | 253 | 271 | 298 | 362 | 371 | 374 | 382 | 472 | 475 | 521 | 575 | 706 | 847 | ||
| Present species | LC377843 | C | A | T | A | A | G | C | T | T | T | A | A | T | T | A | A | C | C | A | A |
| CystoIsospora rivolta | KT184383 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Cystoisospora ohioensis | KT184384 | . | . | . | . | T | . | . | . | . | . | C* | . | . | . | . | . | . | . | . | . |
| Cystoisospora suis | KF854265 | – | – | – | – | – | . | . | . | . | . | C* | . | A* | . | . | . | . | . | – | – |
| Cystoisospora canis | KT184385 | . | . | A | C | . | . | T | A | C | C | . | C* | . | A | C | . | . | A | G | . |
| Cystoisospora felis | KT184386 | T | C | . | C | . | A | T | A | C | C | . | C* | . | . | . | G | T | A | G | T |
| Codon position | 1 | 3 | 3 | 3 | 3 | 3 | 1 | 3 | 3 | 3 | 1 | 1 | 1 | 3 | 3 | 3 | 1 | 1 | 3 | 3 | |
Dot represent matches with nucleotides present in cox1 sequence of Cystoisospora sp. from Asian small-crawed otters (LC377843). Asterisk represent nonsynonymous substitution sites in which amino acid replacement.
3.3. Phylogenetic analyses of Cystoisospora spp.
Phylogenetic analyses using NJ and ML methods with concatenated sequences produced topologically similar trees that supported the monophyletic clade of the genus Cystoisospora (Fig. 3). The present sequence from Asian small-clawed otters was placed within the monophyletic group with C. rivolta detected from a domestic cat (Felis catus) in Canada.
Fig. 3.
Mid-point rooting tree of Cystoisospora spp. inferred from concatenated alignment of two nucleotide sequences (18S and cox1). Nucleotide sequences of Cystoisospora spp. GenBank/DDBJ/EMBL accession numbers for 18S and cox1 are shown next to species names. Nodal support values (NJ/ML) greater than 60% are represented on ML branches. Scale bar represents 0.01 nucleotide substitutions per sites.
4. Discussion
Here, we present the first description of a coccidian from the Asian small-clawed otter. All animals showed typical symptoms of cystoisosporiosis, also known as isosporiosis, including severe diarrhea and oocyst excretion. Although a single treatment with toltrazuril reduced the oocyst shedding, two otters eventually died. The cause of death in these two otters was estimated exhaustion due to diarrhea, followed by aspiration pneumonia.
Morphological observation of the sporulated oocysts from three individuals showed infection with a single coccidian species. The sporulated oocysts contained two sporocysts without a Stieda body, which each harbored four sporozoites, representing typical Cystoisospora features. According to the guideline by Duszynski and Wilber (1997), coccidian species should be compared morphologically in detailed with other species having similar features and belonging to the host family. Tenter et al. (2002) discussed morphological and molecular characters and host-related parameters to be considered for the classification of coccidia. Therefore, the morphological characteristics of Cystoisospora oocysts found in otters were compared with those of other Cystoisospora spp. previously described in carnivores of the family Mustelidae. Table 2 shows a list of 12 valid species of the genus Cystoisospora (syn. Isospora) detected from mustelids. Only a single species of Cystoisospora has been described for otter species (subfamily Lutrinae), i.e., C. lutrae, from the European otter (Lutra lutra) originating from Spain (Torres et al., 2000). However, C. lutrae has significantly larger oocysts. Unidentified Cystoisospora oocysts were reported from the North American otter (Lontra canadensis) (Hoover et al., 1985); however, measurements of oocysts, line drawings, and photomicrographs have not been reported. In Cystoisospora species described from non-otter mustelids (subfamily Mustelinae), the oocysts of C. melis, C. altaica, C. pavlowskyi, and C. laidlawi are much larger (Hoare, 1927; Kotlán and Pospesch, 1933; Svanbaev, 1956; Svanbaev and Rachmatullina, 1971), and the oocysts of C. eversmanni and C. goussevi are much smaller (Svanbaev, 1956; Musaev and Veison, 1983). C. hoogstraali has much larger oocysts and has sporocysts with prominent Stieda bodies (Prasad, 1961). The latter features indicate that C. hoogstraali may an Isospora pseudoparasite passing through the intestine after consumption of birds, reptiles, or feces. The oocysts of C. martessii and C. nivalis are similar in size, whereas the sporocysts of these organisms differ in size (Nukerbaeva, 1981; Musaev and Veison, 1983). Finally, the oocysts of C. africana resemble those of the present species, whereas the sporozoites of C. africana are elongated and have refractile vacuoles at their ends (Prasad, 1961). These features support that this may be a novel Cystoisospora species in mustelids.
Table 2.
Comparative morphology of Cystoispspora spp. recorded from mustelids.
| Cystoisospora species | Type species |
Oocyst |
Description | Sporocyst |
Description | Sporozoite | References |
|---|---|---|---|---|---|---|---|
| Species (Subfamily) | Size | Size | |||||
| C. arctopitheci (Rodhain, 1933) | Eira barbara (Mustelinae) | L: 27.7 (22.7–32.7) | Subspherical to ellipsoidal | L: 17.6 (13.1–20.5) | SR: present | Hendricks, 1974 | |
| W: 24.3 (20.5–27.3) | M: absent | ||||||
| L/W: 1.14 (1.05–1.30) | OR: absent | W: 12.5 (9.8–15.9) | SB: absent | ||||
| PG: absent | |||||||
| C. martessii (Nukerbaeva, 1981) | Martes zibellina (Mustelinae) | L: 25.2–28.0 | Ovoid, short oval to spherical | L: 11.2–16.8 | SR: present | Elongated | Nukerbaeva, 1981 |
| W:16.8–22.4 | M: absent | W: 8.4–11.2 | |||||
| OR: absent | |||||||
| C. melis (Kotlán and Pospesch, 1933) | Meles meles (Mustelinae) | L: 32.8 (25.6–37.8) | Oval | L: 14-16 | SR: present | Round at one end and the other end tapered | Kotlán and Pospesch, 1933, Anwer et al., 2000 |
| W:26.9 (24–29.6) | M: absent | L: 14.2 | |||||
| L/W: 1.22 (1.10–1.57) | OR: absent | W: 12 | SB: absent | W: 4.0 | |||
| PG: absent | |||||||
| C. altaica (Svanbaev and Rachmatullina, 1971) | Mustela altaica (Mustelinae) | L: 33.3 (28.0–33.6) | Oval to spherical | L: 14.0–16.8 | SR: present | Svanbaev and Rachmatullina, 1971 | |
| W: 27.7 (25.2–28.0) | M: absent | ||||||
| L/W: 1.24 (1.11–1.24) | OR: absent | W: 11.1–16.8 | |||||
| PG: absent | |||||||
| C. eversmanni (Svanbaev, 1956) | Mustela eversmanni (Mustelinae) | L: 18.5 (16–20) | Oval | L: 11.5 (10–13.5) | SR: present | Yi-Fan et al., 2012 | |
| W: 14.8 (12–16) | M: absent | ||||||
| L/W: 1.3 (1.1–1.6) | OR: absent | W: 9.8 (9–11) | SB: absent | ||||
| PG: absent | |||||||
| C. pavlowskyi (Svanbaev, 1956) | Mustela eversmanni (Mustelinae) | L: 32.2 (29–36) | Oval | L: 19.5 (18–21) | SR: present | Yi-Fan et al., 2012 | |
| W: 27.3 (26.5–28.5) | M: absent | ||||||
| L/W: 1.2 (1.1–1.4) | OR: absent | W: 14.4 (12–15) | SB: absent | ||||
| PG: absent | |||||||
| C. laidlawi (Hoare, 1927) | Mustela putorius furo (Mustelinae) | L: 34 (32.0–36.8) | Ovoid | L: 20.8 | SR: present (coarse granules) | Sausage shaped | Hoare, 1927 |
| M: absent | |||||||
| W: 29 (27.2–30.4) | OR: absent | W: 14.4 | SB: absent | ||||
| PG: absent | |||||||
| C. goussevi (Musaev and Veison, 1983) | Mustela nivalis (Mustelinae) | L: 22.4 (22–25) | Ovoid | L: 12 (10–13) | SR: present | Elongated | Musaev and Veison, 1983 |
| W: 17.4 (16–19) | M: absent | ||||||
| L/W: 1.35 (1.33–1.37) | OR: present | W: 7 (6–8) | SB: present | ||||
| PG: prebent | |||||||
| C. nivalis (Musaev and Veison, 1983) | Mustela nivalis (Mustelinae) | L: 20.6 (20.0–23.0) | Oval | L: 12.5 (12.0–13.0) | SR: present | Lemon or pear shaped | Musaev and Veison, 1983 |
| W: 18.4 (18.0–21.0) | M: absent | ||||||
| L/W: 1.1 (1.09–1.11) | OR: absent | W: 8.0 (7.0–9.0) | SB: absent | ||||
| PG: absent | |||||||
| C. africana (Prasad, 1961) | Ictonyx lybicus (Mustelinae) | LW: 26 (25–27) | Spherical | LW: 15.5–15.6 | SR: present (prominent) | Elongated | Prasad, 1961 |
| M: absent | L: 13.5 | ||||||
| OR: absent | SB: absent | W: 3.0 | |||||
| PG: absent | |||||||
| C. hoogstraali (Prasad, 1961) | Ictonyx libycus (Mustelinae) | L: 38 (37–40) | Ellipsoid | L: 19.5–21 | SR: present (bulky) | Club-shaped | Prasad, 1961 |
| M: present (small button-like) | |||||||
| W: 33 (32–34) | OR: absent | W: 13-15 | SB: absent | L: 18–19W: 4-6 | |||
| PG: present | |||||||
| C. lutrae (Torres et al., 2000) | Lutra lutra (Lutrinae) | L: 31.2 (27.5–32) | Spherical to subspherical | L: 18.2 (17–19) | SR: present | Spindle-shaped | Torres et al., 2000 |
| W: 29.6 (28–31) | M: absent | L: 12.4 | |||||
| L/W: 1.04 (1.0–1.12) | OR: absent | W: 14.4 (14–16) | W: 2.5 | ||||
| PG: absent | |||||||
| Present species | Aonyx cinereus (Lutrinae) | L: 24.6 (22.0–27.0) | Subspherical to spherical | L: 16.3 (14.0–20.0) | SR: present | Club-shaped | This study |
| W: 21.8 (19.0–25.0) | M: absent | L: 9.6 | |||||
| L/W: 1.1 (1.0–1.3) | OR: absent | W: 12.3 (11.0–15.0) | SB: absent | W: 4.1 | |||
| PG: absent |
Numerous phylogenetic studies have focused on the analysis of 18S, 28S, ITS, and cox1 genes, and 18S and cox1 genes have been commonly used for coccidians belonging to the families Sarcocystidae and Eimeriidae (Carreno et al., 1998; Morrison et al., 2004; Ogedengbe et al., 2011, 2016). The coccidian 18S sequence is phylogenetically useful for distant taxa, but seems to be much less variable than that in other protozoan parasites and to be insufficient for resolving the relationships among closely related coccidian species (Morrison et al., 2004). In contrast, the cox1 sequence provides sufficient sequence divergence for studying phylogenetic relationships among closely related coccidian species. Ogedengbe et al. (2016) made specific cox1 primers for Sarcocystidae groups and suggested that cox1 sequences from sarcocystids (i.e., Toxoplasma, Neospora, and Hammondia) appeared well suited for species delimitation; however, sequences from Cystoisospora spp. showed remarkably limited sequence variation. In this study, because no sequence data exist for Cystopisospora species infecting mustelids, we compared Cystoisospora sequences from other animals. Interestingly, Cystoisospora sp. from otters showed the highest cox1 sequence similarity to C. rivolta. Phylogenetic analyses confirmed the genus Cystoisospora as being a monophyletic clade comprising two morphologically supported clades: (1) one including feline C. felis and canine C. canis, which possess large (>30 μm long) and egg-shaped oocysts (Wenyon, 1923; Lepp and Todd, 1974); and (2) the other including feline C. rivolta, canine C. ohioensis, swine C. suis, and otter Cystoisospora, which possess small oocysts (<30 μm long) (Biester and Murray, 1934; Dubey, 1975, 1979). We cannot molecularly differentiate the present Cystoisospora species from C. rivolta. Furthermore, the sporulated oocysts of the present species morphologically resembles those of C. rivolta from domestic cats, measuring 25.4 (23–29) × 23.4 (20–26) μm in diameter (Dubey, 1979); thus, we have not proposed this as a novel species and have designated this species as Cystoisospora cf. rivolta.
The definitive host specificity of Cystoisospora species is relatively narrow, perhaps at the host genus or family level. Nukerbaeva (1981) reported that C. martessii from sables (Martes zibellina) and C. laidlawi from minks (Mustela vision) were not transmissible between their respective hosts, even if in the same family Mustelidae. Similarly, crossinfection of C. ohioensis from domestic dogs and C. rivolta from domestic cats does not occur among these species (Dubey, 1975). Cystoisospora rivolta was first named from domestic cats. Subsequently, Wenyon (1923) detected Cystoisospora oocysts morphologically similar to those of C. rivolta from domestic dogs and considered C. rivolta from dogs and cats to be identical. Dubey (1975) separated canine C. rivolta on the basis of crossinfection experiments and proposed C. ohioensis. Feline C. rivolta oocysts are commonly found in domestic cats worldwide; however, structurally similar oocysts have also been reported from carnivores belonging to the family Felidae; i.e., cheetahs (Acinonyx jubatus), jungle cats (F. chaus), European wildcats (F. silvestris), clouded leopards (Neofelis nebulosi), leopards (Panthera pardus), lions (Panthera leo), tigers (Panthera tigris), and leopard cats (Prionalilurus bengalensis) (Yakimoff et al., 1933; Penzhorn et al., 1994; Bjork et al., 2000). Recently, Dubey et al. (2015) reported transmission of C. felis-like organisms collected from the bobcat (Lynx rufus), a small wildcat found in North America, to domestic cats by feeding of monozoic cysts in mouse tissues. These studies suggested that feline Cystoisospora are family and not genus specific. In contrast, however, Hendricks (1977) studied the host range of the primate species C. arctopitheci and demonstrated the susceptibility of six genera of New World nonhuman primates, one marsupial species, and four families of carnivores, including tayras (Eira barbar) belonging to the family Mustelidae. The sporulated oocysts from otters morphologically resembled those of C. arctopitheci, measuring 27.7 (22.7–32.7) × 24.3 (20.5–27.3) μm in diameter (Hendricks, 1974), but differed in that they showed an elliptical shape, in contrast to those of the present species, which were mostly spherical.
In conclusion, this is the first description of morphological and molecular characteristics of Cystoisospora sp. detected from Asian short-clawed otters. This coccidian can cause diarrheal disease in juveniles. We conclude that Cystoisospora sp. detected from otters appeared to be highly similar to C. rivolta, but there is still room for discussion of general hypothesis for definitive host specificity of the genus Cystoisospora. Future characterization of Cystoisospora species infecting mustelids is necessary using a combination of morphological, molecular, epidemiological, and crosstransmission experiments.
Conflicts of interest
None.
Acknowledgements
This work was partially supported by JSPS Grant-in-Aid for Young Scientists (Grant number 18K14596) and a research project grant by Nippon Veterinary and Life Science University (to T.T.).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijppaw.2018.07.001.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Anwer M.A., Newman C., Macdonald D.W., Woolhouse M.E.J., Kelly D.W. Coccidiosis in the European badger (Meles meles) from England, and epidemiological study. Parasitology. 2000;120:255–260. doi: 10.1017/s0031182099005491. [DOI] [PubMed] [Google Scholar]
- Barta J.R., Schrenzel M.D., Carreno R., Rideout B.A. The genus Atoxoplasma (Garnham 1950) as a junior objective synonym of the genus Isospora (Schneider 1881) species infecting birds and resurrection of Cystoisospora (Frenkel 1977) as the correct genus for Isospora species infecting mammals. J. Parasitol. 2005;91:726–727. doi: 10.1645/GE-3341.1. [DOI] [PubMed] [Google Scholar]
- Biester B.W., Murray C. Studies in infections enteritis of swine. J. Am. Vet. Med. Assoc. 1934;85:207–219. [Google Scholar]
- Bjork K.E., Averbeck G.A., Stromberg B.E. Parasites and parasite stages of free-ranging wild lions (Panthera leo) of northern Tanzania. J. Zoo Wildl. Med. 2000;31:56–61. doi: 10.1638/1042-7260(2000)031[0056:PAPSOF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Carreno R.A., Schnitzler B.E., Jefferies A.C., Tenter A.M., Johnson A.M., Barte J.R. Phylogenetic analysis of coccidian based on 18S rDNA sequence comparison indicates that Isospora is most closely related to Toxoplasma and Neospora. J. Eukaryot. Microbiol. 1998;45:184–188. doi: 10.1111/j.1550-7408.1998.tb04523.x. [DOI] [PubMed] [Google Scholar]
- Dubey J.P. Isospora ohioensis sp. n. proposed for I. rivolta of the dog. J. Parasitol. 1975;61:462–465. [PubMed] [Google Scholar]
- Dubey J.P. Life cycle of Isospora rivolta (Grassi, 1879) in cats and mice. J. Protozool. 1979;26:433–443. doi: 10.1111/j.1550-7408.1979.tb04650.x. [DOI] [PubMed] [Google Scholar]
- Dubey J.P., Houki A.E., Verma S.K., Calero-Bernal R., Humphreys J.G., Lindsay D.S. Experimental transmission of Cystoisospora felis-like coccidium from bobcat (Lynx rufus) to the domestic cat (Felis catus) Vet. Parasitol. 2015;211:35–39. doi: 10.1016/j.vetpar.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Duszynski D., Wilber P. A guideline for the preparation of species descriptions in the Eimeriidae. J. Parasitol. 1997;83:333–336. [PubMed] [Google Scholar]
- Frenkel J.K. Besnoitia wallacei of cats and rodents: with a reclassification of other cyst-forming isosporoid coccidian. J. Parasitol. 1977;63:611–628. [PubMed] [Google Scholar]
- Hendricks L. A redescription of Isospora arctopitheci Rodhain, 1933 (Protozoa: Eimeriidae) from primates of Panama. Proc. Helminthol. Soc. Wash. 1974;41:229–233. [Google Scholar]
- Hendricks L. Host range characteristics of the primate coccidian, Isospora arctopitheci Rodhain 1933 (Protozoa: Eimeriidae) J. Parasitol. 1977;63:32–35. [PubMed] [Google Scholar]
- Hoare C.A. On the coccidian of the ferret. Ann. Trop. Med. Parasitol. 1927;21:313–320. [Google Scholar]
- Hoover J.P., Bahr R.J., Nieves M.A., Doyle R.T., Zimmer M.A., Lauzon S.E. Clinical evaluation and prerelease management of American river otters in the second year of a reintroduction study. J. Am. Vet. Med. Assoc. 1985;187:1154–1161. [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlán A., Pospesch L. Coccidial infections in the badger Meles taxus L. Parasitology. 1933;25:102–107. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvicerová J., Pakandl M., Hypsa V. Phylogenetic relationships among Eimeria spp. (Apicomplexa, Eimeriidae) infecting rabbits: evolutionary significance of biological and morphological features. Parasitology. 2008;135:443–452. doi: 10.1017/S0031182007004106. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lepp D.L., Todd K.S. Life cycle of Isospora canis Nemeséri, 1959 in the dog. J. Protozool. 1974;21:199–206. doi: 10.1111/j.1550-7408.1974.tb03641.x. [DOI] [PubMed] [Google Scholar]
- Morrison D.A., Bornstein S., Thebo P., Wernery U., Kinne J., Mattsson J.G. The current status of the small subunit rRNA phylogeny of the coccidian (Sporozoa) Int. J. Parasitol. 2004;34:501–514. doi: 10.1016/j.ijpara.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Musaev M.A., Veison A.M. New species of coccidian of the genera Eimeria and Isospora from weasel (Mustela nivalis Lenmans, 1766) Izv. Akad. Nauk Az. SSR Ser. Biol. Nauk. 1983;5:64–70. (In Russian) [Google Scholar]
- Nukerbaeva K.K. Coccidia of sables (Martes zibellina) Inv. Akad. Nauk Kaz. SSR Ser. Biol. Nauk. 1981;0:30–33. (In Russian) [Google Scholar]
- Ogedengbe J.D., Hanner J.D., Barta J.R. DNA barcording identifies Eimeria species and contributes to the phylogenetics of coccidian parasite (Eimeriorina, Apicomplexa, Alveolata) Int. J. Parasitol. 2011;41:843–850. doi: 10.1016/j.ijpara.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Ogedengbe M.E., Ogedengbe J.D., Whale J.C., Elliot K., Juárez-Estrada M.A., Barta J.R. Molecular phylogenetic analyses of tissue coccidia (Sarcocystidae; Apicomplexa) based on nuclear 18s RDNA and mitochondrial COI sequences confirms the paraphyly of the genus Hammondia. Parasitol. Open. 2016;2:e2. [Google Scholar]
- Prasad H. The coccidian of the zorille Ictonyx (Zorilla)capensis Kaup. J. Protozool. 1961;8:55–58. [Google Scholar]
- Penzhorn B.L., Booth L.M., Meltzer D.G. Isospora rivolta recovered from cheetahs. J. S. Afr. Vet. Assoc. 1994;65:2. [PubMed] [Google Scholar]
- Svanbaev S.K. Materials on the fauna of coccidia of wild mammals in western Kazakistan. Tr. Inst. Zool. Akad. Nauk Kaz. SSR. 1956;5:80–191. (in Russian) [Google Scholar]
- Svanbaev S.K., Rachmatullina N.K. The question on coccidian from Fur-bearing animals in Kazakistan. Tr. Inst. Zool. Akad. Nauk Kaz. SSR. 1971;31:165–170. (In Russian) [Google Scholar]
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tenter A.M., Barta J.R., Beveridge I., Duszynski D.W., Mehlhorn H., Morrison D.A., Thompson R.A., Conrad P.A. The conceptual basis for a new classification of the coccidian. Int. J. Parasitol. 2002;32:595–616. doi: 10.1016/s0020-7519(02)00021-8. [DOI] [PubMed] [Google Scholar]
- Tokiwa T., Kojima A., Sasaki S., Kubota R., Ike K. Isospora lunaris n. sp. (Apicomplexa: Eimeriidae) from the domestic Java sparrow in Japan. Parasitol. Int. 2017;66:100–105. doi: 10.1016/j.parint.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Torres J., Modrý D., Fernández J., Slapeta J.R., Koudela B. Isospora lutrae n. sp. (Apicomplexa: Eimeriidae), a new coccidium from the European otter Lutra lutra (L.) (Carnivora: Mustelidae) from Spain. Syst. Parasitol. 2000;47:59–63. doi: 10.1023/a:1006453532286. [DOI] [PubMed] [Google Scholar]
- Wenyon C.M. Coccidiosis of cats and dogs and the status of the Isospora of man. Ann. Trop. Med. Parasitol. 1923;17:231–288. [Google Scholar]
- Yakimoff W.L., Matikaschwili I.L., Rastegaïeff E.F., Lewkowitsch E.N. Coccidia of the felidae. Parasitology. 1933;25:389–391. [Google Scholar]
- Yi-Fan C., Le Y., Yin D., Jiang-Hui B., Duszynski D. Emendation of 2 Isospora species (Apicoimplexa: Eimeriidae) infection the steppe polecat, Mustela eversmanii lesson, 1827, in China, to the genus Cystoisospora (Apicomplexa: Sarcocystidae) Comp. Parasitol. 2012;79:147–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.