Abstract
Non-denaturing FISH (ND-FISH) technology has been widely used to study the chromosomes of Triticeae species because of its convenience. The oligo probes for ND-FISH analysis of wheat (Triticum aestivum L.) chromosomes are still limited. In this study, the whole genome shotgun assembly sequences (IWGSC WGA v0.4) and the first version of the reference sequences (IWGSC RefSeq v1.0) of Chinese Spring (T. aestivum L.) were used to find new tandem repeats. One hundred and twenty oligo probes were designed according to the new tandem repeats and used for ND-FISH analysis of chromosomes of wheat Chinese Spring. Twenty nine of the 120 oligo probes produce clear or strong signals on wheat chromosomes. Two of the 29 oligo probes can be used to conveniently distinguish wheat A-, B-, and D-genome chromosomes. Sixteen of the 29 oligo probes only produce clear or strong signals on the subtelomeric regions of 1AS, 5AS, 7AL, 4BS, 5BS, and 3DS arms, on the telomeric regions of 1AL, 5AL, 2BS, 3BL, 6DS, and 7DL arms, on the intercalary regions of 4AL and 2DL arms, and on the pericentromeric regions of 3DL and 6DS arms. Eleven of the 29 oligo probes generate distinct signal bands on several chromosomes and they are different from those previously reported. In addition, the short and long arms of 6D chromosome have been confirmed. The new oligo probes developed in this study are useful and convenient for distinguishing wheat chromosomes or specific segments of wheat chromosomes.
Keywords: wheat, chromosome, tandem repeats, ND-FISH, oligo probe
Introduction
Genomic in situ hybridization (GISH) and Fluorescent in situ hybridization (FISH) techniques are often used to identify plant chromosomes (Kato et al., 2005). GISH is extensively used to discriminate the genomes between two distant genera and within the same genus of the Triticeae tribe (Zhao et al., 2011; Fu et al., 2014; Yang et al., 2017; Grewal et al., 2017). Multicolor GISH has been successfully used to identify the A, B, and D genomes of common wheat (Mukai et al., 1993; Han et al., 2003, 2004; Zhao et al., 2011). This technology is useful for detecting the genomic constitution and chromosomal alterations of common wheat or synthetic wheat (Kato et al., 2005; Zhao et al., 2011). FISH can physically locate genes or repetitive DNA sequences on individual chromosomes of common wheat and its relatives (Danilova et al., 2012, 2017; Komuro et al., 2013). Therefore, FISH technology using genes and repetitive DNA sequences as probes is often used to establish chromosomal landmarks of the Triticeae tribe (Cuadrado and Schwarzacher, 1998; Yuan and Tomita, 2009; Wang et al., 2010; Linc et al., 2012; Cuadrado et al., 2013; Molnár et al., 2014; Danilova et al., 2017). Tandem repeats including pAs1, pSc119.2, 5S rDNA, 45S rDNA, and microsatellites such as (AAG)n and (AAC)n are the most commonly used probes for FISH analysis in the Triticeae species (Pedersen and Langridge, 1997; Cuadrado and Schwarzacher, 1998; Sepsi et al., 2008; Yuan and Tomita, 2009; Wang et al., 2010; Linc et al., 2012; Carvalho et al., 2013; Cuadrado et al., 2013; Molnár et al., 2014; Badaeva et al., 2015; Danilova et al., 2017). Some new tandem repeats such as pTa535, pTa713, pTa465, pTak566, pTas120, and pTas126 have been cloned from wheat (Triticumaestivum L.), and they are also valuable probes for wheat chromosome identification (Komuro et al., 2013). Some repetitive DNA sequences can replace the multicolor GISH to simultaneously distinguish A-, B-, and D- genome chromosomes of wheat (Komuro et al., 2013). Although the multicolor GISH using total genomic DNA as probes and the FISH using the repetitive DNA sequences as probes can unambiguously identify the different genome chromosomes or individual chromosome of wheat, their procedures are time-consuming because of the preparation of probe sequences, the labeling of DNA sequences, and the denaturing of chromosomes and probes.
Since Cuadrado et al. (2009) reported the non-denaturing FISH (ND-FISH) technology for the investigation of plant telomeres, ND-FISH assay has been widely used to study the chromosomes of Triticeae species because of its convenience (Cuadrado and Jouve, 2010; Carmona et al., 2013; Carvalho et al., 2013; Cuadrado et al., 2013; Cabo et al., 2014; Delgado et al., 2017). In these previous studies, simple sequence repeats (SSRs) were often used as probes for ND-FISH analysis. In fact, non-SSR oligonucleotide (oligo) probes can also be used for ND-FISH assays of plant chromosomes (Fu et al., 2015; Delgado et al., 2016, 2017; Li et al., 2016; Li J. et al., 2017; Du et al., 2017; Jiang et al., 2017; Kirov et al., 2017; Zhu et al., 2017). ND-FISH technology using oligo probes can also replace GISH technology to discriminate rye (Secale cereale L.) and Dasypyrum villosum chromosomes in wheat backgrounds (Fu et al., 2015; Xiao et al., 2017). Although some non-SSR oligo probes such as Oligo-pSc119.2, Oligo-pTa535, Oligo-k566, and Oligo-713 can be used for ND-FISH analysis to identify individual wheat chromosome (Fu et al., 2015; Tang et al., 2016; Zhao et al., 2016; Du et al., 2017), they are not chromosome-specific or chromosome segment-specific. Therefore, chromosome-specific or chromosome segment-specific oligo probes are needed. In this study, some tandem repeats were obtained from the genomic sequences of T. aestivum cv. Chinese Spring that were downloaded from the International Wheat Genome Sequencing Consortium (IWGSC) and these tandem repeats were used to design new oligo probes for ND-FISH analysis of wheat chromosomes.
Materials and Methods
Plant Materials
Common wheat cv. Chinese Spring (T. aestivum L.) that was kept in our laboratory was used in this study.
Designing of New Oligo Probes
The whole genome shotgun assembly sequences (IWGSC WGA v0.4) and the first version of the reference sequences (IWGSC RefSeq v1.0) of wheat Chinese Spring were downloaded from the IWGSC1. Tandem Repeat Finder (TRF, version 4.09) (Benson, 1999) was used to search tandem repeats from the IWGSC WGA v0.4 and IWGSC RefSeq v1.0 (parameters: Match = 2, Mismatch = 7, Indel = 7, Probability of match = 80, Probability of indel = 10, Min score = 50, and Max period = 2000). Then, two strategies were used to filter these tandem repeats annotated by the TRF. The first, the tandem repeats were filtered using an in-house Python script in which the parameters including period size (>6), copy number (>10), and percent matches (>70). The consensus monomer sequences with identity >75% were clustered using CD-HIT (Li and Godzik, 2006) and the consensus sequences of each cluster were kept. The second, the tandem repeats annotated by the TRF were firstly divided into five classes according to the period size (6–30, 30–100, 100–200, 200–300, >300) using an in-house Python script. Each class of tandem repeats was subsequently clustered using CD-HIT (Li and Godzik, 2006) with the parameters including identity >75% and copy number >100. Then, the consensus sequences of each cluster were obtained.
Finally, the consensus sequences obtained through the two strategies were used for Nucleotide BLAST searches against the Nucleotide collection (nr/nt) database using blastn tool in National Center for Biotechnology Information. The consensus sequences that have the identity ≥70% with the published tandem repeats such as 45S rDNA, 5S rDNA, pSc119.2, pAs1, pTa-535, pTa-713, pTa-86, pTa-465, pTa-k566, pTa-s120, pTa-s126, pTa-k288, and pTa-k229 etc. were omitted, and the rest ones were used for designing oligo probes. The new oligo probes that can work are listed in Supplementary Table S1.
ND-FISH Analysis
The oligo probes listed in Supplementary Table S1 were synthesized by Tsingke Biological Technology Co. Ltd. (Beijing, China). The oligonucleotide sequences were 5′- or 3′-end-labeled with 6-carboxyfluorescein (6-FAM) or 6-carboxytetramethylrhodamine (TAMRA) (Supplementary Table S1). In addition, oligo probes Oligo-pSc119.2-1 and Oligo-pTa535-1 developed by Tang et al. (2014) were used to identify individual wheat chromosome. Probe Oligo-pSc119.2-1 was 5′-end-labeled with TAMRA, and probe Oligo-pTa535-1 was 5′-end-labeled with Cyanine Dye 5 (Cy5). The methods described by Han et al. (2006) were used to prepare chromosome spreads of wheat Chinese Spring. The ND-FISH analysis was performed according to the methods described by Fu et al. (2015) with some modification. That is, for the oligo probes Oligo-B and Oligo-D, when the probe mixture was dropped at the cell spreads, the room temperature should be kept at above 28°C, and the slides were immediately put in a moist box that was incubated at 42°C in advance. The high room temperature and the immediate incubating at 42°C can effectively prevent the probe Oligo-B from hybridizing with A-genome chromosomes. For new developed oligo probes, the amount of probe for each slide is listed in Supplementary Table S1. For each probe, three ND-FISH experiments were repeated. Images were made and processed according to the methods described by Duan et al. (2017).
Confirmation of the Orientation of 6D Chromosome Arms
The consensus sequence of the oligo probe that only produced hybridization signal on the telomeric region of one arm of 6D chromosome was aligned using blastn tool in B2DSC2 against IWGSC RefSeq v1.0 of wheat Chinese Spring, and the chromosome plot was drawn using Plot tool in B2DSC.
Results
Obtaining the Sequences of Oligo Probes
One hundred and fourteen new tandem repeats were detected from whole genome shotgun assembly sequences (IWGSC WGA v0.4) and the first version of the reference sequences (IWGSC RefSeq v1.0) of wheat Chinese Spring. According to these tandem repeats, 120 oligo probes were designed and used for ND-FISH analysis of the root tip metaphase chromosomes of wheat Chinese Spring. Twenty-seven of the 120 oligo probes produced strong or distinct signal bands on wheat chromosomes, and two of the 120 oligo probes produced clear signals mainly on the whole B- or D-genome chromosomes. The ND-FISH signals of the 29 oligo probes are reproducible. The information of the 29 probes is listed in Supplementary Table S1.
Chromosome Segment-Specific Probes
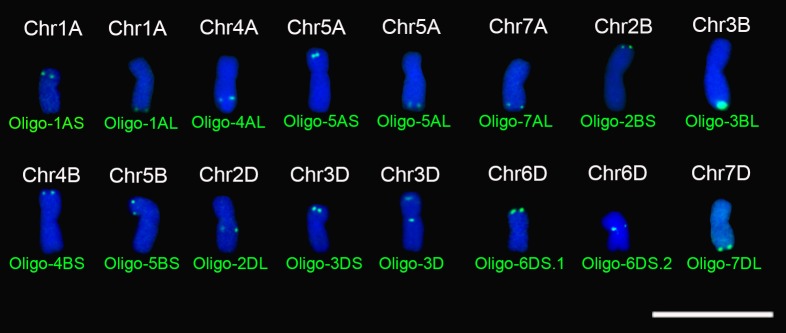
Combined with oligo probes Oligo-pSc119.2-1 and Oligo-pTa535-1, all the new designed oligo probes were used for ND-FISH analysis of chromosomes of wheat Chinese Spring. The signal patterns of Oligo-pSc119.2-1 and Oligo-pTa535-1 can help for the recognition of individual wheat chromosome (Supplementary Figures S1–S12). Sixteen of the 29 new oligo probes only produced clear or strong signals on specific segments of wheat chromosomes (Table 1, Figure 1 and Supplementary Figures S1–S8). Probes Oligo-1AS, Oligo-5AS, Oligo-7AL, Oligo-4BS, Oligo-5BS, and Oligo-3DS only generated hybridization signals on the subtelomeric regions of 1AS, 5AS, 7AL, 4BS, 5BS, and 3DS arms, respectively (Table 1, Figure 1 and Supplementary Figures S1–S3). Probes Oligo-1AL, Oligo-5AL, Oligo-2BS, Oligo-3BL, Oligo-6DS.1, and Oligo-7DL only produced strong or distinct hybridization signals on the telomeric regions of 1AL, 5AL, 2BS, 3BL, 6DS, and 7DL arms, respectively (Table 1, Figure 1 and Supplementary Figures S4–S6). The signals of probes Oligo-4AL and Oligo-2DL only occurred on the intercalary regions of 4AL and 2DL arms, respectively (Table 1, Figure 1 and Supplementary Figure S7). The signal of probe Oligo-6DS.2 only appeared on the pericentromeric region of 6DS arm (Table 1, Figure 1 and Supplementary Figure S8), and probe Oligo-3D only produced signals on the subtelomeric region of 3DS arm and the pericentromeric region of 3DL arm (Table 1, Figure 1 and Supplementary Figure S8).
Table 1.
Signal patterns of oligo probes developed in this study.
| Probe | Nucleotide sequences | Signal location on chromosome | Probe | Nucleotide sequences | Signal location on chromosome |
|---|---|---|---|---|---|
| Oligo-1AS | 5′AAAAACGCATGTCTTTAGCATTCAAAA AATGAAAAACGGTTTTTCTTGTTAAACAA3′ |
Subtelomeric region of 1AS | Oligo-7DL | 5′CATCATTTAGGTTCTTATCGACACCG AGGCACCCTAAAAGCCTAAGCTTT3′ |
Telomeric region of 7DL |
| Oligo-1AL | 5′TTCATTTTTACTCAACTCAAACTAT GGCACATACCAGGTACATCCAAA3′ |
Telomeric region of 1AL | Oligo-119 | 5′ATTTTGAGCTAGCTAAGCATATTAAGTCAT TTTGGAGCAAAAAAATGCTAAGTATAGGT3 |
Intercalary regions of 3BS, 6BL, and 7BS; pericentromeric region of 4BL |
| Oligo-4AL | 5′TAACTACATGCATGAATATCCTATCAC GCCATGAAGCTAGCTAACCAACTAACC3′ |
Intercalary region of 4AL | Oligo-18 | 5′GTAGCAGTAGCAGTAGTA3′ | Pericentromeric regions of 3BL and 7BL; centromeric region of 2B; intercalary regions of 1AL, 7AL, and 7BL |
| Oligo-5AS | 5′AACTTTTTTCAAATTCGATG3′ | Subtelomeric region of 5AS | Oligo-44 | 5′TAGCTCTACAAGCTAGTTCAAA TAATTTTACACTAGAGTTGAAC3′ |
Intercalary regions of 3AL, 5AL, and 5BL; telomeric region of 7AS; subtelomeric region of 7AL; pericentromeric region of 5DS |
| Oligo-5AL | 5′ATCTATCTATAGTGATACTCCTTTGTTT GTACTAGTAATTTTGTTTGTACCCTTTC3′ |
Telomeric region of 5AL | Oligo-42 | 5′CTCGCTCGCCCAGCTGCTGCT ACTCCGGCTCTCGCTCGATCG3′ |
Pericentromeric regions of 1AS, 6BL, and 7BL; centromeric region of 2B; intercalary regions of 3BS, 6BS, and 7BL; centromeric and pericentromeric regions of 5B |
| Oligo-7AL | 5′CCTTCTCCATCATTGGTGCCATGACG GAGATGTTCTTATCAATGTAAGAGACC3′ |
Subtelomeric region of 7AL | Oligo-45 | 5′CGGCCGCTCCGCGCGTCGCCATC GGTTGGTCACCTCATCACCACT3′ |
Pericentromeric regions of 3AS, 6AS, 4DS, 5DS, and 7DS; telomeric region of 5AL |
| Oligo-2BS | 5′GCGTGGCCAGCCACGGCGGAACCAAC CCAATCTCGCAAAAGCGACCCCATCAC3′ |
Telomeric region of 2BS | Oligo-428 | 5′AAATGAACTGGCGAGCGGAATCGTTTGGA ACACAAAGAACCATGAAACGTGACTCTCTC3′ |
Pericentromeric regions of 2AS and 5DS; intercalary regions of 1DS, 1DL, 2DL, 4DL, and 6DS; subtelomeric regions of 2DS and 5DL |
| Oligo-3BL | 5′GCCTGAAAATTACAACTTTTCGACACCGC CACGTGTAAGCCCGACCCAAACACAACCC3′ |
Telomeric region of 3BL | Oligo-440.1 | 5′CGAGAGGATCGCCGAGGGGTCGG GAGGTTGCCTAGTGTCAAAGGATTCAA3′ |
Subtelomeric regions of 2AS, 7AS, and 7AL |
| Oligo-4BS | 5′GGCGGTGGGAGTGTGTTTGTCGCGCCG CAACGCCTTCTCTGTTCCTCTACGCAC3′ |
Subtelomeric region of 4BS | Oligo-440.2 | 5′CGACCCCTCGGCGATCCTCGACCCCTCGA ACCCTCGACGACCCTGGAACCCTCGACCCC3′ |
Intercalary regions of 3BS and 5BS; pericentromeric region of 5BL |
| Oligo-5BS | 5′GTAGCAAGCATAGATGCTCTTCCACACGGT TTCTCTCAAGAACCGACCAATGCAGAAAC3′ |
Subtelomeric region of 5BS | Oligo-37 | 5′CTATATGATGAATTGCTA TGAGCTGCTCCTGCTGCTG3′ |
Pericentromeric region of 1AL; pericentromeric and intercalary regions of 7BL |
| Oligo-2DL | 5′AAGAGAATTGCTCTGTTAA TACGAATCAATGCAACCG3′ |
Intercalary region of 2DL | Oligo-60 | 5′CCTCTCCCTCTTCTCCATTGCAAAACC ACCCCCTCTCCTCTCCATCATCTCCATT3′ |
Intercalary regions of 3BS and 6BS; pericentromeric regions of 5BS and 5BL |
| Oligo-3DS | 5′CCTCGCGGGGGCGCGTCAGCGCACCCGCT GGGTGTAGCCCCCGAGATTCGGGCCGACTG3′ |
Subtelomeric region of 3DS | Oligo-107 | 5′ATTTCAGTACTGAAATACAACACCAC GATGACCCGACTAGTTCTATATCACTAA3′ |
Telomeric regions of 3DS, 4DS, and 4DL; intercalary region of 4AL |
| Oligo-3D | 5′GGCGGTTGACACCATGGCGTGGAGAT GGAGATCACCACGAGGACAACCATG3′ |
Subtelomeric region of 3DS and pericentromeric region of 3DL | Oligo-B | 5′GGTTCAGGAATAGCCT CAGGAATTGGCTCAATT3′ |
Mainly on the whole B-genome chromosomes |
| Oligo-6DS.1 | 5′TTTTAGCTCCCATTTAACACACC AAACTTTGACTTCCTTTTTCTTTT3′ |
Telomeric region of 6DS | Oligo-D | 5′TACGGGTGCCAAACGAGTGTCTGAA AGACTCCTCGAGAGGAAAATGCGAA3′ |
Mainly on the whole D-genome chromosomes |
| Oligo-6DS.2 | 5′GAAGGGCCCCACATGTCATACTCTCACAG GGGACCATGCCCCACCTGTGTCATCCCC3′ |
Pericentromeric region of 6DS |
FIGURE 1.
Signal patterns of the 16 oligo probes that produce signals on specific regions of metaphase chromosomes of wheat Chinese Spring. Chromosomes were counterstained with DAPI (blue). Scale bar: 30 μm.
Probes Producing Signals on Several Chromosomes
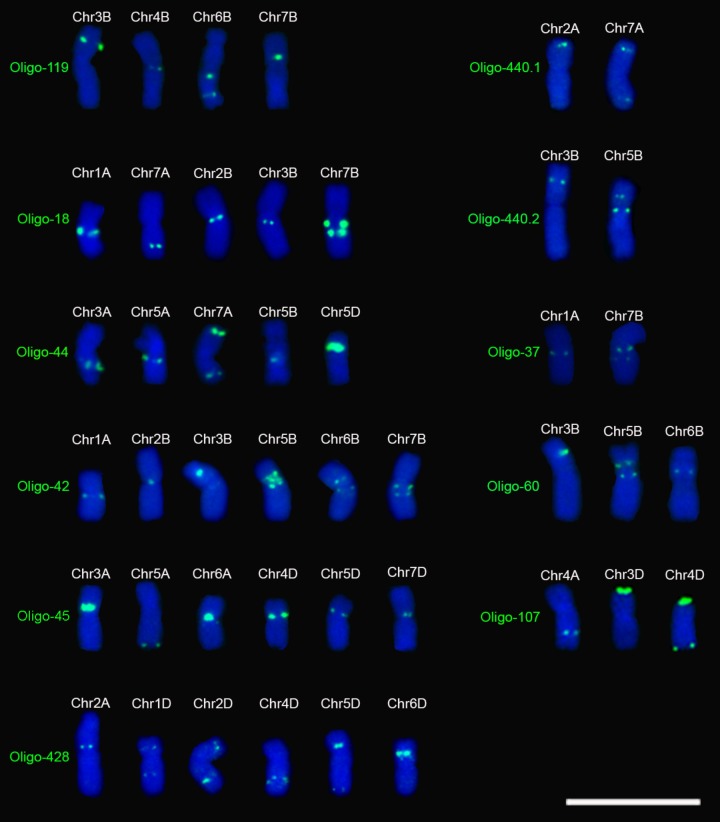
Eleven of the 29 oligo probes generated distinct signal bands on several chromosomes and they are different from those previously reported (Table 1, Figure 2 and Supplementary Figures S9–S12). All three of the probes Oligo-440.1, Oligo-440.2, and Oligo-37 produced signals on two wheat chromosomes (Table 1, Figure 2 and Supplementary Figure S9). Both of the probes Oligo-60 and Oligo-107 generated signals on three chromosomes, and four chromosomes carried the signals of Oligo-119 (Table 1, Figure 2 and Supplementary Figure S10). Both of the probes Oligo-18 and Oligo-44 produced signals on five chromosomes (Table 1, Figure 2 and Supplementary Figure S11). All of the three probes Oligo-42, Oligo-45 and Oligo-428 generated signals on six chromosomes (Table 1, Figure 2 and Supplementary Figure S12).
FIGURE 2.
Signal patterns of the 11 oligo probes that produce signals on several metaphase chromosomes of wheat Chinese Spring. Chromosomes were counterstained with DAPI (blue). Scale bar: 30 μm.
B- and D-Genome Specific Probes
Probe Oligo-B mainly produced signals on the whole B-genome chromosomes, and probe Oligo-D mainly generated signals on the whole D-genome chromosomes (Figure 3). The combination of the two probes Oligo-B and Oligo-D can be used to distinguish wheat A-, B-, and D-genome chromosomes (Figure 3). Therefore, it is convenient to use the two oligo probes and ND-FISH analysis to identify wheat A-, B-, and D-genome chromosomes.
FIGURE 3.
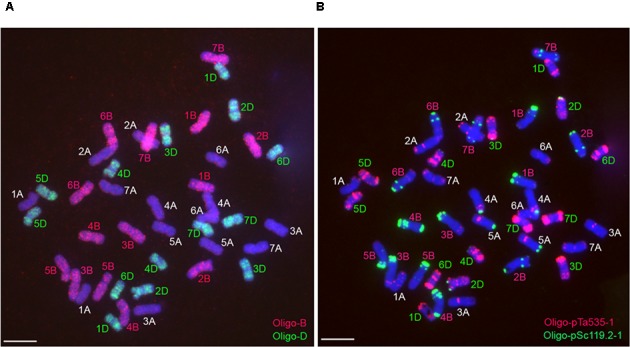
Sequential ND-FISH analysis of the metaphase chromosomes of wheat Chinese Spring. (A) Using probes Oligo-B (red) and Oligo-D (green) to analyze root tip metaphase chromosomes of wheat Chinese Spring. (B) Using probes Oligo-pTa535-1 (red) and Oligo-pSc119.2-1 (green) to analyze the same cell in (A) after rising the slide. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
The Determination of Short and Long Arms of 6D Chromosomes
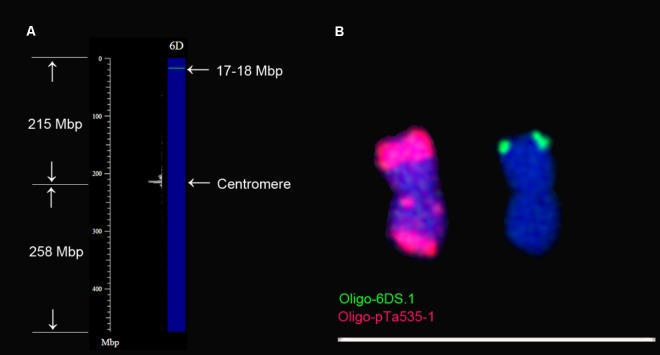
To determine that the arm of 6D containing Oligo-6DS.1 signal is short or long arm, the consensus sequences of probe Oligo-6DS.1 was used for Nucleotide BLAST searches against the IWGSC RefSeq v1.0 of wheat Chinese Spring using blastn tool in B2DSC, and chromosome plots were drawn to view the locations of the consensus sequence on chromosomes (Figure 4A). The consensus sequence of Oligo-6DS.1 hits the sequences between 17 and 18 Mbp regions of 6D chromosome (Figure 4A). From Figure 4A, it can be noted that the 17–18 Mbp region should be located on the short arm of 6D chromosome (6DS). Therefore, the signal of probe Oligo-6DS.1 should be located on the telomeric region of 6DS arm (Figures 4A,B). The 6D chromosome can be identified using probe Oligo-pTa535-1 (Figure 4B). Probe Oligo-pTa535-1 produces a very strong signal band on one arm of 6D chromosome, and two clear signal bands on the other arm (Figure 4B). According to the Figures 4A,B, therefore, the arm carrying one signal band of Oligo-pTa535-1 should be the 6DS arm.
FIGURE 4.
Confirmation of the orientation of 6D chromosome. (A) 6D chromosome plot is drawn using Plot tool in B2DSC. The green line on the chromosome plot indicates that the consensus sequence of Oligo-6DS.1 hits the sequences between the 17 and 18 Mbp regions on 6DS arm. (B) The location of the signal of oligo-6DS.1 is on the same arm with the one that contains one signal band of Oligo-pTa535-1. Scale bar: 70 μm.
Discussion
Developing New Oligo Probes for ND-FISH Analysis of Common Wheat Chromosomes
ND-FISH technology has provided a convenient and efficient way to identify the chromosomes of Triticeae species (Cuadrado and Jouve, 2010; Carmona et al., 2013; Carvalho et al., 2013; Cuadrado et al., 2013; Cabo et al., 2014; Tang et al., 2014; Fu et al., 2015; Delgado et al., 2016, 2017; Li et al., 2016; Li J. et al., 2017; Li H. et al., 2017; Du et al., 2017; Duan et al., 2017; Jiang et al., 2017; Xiao et al., 2017). The key step for a successful ND-FISH analysis is to obtaining suitable oligo probes. Most of the previously reported oligo probes that produce signals on wheat chromosomes were developed from tandem repeats. It is easy to obtain SSR oligo probes. For developing new non-SSR oligo probes, the new repetitive DNA sequences must be obtained firstly. It has been reported that DNase-I-digested genomic TA cloning procedure is highly effective to get FISH-positive repeated sequences of barley (Hordeum vulgare L.) (Kato, 2011). This procedure is equally valid for common wheat (Komuro et al., 2013). With the development of next generation sequencing technology, more and more genomes of species have been successfully sequenced, and this provides a convenient way to search new FISH-positive repeated sequences. Using the next generation sequencing data of Allium fistulosum, two FISH-positive tandem repeats HAT58 and CAT36 were detected (Kirov et al., 2017). Based on the reference genome sequences of cucumber (Cucumis sativus), a bioinformatic pipeline was used to develop bulked oligo probes, in which repetitive DNA sequences were omitted (Han et al., 2015). The bulked oligo probes can be used to identify single cucumber chromosome and to paint homeologous chromosomes in other Cucumis species (Han et al., 2015). The same way was used to develop oligo probes that can build a bar code or banding pattern to uniquely label each of the 12 chromosomes of potato (Braz et al., 2018). The released whole genome shotgun assembly sequences (IWGSC WGA v0.4) and the first version of the reference sequences (IWGSC RefSeq v1.0) of wheat Chinese Spring provide a good resource for searching new tandem repeats of common wheat and designing new ND-FISH-positive oligo probes. In the present study, 29 ND-FISH-positive oligo probes are obtained from the reference genome sequence of wheat Chinese Spring. Another excellent method, graph-based approach, was also used to search repetitive sequences from next-generation sequencing data (Novák et al., 2010). In this method, two-step analysis including partitioning the data into clusters of overlapping reads representing individual repeated elements and further characterization of these clusters was used to find repetitive elements including tandem and dispersed repeats (Novák et al., 2010). The graph-based approach mainly focuses on distinguishing basic types of repeats by graph representation and investigating sequence variability within repeat families (Novák et al., 2010). In this study, tandem repeats were focused on and the software Tandem Repeats Finder (Benson, 1999) was used to search tandem repeats from wheat genomic sequences. Then all tandem repeats are clustered into clusters by CD-HIT program that meet 75% similarity threshold, and the consensus sequence of each cluster that has not been reported was selected to design oligonucleotide probe. Therefore, in this study, the dispersed repeats were excluded firstly and the tandem repeats that are suitable for designing oligo probes can be effectively found.
The Applications of the New Oligo Probes Developed in This Study
Simple sequence repeat and non-SSR oligo probes such as (AAG)n, (AAC)n, Oligo-pSc119.2, Oligo-pTa535, Oligo-pTa71, Oligo-s120, Oligo-275, Oligo-k566, and Oligo-713 have already been used to identify individual wheat chromosome (Cuadrado and Schwarzacher, 1998; Cuadrado et al., 2000; Carvalho et al., 2013; Mirzaghaderi et al., 2014; Tang et al., 2014, 2016; Fu et al., 2015; Zhang et al., 2015; Li et al., 2016; Li J. et al., 2017; Li H. et al., 2017; Yang et al., 2016; Zhao et al., 2016, 2018; Delgado et al., 2017; Du et al., 2017; Duan et al., 2017; Jiang et al., 2017; Xiao et al., 2017). However, these oligo probes produce their signals on several wheat chromosomes and they are not chromosome-specific or chromosome segment-specific. Generally, using these oligo probes, wheat chromosomes can be well distinguished only when they are intact. Some segments of wheat chromosomes are difficult to be identified if they are broken off from their original chromosomes because of the lack of chromosome-specific or chromosome segment-specific probes. Bulked oligo probes that are non-repetitive DNA sequences can be used to identify a single entire chromosome and special segment of a chromosome (Han et al., 2015). In the present study, 16 oligo probes that were derived from tandem repeats are chromosome segment-specific and they can also be used to recognize special segments of some wheat chromosomes. In addition, a method named fluorescent in situ hybridization in suspension (FISHIS) has been developed to effectively isolate individual chromosome of wheat and its relatives (Giorgi et al., 2013; Lucretti et al., 2014). In this method, synthetic and fluorescence-labeled SSR oligo probes were used, and heat denaturation of chromosomes was avoided (Giorgi et al., 2013; Lucretti et al., 2014). Therefore, the 16 oligo probes developed in this study might be helpful for the flow sorting and isolation of individual wheat chromosome using FISHIS technology because these oligo probes are chromosome segment-specific and they can be used for ND-FISH analysis.
It has already been reported that chromosomal rearrangements are ubiquitous among polyploidy wheat accessions (Badaeva et al., 2007). Multicolor GISH technology can be used to effectively detect the reciprocally translocated chromosomes involving the A, B, and D genome of wheat (Mukai et al., 1993; Han et al., 2004; Kato et al., 2005). In this study, the combination of the oligo probes Oligo-B and Oligo-D can easily distinguish the A, B, and D genomes of common wheat through ND-FISH analysis. Therefore, the two oligo probes can replace the multicolor GISH to fastly identify some reciprocal translocations among the three genomes of common wheat.
For the other new oligo probes in this study, they produced the FISH signal patterns that are different from those previously reported (Pedersen and Langridge, 1997; Sepsi et al., 2008; Komuro et al., 2013; Tang et al., 2014; Du et al., 2017). The FISH signal patterns built in the present study provide one more new chromosomal landmarks of common wheat.
The Orientation of 6D Chromosome
Tandem repeats pAs1 mainly produce signal bands on D-genome chromosomes, and the arm of 6D chromosome that contains one signal band of pAs1 was regarded as the long arm (Mukai et al., 1993; Pedersen and Langridge, 1997). Tandem repeats pTa-535 produce the same signal pattern as the one produced by pAs1 on 6D chromosome (Komuro et al., 2013). Again, the arm that contains one signal band of pTa-535 was regarded as the 6DL arm (Komuro et al., 2013). Oligo probe Oligo-pTa535 can replace the role of pTa-535 to identify wheat chromosomes (Tang et al., 2014). The arm of 6D chromosome that carries one signal band of Oligo-pTa535 was regarded as 6DS arm (Tang et al., 2014). In the present study, the reference sequences of wheat Chinese Spring (IWGSC RefSeq v1.0) combined with ND-FISH analysis and blastn tool in B2DSC was used to confirm the orientation of 6D chromosome, and the results indicate that the arm carrying one signal band of Oligo-pTa535-1 should be the 6DS arm. That is, the arm of 6D chromosome that contains one signal band of pAs1 or pTa-535 should be 6DS arm. B2DSC was established by Dr. Zujun Yang, Center for Informational Biology, University of Electronic Science and Technology of China. It provides a view of the distribution of the homologous sequences of oligonucleotide probes on chromosomes, mainly for wheat research. Based on the newly released IWGSC RefSeq v1.0 of wheat Chinese Spring, the general distribution patterns of repeated sequences in the genome can be viewed through drawing a chromosome plot.
Conclusion
The whole genome shotgun assembly sequences (IWGSC WGA v0.4) and the first version of the reference sequences (IWGSC RefSeq v1.0) of wheat Chinese Spring were used to develop new oligo probes for ND-FISH analysis of wheat chromosomes. Two oligo probes can be used to identify wheat A-, B-, and D-genome chromosomes, 16 oligo probes can be used to identify specific segments of wheat chromosomes, and 11 oligo probes give new chromosomal landmarks of common wheat. These oligo probes are useful and convenient for distinguishing wheat chromosomes or specific segments of wheat chromosomes. In addition, the short and long arms of 6D chromosome have been confirmed.
Author Contributions
SF and ZT designed the study, analyzed the data, and wrote the manuscript. SF, ZT, ST, TL, and ZY analyzed the genomic sequences of wheat Chinese Spring and designed the oligo probes. LQ, GL, WZ, and JZ performed the experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. We gratefully acknowledge the financial support from the National Key Research and Development Program of China (No. 2016YFD0102001) and National Natural Science Foundation of China (Nos. 31471498 and 31470409).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01104/full#supplementary-material
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-1AS (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-5AS (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-7AL (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-4BS (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-5BS (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-3DS (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-1AL (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-5AL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-2BS (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-3BL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-6DS.1 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-7DL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-4AL (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-2DL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-3D (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-6DS.2 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-440.1 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-440.2 (green) as probes. (E,F) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-37 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of what Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-60 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-107 (green) as probes. (E,F) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-119 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-18 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-44 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-42 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-45 (green) as probes. (E,F) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-448 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
Information of the new oligo probes.
References
- Badaeva E. D., Amosova A. V., Goncharov N. P., Macas J., Ruban A. S., Grechishnikova I. V., et al. (2015). A set of cytogenetic markers allows the precise identification of all A-genome chromosomes in diploid and polyploidy wheat. Cytogenet. Genome Res. 146 71–79. 10.1159/000433458 [DOI] [PubMed] [Google Scholar]
- Badaeva E. D., Dedkova O. S., Gay G., Pukhalskyi V. A., Zelenin A. V., Bernard S., et al. (2007). Chromosomal rearrangements in wheat: their types and distribution. Genome 50 907–926. 10.1139/G07-072 [DOI] [PubMed] [Google Scholar]
- Benson G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braz G. T., He L., Zhao H., Zhang T., Semrau K., Rouillard J. M., et al. (2018). Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208 513–523. 10.1534/genetics.117.300344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabo S., Carvalho A., Martin A., Lima-Brito J. (2014). Structural rearrangements detected in newly-formed hexaploid tritordeum after three sequential FISH experiments with repetitive DNA sequences. J. Genet. 93 183–188. 10.1007/s12041-014-0328-5 [DOI] [PubMed] [Google Scholar]
- Carmona A., Friero E., Bustos A. D., Jouve N., Cuadrado A. (2013). Cytogenetic diversity of SSR motifs within and between Hordeum species carrying the H genome: H. vulgare L. and H. bulbosum L. Theor. Appl. Genet. 126 949–961. 10.1007/s00122-012-2028-y [DOI] [PubMed] [Google Scholar]
- Carvalho A., Guedes-Pinto H., Lima-Brito J. (2013). Polymorphism of the simple sequence repeat (AAC)5 in the nucleolar chromosomes of old Portuguese wheat cultivars. J. Genet. 92 583–586. 10.1007/s12041-013-0291-6 [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Schwarzacher T. (1998). The chromosomal organization of simple sequence repeats in wheat and rye genome. Chromosoma 107 587–594. 10.1007/s004120050345 [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Schwarzacher T., Jouve N. (2000). Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor. Appl. Genet. 101 711–717. 10.1007/s001220051535 [DOI] [Google Scholar]
- Cuadrado Á., Carmona A., Jouve N. (2013). Chromosomal characterization of the three subgenomes in the polyploids of Hordeum murinum L.: new insight into the evolution of this complex. PLoS One 8:e81385. 10.1371/journal.pone.0081385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado Á. H., Golczyk N. (2009). A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res. 17 755–762. 10.1007/s10577-009-9060-z [DOI] [PubMed] [Google Scholar]
- Cuadrado Á., Jouve N. (2010). Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 119 495–503. 10.1007/s00412-010-0273-x [DOI] [PubMed] [Google Scholar]
- Danilova T. V., Akhunova A. R., Akhunov E. D., Friebe B., Gill B. S. (2017). Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant J. 92 317–330. 10.1111/tpj.13657 [DOI] [PubMed] [Google Scholar]
- Danilova T. V., Friebe B., Gill B. S. (2012). Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121 597–611. 10.1007/s00412-012-0384-7 [DOI] [PubMed] [Google Scholar]
- Delgado A., Carvalho A., Martín A. C., Martín A., Lima-Brito J. (2016). Use of the synthetic Oligo-pTa535 and Oligo-pAs1 probes for identification of Hordeum chilense-origin chromosomes in hexaploid tritordeum. Gene Resour. Crop Evol. 63 945–951. 10.1007/s10722-016-0402-3 [DOI] [Google Scholar]
- Delgado A., Carvalho A., Martín A. C., Martín A., Lima-Brito J. (2017). Genomic reshuffling in advanced lines of hexaploid tritordeum. Gene Resour. Crop Evol. 64 1331–1353. 10.1007/s10722-016-0439-3 [DOI] [Google Scholar]
- Du P., Zhuang L., Wang Y., Yuan L., Wang Q., Wang D., et al. (2017). Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60 93–103. 10.1139/gen-2016-0095 [DOI] [PubMed] [Google Scholar]
- Duan Q., Wang Y. Y., Qiu L., Ren T. H., Li Z., Fu S. L., et al. (2017). Physical location of new PCR-based markers and powdery mildew resistance gene(s) on rye (Secale cereale L.) chromosome 4 using 4R dissection lines. Front. Plant Sci. 8:1716. 10.3389/fpls.2017.01716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S. L., Chen L., Wang Y. Y., Li M., Yang Z. J., Qiu L., et al. (2015). Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 5:10552. 10.1038/srep10552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S. L., Ren Z. L., Chen X. M., Yan B. J., Tan F. Q., Fu T. H. (2014). New wheat-rye 5DS-4RS.4RL and 4RS-5DS.5DL translocation lines with powdery mildew resistance. J. Plant Res. 127 743–753. 10.1007/s10265-014-0659-6 [DOI] [PubMed] [Google Scholar]
- Giorgi D., Farina A., Grosso V., Gennaro A., Ceoloni C., Lucretti S. (2013). FISHIS: Fluorescence in situ hybridization in suspension and chromosome flow sorting made easy. PLoS One 8:e57994. 10.1371/journal.pone.0057994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S., Yang C., Edwards S. H., Scholefield D., Ashling S., Burridge A. J., et al. (2017). Characterisation of Thinopyrum bessarabicum chromosomes through genome-wide introgressions into wheat. Theor. Appl. Genet. 131 389–406. 10.1007/s00122-017-3009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Fedak G., Benabdelmouna A., Armstrong K., Ouellet T. (2003). Characterization of six wheat × Thinopyrum intermedium derivatives by GISH, RFLP and multicolor GISH. Genome 46 490–495. 10.1139/g03-032 [DOI] [PubMed] [Google Scholar]
- Han F., Liu B., Fedak G., Liu Z. (2004). Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 109 1070–1076. 10.1007/s00122-004-1720-y [DOI] [PubMed] [Google Scholar]
- Han F. P., Lamb J. C., Birchler J. A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. U.S.A. 103 3238–3243. 10.1073/pnas.0509650103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang T., Thammapichai P., Weng Y., Jiang J. (2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200 771–779. 10.1534/genetics.115.177642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Xiao Z. Q., Fu S. L., Tang Z. X. (2017). FISH karyotype of 85 common wheat cultivars/lines displayed by ND-FISH using oligonucleotide probes. Cereal Res. Commun. 45 549–563. 10.1556/0806.45.2017.049 [DOI] [Google Scholar]
- Kato A. (2011). High-density fluorescence in situ hybridization signal detection on barley (Hordeum vulgare L.) chromosomes with improved probe screening and reprobing procedures. Genome 54 151–159. 10.1139/G10-098 [DOI] [PubMed] [Google Scholar]
- Kato A., Vega J. M., Han F. P., Lamb J. C., Birchler J. A. (2005). Advances in plant chromosome identification and cytogenetic techniques. Curr. Opin. Plant Biol. 8 148–154. 10.1016/j.pbi.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Kirov I. V., Kiseleva A. V., Laere K. V., Roy N. V., Khrustaleva L. I. (2017). Tandem repeats of Allium fistulosum associated with major chromosomal landmarks. Mol. Genet. Genomics 292 453–464. 10.1007/s00438-016-1286-9 [DOI] [PubMed] [Google Scholar]
- Komuro S., Endo R., Shikara K., Kato A. (2013). Genomic and chromosomal distribution patterns of various prepeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56 131–137. 10.1139/gen-2013-0003 [DOI] [PubMed] [Google Scholar]
- Li G., Gao D., Zhang H., Li J., Wang H., La S., et al. (2016). Molecular cytogenetic characterization of Dasypyrum breviaristatum chromosomes in wheat background revealing the genomic divergence between Dasypyrum species. Mol. Cytogenet. 9:6. 10.1186/s13039-016-0217-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lang T., Li B., Yu Z., Wang H., Li G., et al. (2017). Introduction of Thinopyrum intermedium ssp. trichophorum chromosomes to wheat by trigeneric hybridization involving Triticum, Secale and Thinopyrum genera. Planta 245 1121–1135. 10.1007/s00425-017-2669-9 [DOI] [PubMed] [Google Scholar]
- Li H., Deal K. R., Luo M. C., Ji W., Distelfeld A., Dvorak J. (2017). Introgression of the Aegilops speltoides Su1-Ph1 suppressor into wheat. Front. Plant Sci. 8:2163. 10.3389/fpls.2017.02163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Linc G., Sepsi A., Molnár-Láng M. (2012). A FISH karyotype to study chromosome polymorphisms for the Elytrigia elongata E genome. Cytogenet. Genome Res. 136 138–144. 10.1159/000334835 [DOI] [PubMed] [Google Scholar]
- Lucretti S., Giorgi D., Farina A., Grosso V. (2014). “FISHIS: a new way in chromosome flow sorting makes complex genomes more accessible,” in Genomics of Plant Genetic Resources, eds Tuberosa R., Graner A., Frison E. (Dordrecht: Springer; ), 319–348. 10.1007/978-94-007-7572-5_13 [DOI] [Google Scholar]
- Mirzaghaderi G., Houben A., Badaeva E. (2014). Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol. Cytogenet. 7:91. 10.1186/s13039-014-0091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár I., Kubaláková M., Šimková H., Farkas A., Cseh A., Megyeri M., et al. (2014). Flow cytometric chromosome sorting from diploid progenitors of bread wheat, T. urartu, Ae. speltoides and Ae. tauschii. Theor. Appl. Genet. 127 1091–1104. 10.1007/s00122-014-2282-2 [DOI] [PubMed] [Google Scholar]
- Mukai Y., Nakahara Y., Yamamoto M. (1993). Simultaneous discrimination of the three genomes in hexaploid wheat by multicolour fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36 489–494. 10.1139/g93-067 [DOI] [PubMed] [Google Scholar]
- Novák P., Neumann P., Macas J. (2010). Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378. 10.1186/1471-2105-11-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen C., Langridge P. (1997). Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40 589–593. 10.1139/g97-077 [DOI] [PubMed] [Google Scholar]
- Sepsi A., Molnár I., Szalay D., Molnár-Láng M. (2008). Characterization of a leaf rust-resistant wheat-Thinopyrum ponticum partial amphipliod BE-1, using sequential multicolor GISH and FISH. Theor. Appl. Genet. 116 825–834. 10.1007/s00122-008-0716-4 [DOI] [PubMed] [Google Scholar]
- Tang S. Y., Qiu L., Xiao Z. Q., Fu S. L., Tang Z. X. (2016). New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 7:118. 10.3390/genes7120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z. X., Yang Z. J., Fu S. L. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55 313–318. 10.1007/s13353-014-0215-z [DOI] [PubMed] [Google Scholar]
- Wang Q., Xiang J., Gao A., Yang X., Liu W., Li X., et al. (2010). Analysis of chromosomal structural polymorphisms in the St, P, and Y genomes of Triticeae (Poaceae). Genome 53 241–249. 10.1139/g09-098 [DOI] [PubMed] [Google Scholar]
- Xiao Z., Tang S., Qiu L., Tang Z., Fu S. (2017). Oligonucleotides and ND-FISH displaying different arrangements of tandem repeats and identification of Dasypyrum villosum chromosomes in wheat backgrounds. Molecules 22:E973. 10.3390/molecules22060973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. R., Zhang H. Q., Chen W. H., Kang H. Y., Wang Y., Sha L. N., et al. (2017). Genomic constitution and intergenomic translocations in the Elymus dahuricus complex revealed by multicolor GISH. Genome 60 510–517. 10.1139/gen-2016-0199 [DOI] [PubMed] [Google Scholar]
- Yang E., Li G., Li L., Zhang Z., Yang W., Peng Y., et al. (2016). Characterization of stripe rust resistance genes in the wheat cultivar Chuanmai45. Int. J. Mol. Sci. 17:E601. 10.3390/ijms17040601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W. Y., Tomita M. (2009). Centromeric distribution of 350-family in Dasypyrum villosum and its application to identifying Dasypyrum chromatin in the wheat genome. Hereditas 146 58–66. 10.1111/j.1601-5223.2009.02110.x [DOI] [PubMed] [Google Scholar]
- Zhang J., Jiang Y., Guo Y., Li G., Yang Z., Xu D., et al. (2015). Identification of novel chromosomal aberrations induced by 60Co-γ-irradiation in wheat-Dasypyrum villosum lines. Int. J. Mol. Sci. 16 29787–29796. 10.3390/ijms161226134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ning S., Yi Y., Zhang L., Yuan Z., Wang J., et al. (2018). Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genomics 19:3. 10.1186/s12864-017-4384-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ning S., Yu J., Hao M., Zhang L., Yuan Z., et al. (2016). Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed. Sci. 66 522–529. 10.1270/jsbbs.16011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Xu L. Y., Zhu B., Li M. J., Zhang H. K., Qi B., et al. (2011). Chromosomal and genome-wide molecular changes associated with initial stages of allohexaploidization in wheat can be transit and incidental. Genome 54 692–699. 10.1139/G11-028 [DOI] [PubMed] [Google Scholar]
- Zhu M., Du P., Zhuang L., Chu C., Zhao H., Qi Z. (2017). A simple and efficient non-denaturing FISH method for maize chromosome differentiation using single-strand oligonucleotide probes. Genome 60 657–664. 10.1139/gen-2016-0167 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-1AS (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-5AS (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-7AL (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-4BS (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-5BS (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-3DS (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-1AL (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-5AL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-2BS (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-3BL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-6DS.1 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-7DL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-4AL (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-2DL (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-3D (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-6DS.2 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-440.1 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-440.2 (green) as probes. (E,F) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-37 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of what Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-60 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-107 (green) as probes. (E,F) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-119 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-18 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-44 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
ND-FISH analysis of root tip metaphase chromosomes of wheat Chinese Spring. (A,B) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-42 (green) as probes. (C,D) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-45 (green) as probes. (E,F) ND-FISH analysis of the same cell using Oligo-pTa535-1 (red), Oligo-pSc119.2-1 (green), Oligo-448 (green) as probes. Chromosomes were counterstained with DAPI (blue). Scale bar: 10 μm.
Information of the new oligo probes.