Abstract
Oncolytic virus (OV) therapy is an emerging cancer treatment that uses replicating viruses to infect and kill tumor cells and incite anticancer immunity. While the approach shows promise, it currently fails most patients, indicating strategies to improve OV activity are needed. Developing these will require greater understanding of OV biology, particularly in the context of OV delivery and clearance, the infection process within a complex tumor microenvironment, and the modulation of anticancer immunity. To help achieve this, we have established a technique for high-resolution 4D imaging of OV-host interactions within intact tissues of live mice using intravital microscopy (IVM). We show that oncolytic vesicular stomatitis virus (VSV) directly labeled with Alexa Fluor dyes is easily visualized by single- or multiphoton microscopy while retaining bioactivity in vivo. The addition of fluorophore-tagged antibodies and genetically encoded reporter proteins to image target cells and the virus infection enables real-time imaging of dynamic interactions between VSV and host cells in blood, tumor, and visceral organs of live mice. The method has sufficient in vivo resolution to observe leukocytes in blood binding to and transporting VSV particles, foci of VSV infection spreading through a tumor, and antigen-presenting cells in the spleen interacting with and being infected by VSV. Visualizing OV-host interactions by IVM represents a powerful new tool for studying OV therapy.
Keywords: oncolytic virus, intravital microscopy, virus-host interactions, tumor microenvironment, secondary lymphoid organs
Introduction
Oncolytic virus therapy (OVT) is an approach to treating cancer in which patients are infused with live, replicating viruses that seek out, infect, and kill tumor cells while simultaneously eliciting an immune response toward the cancer.1, 2 After decades of research and development, the treatment has finally been validated in human clinical trials. In 2015, the U.S. Food and Drug Administration approved an engineered herpes simplex virus called Imlygic for treating metastatic melanoma,3 and wild-type reovirus and genetically attenuated poliovirus have recently been granted orphan drug and breakthrough therapy designation, respectively, for treating various cancers.4, 5 Numerous other OVs are currently being tested in phase 1, 2, and 3 clinical trials, as monotherapy or in combination with conventional cancer drugs and other immunotherapies. It is widely anticipated that additional OV and combination therapies will be licensed for treating a variety of cancers in the coming decade.
Yet despite the recent approvals and ongoing excitement, OVT still fails most patients.6, 7, 8 More effective OV strains, combination therapies, and dosing regimens are required to maximize this drug class’s potential. Developing these, however, will be predicated upon a deeper understanding of the mechanisms governing success or failure of OVT. Elucidating these mechanisms has been difficult, partly because methods for studying the dynamic interactions between OV and host cells in situ are lacking. For example, determining how OV travels through the bloodstream and migrates across tumor endothelia for delivery to cancer cells,9 and the barriers that impede it from doing so, would benefit tremendously from a method that enables real-time visualization of OV particles, leukocytes, and stromal and cancer cells in the blood vessels and tumor microenvironment of live mice. Similarly, understanding how engineered OVs boost anticancer T cell responses in secondary lymphoid organs10 would be aided by in vivo imaging of virus-immune cell interactions and infections in the spleen and tumor-draining lymph nodes (LNs). So far, high-resolution visualization of OV-host interactions in situ has been limited to snapshots in time provided by histological analyses.
Intravital microscopy (IVM) is a powerful technique for studying dynamic cellular processes within intact tissues of live animals.11, 12 IVM uses confocal or multiphoton microscopes equipped with powerful lasers to excite and visualize fluorescently labeled structures in situ. The technique provides sub-micron resolution and has been increasingly used to elucidate biological, pathological, and therapeutic mechanisms of action in mouse models. For example, our understanding of organ function,13, 14 cancer biology,15, 16 and cellular therapies17, 18 has benefited tremendously from IVM studies in mice. Likewise, our appreciation of virus-host interactions owes considerable resolution to IVM.19, 20 IVM studies have revealed critical insights into the pathogenesis of HIV,21 influenza,22 and West Nile,23 among other viruses,24, 25 and the host’s immune response to virus infections.26 Being increasingly accessible to modern day biomedical laboratories, IVM represents a powerful tool for understanding how viruses naturally infect host cells and are cleared by, or evade, the host’s immune system.
To enable high-resolution studies of the dynamic interactions between OVs and host cells in vivo, we developed an IVM method for simultaneous, real-time visualization of bioactive OV particles, cancer and normal cells, and the OV infection in tumor-bearing mice. We show that individual OVs, cellular infections, and host responses can be imaged, tracked, and measured in blood, tumor, and visceral organs of live mice using confocal and multiphoton IVM. Preliminary studies revealed interactions between OVs and intravascular leukocytes, OV infection of mouse tumors, and OV interactions with immune cells in the spleen. While we developed our method to visualize oncolytic rhabdoviruses (vesicular stomatitis virus [VSV]ΔM51 and MarabaMG1), the technique can easily be adapted to image other OVs. Our method sets the stage for comprehensive high-resolution studies of OV-host interactions in their native therapeutic environment.
Results
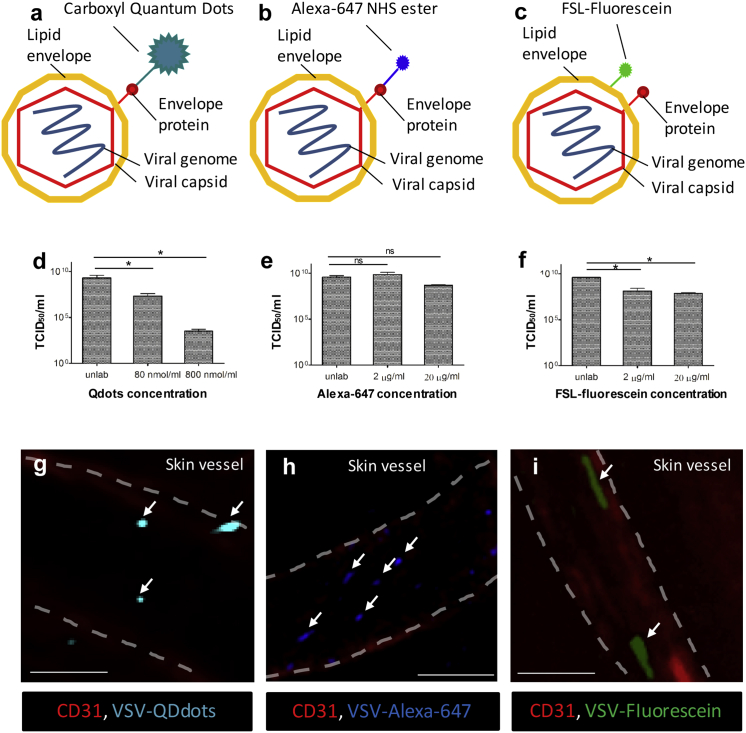
To develop an IVM method for studying OV-host interactions in vivo, we first sought to identify an OV labeling strategy that would support the visualization of individual virus particles without affecting their bioactivity. Potential strategies included engineering fluorescently tagged viruses (genetic approaches),27, 28, 29 growing viruses in cells conditioned to transfer fluorescence (cell-assisted approaches),30 and directly labeling virus with fluorescent chemical dyes (direct-labeling approaches).31, 32 Because (1) compact viruses often lose structure and/or infectivity when genetically tagged with fluorescent protein (FP),28, 33 (2) most cell-assisted strategies only work for enveloped viruses,34 and (3) many clinically advanced OVs are highly compacted (e.g., VSV, Maraba, and poliovirus) or non-enveloped (e.g., reovirus), we opted to test direct-labeling over genetic or cell-assisted approaches for their ability to brightly and inertly label OV for IVM imaging. Of amine-linked quantum dots (Qdots; Figure 1A), amine-linked Alexa Fluor (AF; Alexa; Figure 1B), and lipophilic FSL (Function-Spacer-Lipid)-fluorescein (Figure 1C) technologies, each successfully labeled the outer surface of an oncolytic strain of VSV (VSVΔM51, hereafter referred to as VSV),35 a small enveloped oncolytic rhabdovirus currently under pre-clinical development. While Qdot and FSL-fluorescein labeling significantly reduced VSV infectivity in vitro, as measured by TCID50, AF technology had minimal impact (Figures 1D–1F). All three strategies labeled VSV with sufficient fluorescence signal to enable visualization within superficial skin vessels of live animals using confocal IVM (Figures 1G and 1H).
Figure 1.
Labeling Strategies for IVM Tracking of OVs In Vivo
(A–C) Schematic representation of labeling strategies. VSV was labeled using carboxyl quantum dots (Qdots) and Alexa-647 NHS ester (AF647) that covalently bind to superficial amino groups (A and B) or FSL-fluorescein incorporating into lipid membrane (C). (D–F) Virus titers after labeling with low and high concentration of Qdots (D), AF647 (E), and FSL-fluorescein (F) were measured by TCID50 (n = 3). Values represent mean ± SD; *p < 0.05; ns, non-significant. (G–I) In vivo visualization of labeled virus particles within skin blood vessels (red, CD31) using intravital confocal microscopy, immediately following i.v. injection of VSV (5 × 108 PFU) labeled with a high concentration of Qdots (G, 800 nM/mL), AF647 (H, 20 μg/mL), and FSL-fluorescein (I, 20 μg/mL). Vasculature is delineated by white dashed lines, and arrows indicate virus particles. Scale bars, 50 μm; representative images of two independent experiments.
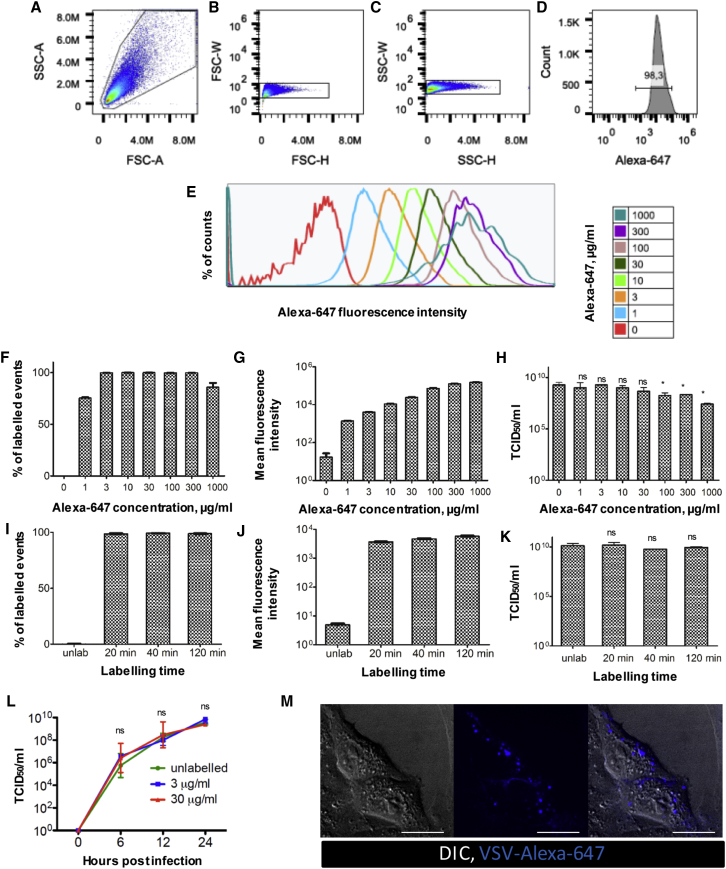
Given their brightness of labeling and minimal impact on VSV infectivity, AF dyes were selected for further optimization and analyses (Figure 2). Flow virometry confirmed that greater than 98% of VSV particles were labeled by AF647 (VSV-AF647; Figures 2A–2F), even at low dye concentrations. AF labeling conformed to a single tight histogram peak, suggesting that single virus particles of relatively uniform AF labeling were being detected (Figures 2D and 2E). Increasing the concentration of AF dye increased the mean fluorescence intensity (MFI) of labeled virions (Figures 2E and 2G) without significantly impacting upon virus infectivity up to 100 μg/mL, with higher concentrations slightly impairing infectivity (Figure 2H). Increasing the labeling time did not affect the percentage of labeled virions (Figure 2I), labeling brightness (Figure 2J), or viral infectivity (Figure 2K). Collectively, these experiments indicated to us that a labeling concentration of 30 μg/mL AF647 for 20 min provided an optimal balance between brightness of fluorescence signal and impact on viral infectivity. Indeed, this labeling approach engendered no measurable effect on the virus’s life cycle in vitro, as determined by single-step growth curve analyses (Figure 2L). Moreover, VSV-AF647 could be observed interacting with and binding to live Vero cells in culture (Figure 2M), demonstrating a dynamic association (Video S1) with the cell membrane that has been previously reported.36
Figure 2.
In Vitro Optimization of VSV Labeling with AF647
(A–K) VSV was labeled with serial dilutions of AF647 using a fixed incubation time of 20 min (A–H) or with 30 μg/mL AF647 for variable incubation times (I–K). Labeled virions were analyzed by flow cytometry gating on FSC-A/SSC-A (A) and gating on FSC-H/FSC-W (B) and SSC-H/SSC-W (C) to exclude doublets, followed by measurement of percentage of events positive in AF647 channel (D) and fluorescence intensity (E). Values are reported as the percentage of labeled events (F and I) and mean fluorescence intensity (G and J) ± SD (n = 3). The ability of labeled VSV to infect and replicate was measured in vitro and is reported as TCID50 (H and K). Values represent mean ± SD (n = 3); *p < 0.05; ns, non-significant in comparison to unlabeled control. To further compare the effect of labeling on infection, Vero cells were infected with MOI = 1 by labeling with 3 or 30 μg/mL AF647 or unlabeled control. (L) At 6, 12, and 24 hpi, supernatant was collected and titered by TCID50 assay. Values are reported as the mean ± SD (n = 3). (M) Vero cells were infected with VSV (MOI = 50) labeled with 10 μg/mL AF647 and immediately imaged by confocal microscopy (see also Video S1). Scale bars, 5 μm; DIC, differential interference contrast; representative images of three independent experiments.
Arrows indicate co-localization of multiple virus particles on the cell surface. Images are captured at 1 frame/s for a 420s interval.
To determine if VSV could be visualized using other AF dyes, similar experiments were performed using AF555 (Figure S1). Nearly identical labeling results were achieved with this fluorophore (Figures S1A–S1H). VSV-AF555 could be easily visualized with confocal (Figure S1I; Video S2) and multiphoton imaging modalities (Figure S1J; Video S2). Additionally, VSV-AF555 and VSV-AF647 could be simultaneously visualized, and individually discerned, when injected intravenously (i.v.) into the same animal (Figure S1K). Moreover, particle size analyses using dynamic light scattering (DLS) identified the majority of AF555-labeled VSV particles to have a mean diameter of ∼200 nm (Figures S1L and S1M), consistent with the size of an unlabeled VSV particle.37 We also tested the AF labeling and imaging approach for other OVs, including another rhabdovirus (MarabaMG1, hereafter referred to as Maraba)38, 39 (Figure S2) and a non-enveloped RNA virus (Reovirus)40 (Figure S3). Like VSV, Maraba could be efficiently and brightly labeled with AF647 (Figures S2E–S2G) with negligible impact on its infectivity in vitro (Figure S2H). Reovirus could also be labeled with AF647 (Figures S3E–S3G). While a significant drop in reovirus productivity was observed when using high AF concentrations (Figure S3H), lower concentrations could efficiently, and brightly, label reovirus without affecting its growth. Both Maraba and reovirus could be easily visualized in skin vasculature by confocal IVM (Figure S4).
Using resonant scanning confocal microscopy followed by imaging the same vessel with multiphoton microscopy. Blue, CD11b; green, Ly6g; cyan, collagen.
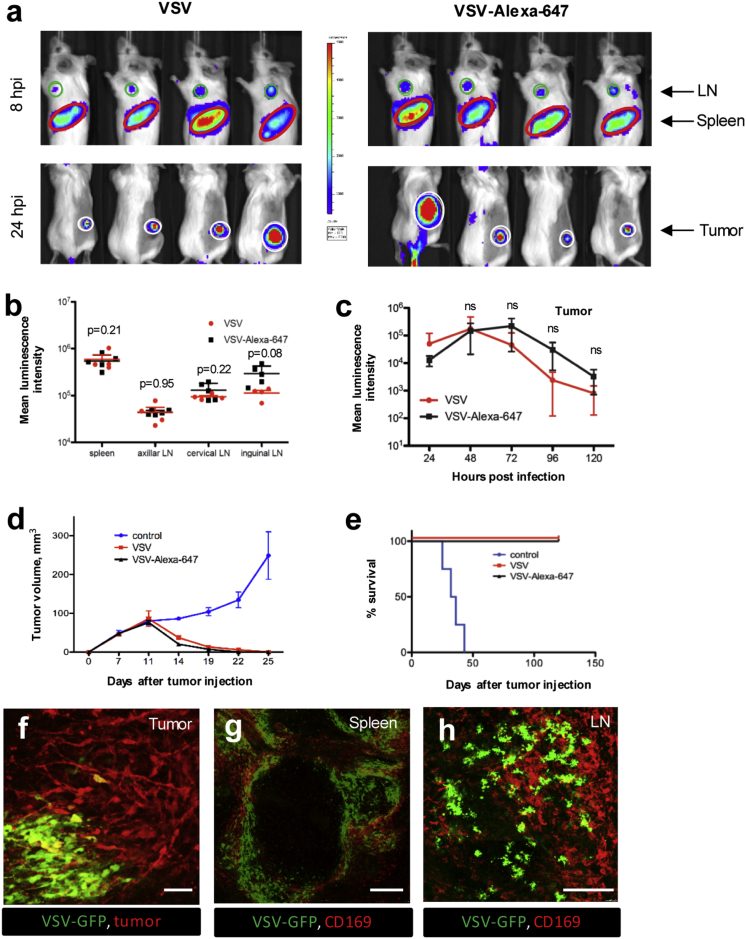
The CT-26 mouse model system was then used to evaluate the impact of AF labeling on virus bioactivity in vivo. This tumor system was chosen because of its high responsiveness to OV therapy35, 38 and because it is grown in fully immunocompetent mice. Visualization of VSV-delivered Firefly Luciferase (Fluc) by whole-body bioluminescence imaging revealed that treatment of CT-26 tumor-bearing mice with either VSVFluc or VSVFluc-AF647 resulted in a similar infection pattern of the tumor, axillary LN, and spleen at both 8 hr post-infection (hpi; Figure 3A, top row) and 24 hpi (Figure 3A, bottom row). Consistent with this, quantification of bioluminescent signal confirmed that AF647 labeling had no bearing on the VSV infection in vivo (Figures 3B and 3C). AF647 labeling also had no impact on VSV-mediated tumor regression (Figure 3D) and overall survival (Figure 3E). IVM of red fluorescent protein (RFP)-expressing CT-26 tumors demonstrated clear infection of CT-26 cells by AF647-labeled VSV genetically encoding a GFP transgene (VSVGFP; Figure 3F). Furthermore, fluorescence microscopy of ex vivo spleen (Figure 3G) and LN (Figure 3H) tissues revealed widespread infection of host cells by VSVGFP-AF647, including CD169+ macrophages known to support infection of wild-type VSV.41 Collectively, these data establish AF labeling as an effective strategy for multiplex visualization of bioactive OV particles in live mice using IVM.
Figure 3.
In Vivo Evaluation of VSV-AF647 Functionality
(A) 5 × 108 PFU unlabeled VSVFluc (left panel) or VSVFluc labeled with 30 μg/mL AF647 (right panel) was injected i.v. into CT-26-bearing mice (n = 4), and at 8–120 hpi Fluc activity was measured by bioluminescence in LNs (upper row, green circles), spleen (upper row, red ovals), and tumors (lower panel, white circles). (B) Luminescence intensity for LNs and spleen at 8 hpi, values represent mean bioluminescence intensity ± SD (n = 4). (C) Luminescence intensity for tumors at 24–120 hpi, values represent mean bioluminescence intensity ± SD (n = 4). (D and E) Tumor measurements (D) and Kaplan-Meier survival plots (E) for animals treated with 5 × 108 PFU unlabeled VSV, VSV labeled with 30 μg/mL AF647, or PBS-treated control group. Values represent mean ± SD (n = 4). (F) Intravital imaging of CT-26RFP tumors (red) infected with VSVGFP (5 × 108 PFU) labeled with 30 μg/mL AF647, 24 hr post i.v. delivery. Scale bars, 50 μm; representative image of three independent experiments. (G) Confocal imaging of CD169+ metallophilic macrophages (red) infected by VSVGFP (5 × 108 PFU) labeled with 30 μg/mL AF647 in spleen ex vivo, 8 hpi. Scale bars, 250 μm; representative image of three independent experiments. (H) Confocal imaging of CD169+ subcapsular sinus macrophages (red) infected by VSVGFP (5 × 108 PFU) labeled with 30 μg/mL AF647 in inguinal LN ex vivo, 8 hpi. Scale bars, 250 μm; representative image of three independent experiments.
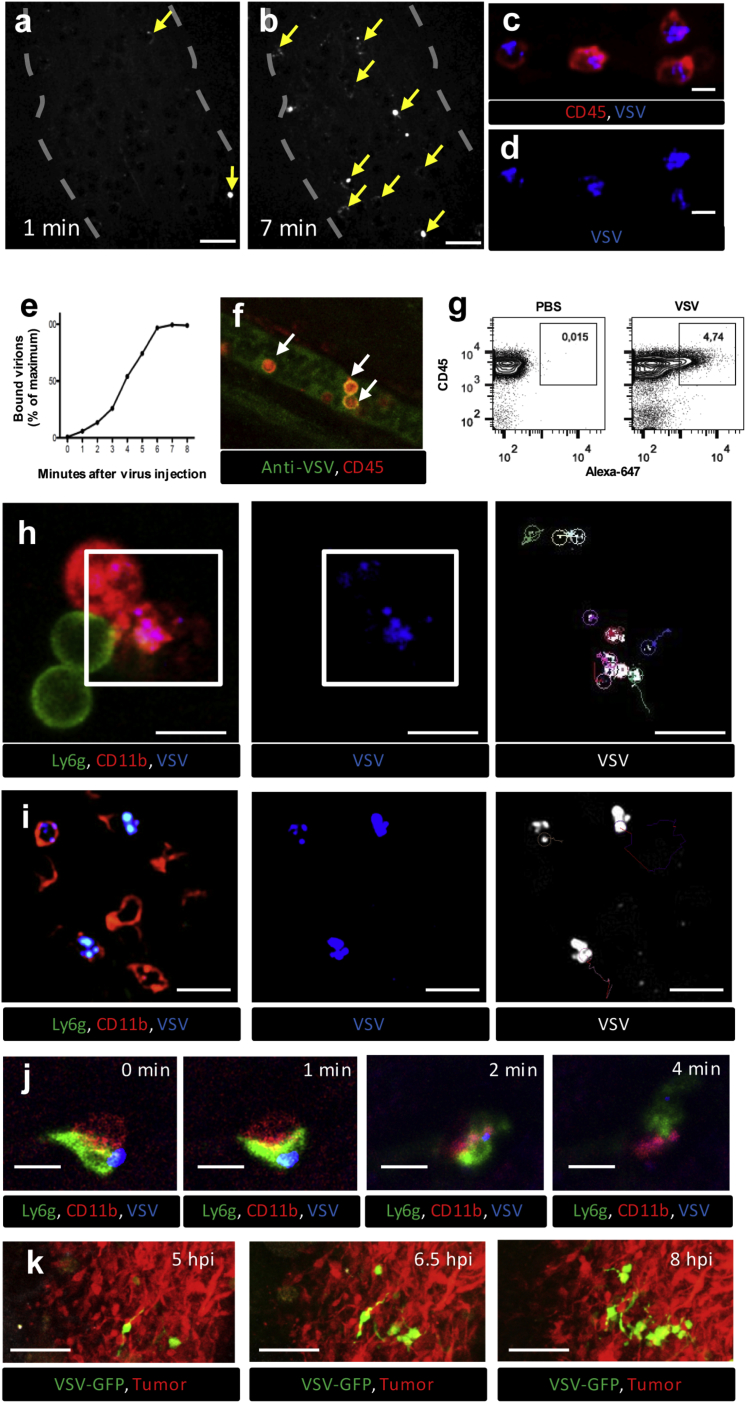
We next sought to determine whether IVM could effectively identify, characterize, and track the interactions between OV and host cells in situ. Because the primary goal of OV therapy is to deliver virus to cancer cells for direct oncolysis,1, 42 we began these experiments by imaging for VSV within the tumor. Immediately following i.v. injection, VSV-AF647 was easily visualized in CT-26 tumor vessels (Figures 4A and 4B; Video S3). Strikingly, VSV was commonly observed binding to, or being captured by, intravascular cells within the tumor, often forming a halo of virions around unlabeled cells. The addition of fluorescently labeled antibody enabled the identification of OV-bound cells as CD45+ leukocytes (Figures 4C and 4D), many of which expressed the monocyte marker CD11b (e.g., Figures 4H and 4I; Video S4). Similarly, labeled Maraba virus and reovirus were also commonly observed interacting with intravascular leukocytes expressing monocyte (CD11b) or granulocyte (Ly6g) cell surface markers (Figure S4). VSV particles were also found co-localized with vascular endothelial cells (Figure S5). Time-lapse imaging allowed for OV-cell interactions to be quantified over time, which demonstrated VSV binding to intravascular leukocytes within the first 7–8 min post-injection (Figures 4B and 4E).
Figure 4.
IVM Imaging of OV-Host Interactions in Mouse Tumors
(A and B) 5 × 108 PFU VSV labeled with 30 μg/mL AF647 were injected i.v. while visualizing superficial CT-26 tumor vessels by confocal IVM. Arrows indicate virus particles (white) bound to leukocytes (shadows) at 1 (A) and 7 (B) min following virus injection (see also Video S3). Scale bars, 20 μm; representative images of three independent experiments. (C and D) IVM of CD45 cells (red) bound by VSV-AF647 (blue) in the CT-26 tumor vessels (see also Video S4) (C, merged channels; D, AF647 channel only). Scale bars, 10 μm; representative images of three independent experiments. (E) Dynamics of virus binding by CD45+ cells inside the CT-26 tumor vessels during first 8 min after i.v. injection. Values represent the percentage of maximum virus binding by CD45+ cells within the field of view (representative plot of three independent experiments). (F) Confirmation of VSV binding to CD45 leukocytes using VSV antibody. CT-26-bearing mice were treated i.v. with unlabeled VSV (5 × 108 PFU) and, 5 min later, with antibody targeting VSV (green) and CD45 (red). Tumor vessels were imaged using confocal IVM. White arrows indicate VSV-bound CD45 leukocytes. Scale bars, 20 μm; representative image of two independent experiments. (G) Flow cytometry analysis of blood collected 5 min after i.v. injection of PBS (left panel) or AF647-labeled VSV (right panel) (representative plot of three independent experiments). (H) Tracking of individual viral particles 5 min after i.v. injection of AF647-labeled VSV. Time-lapse imaging was collected for 120 s, and individual virus particle trajectories on the surface of CD11b+/Ly6g− cells in vessels were reconstructed using ImageJ (see also Video S5) (left, merged channels [red, CD11b; green, Ly6g; blue, VSV]; middle, AF647 channel only; right [selected area from left and middle], viral trajectories). Scale bars, 10 μm; representative images of three independent experiments. (I) Tracking of intravascular leukocytes 5 min after i.v. injection of AF647-labeled VSV. Time-lapse imaging was collected for 200 s, and the trajectories of intravascular leukocytes bound by virus were reconstructed using ImageJ (see also Video S6) (left, merged channels [red, CD11b; green, Ly6g; blue, virus]; middle, AF647 channel only; right, cell trajectories). Scale bars, 20 μm; representative images of three independent experiments. (J) Tracking of individual viral particles 30 min after i.v. injection of AF647-labeled VSV. Time-lapse imaging of virus (blue) transfer from monocyte (red, CD11b) to neutrophil (green, Ly6g) within the blood vessel (gray) 30 min after i.v. injection of AF647-labeled VSV (see also Video S7). Scale bars, 10 μm; representative images of three independent experiments. (K) IVM of VSV spreading through CT-26 tumors. CT-26RFP-bearing mice were treated i.v. with VSVGFP (5 × 108 PFU). Growing foci of VSV infection were imaged by IVM between 5 and 8 hpi. Green, VSV; red, CT-26 tumor cells; scale bars, 100 μm; representative images of three independent experiments.
Arrows indicate examples of leukocytes bound by many virus particles, forming a “halo” around the cell surface. Images are captured at 1 frame/6 s for a 420 s interval.
Arrow indicates multiple virus particles co-localizing on the cell surface. Images are captured at 1 frame/6s for a 420 s interval. Green, Ly6g.
To ensure the observed interactions were not due to AF647 conjugation, unlabeled VSV was injected i.v. followed by fluorescein isothiocyanate (FITC)-conjugated anti-VSV antibodies. This approach demonstrated VSV binding to leukocytes irrespective of AF647 labeling (Figure 4F). Similarly, to ensure viral binding was not due to an artifact associated with antibody-labeled cells, animal surgery, or IVM, blood samples collected 5 min following i.v. delivery of vehicle (PBS) or AF647-labeled VSV were analyzed by flow cytometry to confirm the association of labeled VSV particles with peripheral blood leukocytes (CD45+ cells; Figure 4G). These data demonstrate that IVM has sufficient visual resolution to image OV-cell interactions within the tumor. They also identify the capture of OV by intravascular leukocytes as a common, early event in tumor vessels following i.v. delivery.
Closer observation and tracking of labeled OV enabled the study of its behavior when interacting with intravascular leukocytes (Figure 4H; Video S5). Individual virions were found to be either loosely associated with leukocytes; randomly surfing across the cell surface with a Brownian-like motion (Video S5, trajectory 1); or tightly associated with the leukocytes, demonstrating limited movement along the cell surface and appearing to vibrate back and forth in a single location (Video S5, trajectory 2). Similar behaviors were observed by VSV bound to endothelial cells (Video S5). These different viral behaviors have previously been reported in vitro, and they are believed to reflect either the non-specific association of virus with cells or the high-affinity interactions between virions and specific cellular receptors.43, 44 The leukocytes bound by virus also displayed a range of behaviors, with some remaining stationary for long periods of time, sometimes probing the local environment, while others were seen crawling along the blood vessel wall (Figure 4I; Video S6). Time-lapse imaging also revealed the occasional transfer of VSV from one leukocyte to another (Figure 4J; Video S7) and growing foci of infection within the tumor from its earliest, single-cell stage (Figure 4K). Collectively, these data demonstrate the capacity of IVM to characterize OV dynamics and monitor OV-host interactions, over time, within the tumor microenvironment in live mice.
Using IVM 5 minutes after i.v. injection of virus.
Prominent examples are indicated by arrows. Red, CD11b; Blue, VSV-AF647; Green, Ly6g. Each of 3 videos were captured with 1 frame/6s acquisition rate for a 300 s interval.
30 minutes after virus injection.
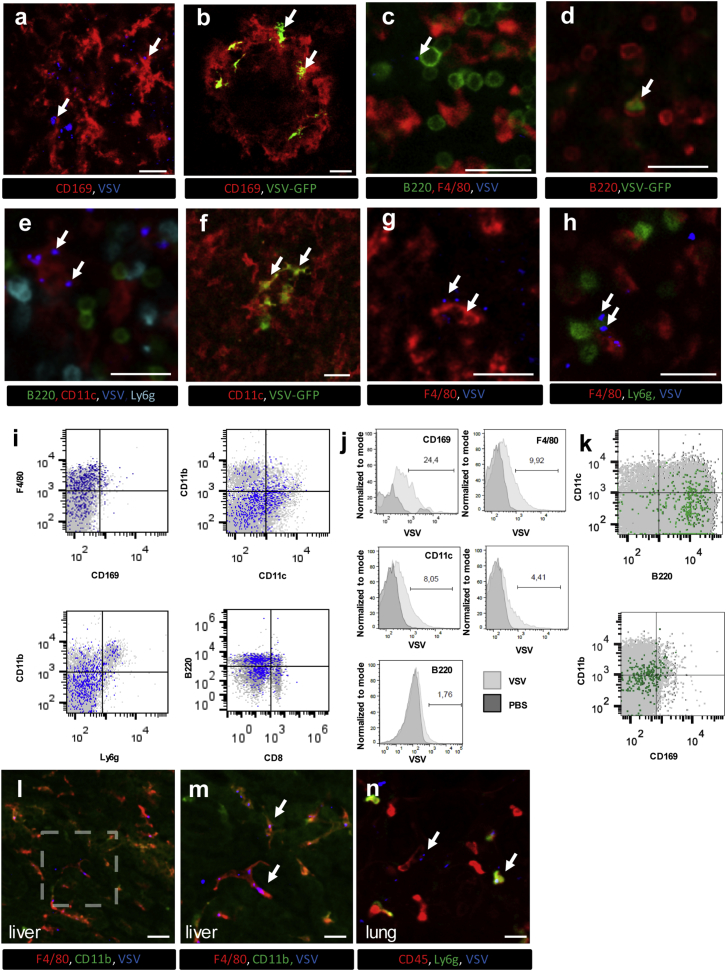
In addition to killing cancer cells directly, OVs can also boost anticancer T cell responses by delivering tumor antigens to B cells and dendritic cells (DCs) residing in the spleen, through a largely undefined mechanism.45, 46, 47, 48 We therefore sought to determine whether IVM could be used to visualize and study OV-host interactions in the spleen. Indeed, VSV could be observed binding to and infecting various leukocyte populations in both red and white pulp after i.v. injection (Figures 5A–5H). AF647-labeled VSV could be seen interacting with marginal zone CD169+ metallophilic macrophages (Figure 5A), as well as B cells (Figure 5C), DCs (Figure 5E), macrophages (Figure 5G), and neutrophils (Figure 5H) within the outer marginal zone and red pulp regions of the spleen. Visualization of the GFP transgene showed VSV infection of CD169+ macrophages, B cells, and DCs (Figures 5B, 5D, and 5F; Videos S8, S9, S10, and S11). Interestingly, we were unable to detect infection of CD169neg macrophages or neutrophils despite frequent interaction between these cells and VSV. Flow cytometry analysis confirmed the virus-cell associations identified by IVM (Figures 5I and 5J) and, at later time points, viral infection of B cells and CD169+ macrophages (Figure 5K). Collectively, these data show that IVM can be used to track OV-host interactions in the spleen, both within the red pulp region and the follicle itself. Visualizing additional tissues, IVM of the liver demonstrated viral interaction with and capture by the resident macrophage population, the Kupffer cell (Figures 5L and 5M; Video S12), and IVM of the lung showed occasional capture of virions by leukocytes within the lung vasculature (Figure 5N; Video S12). When taken together, our data demonstrate the ability of IVM to track viral dynamics in various intact mouse organs and visualize OV-host interactions in real time in vivo.
Figure 5.
IVM Imaging of OV-Host Interactions in Mouse Spleen and Other Visceral Organs
(A) Confocal microscopy of the spleen extracted 30 min after i.v. injection of 5 × 108 PFU VSV-AF647 revealed interactions between labeled virions (blue, marked by arrows) and CD169+ macrophages (red) 10 min following (representative image of three independent experiments). (B) Multiphoton IVM of the spleen 8 hpi shows infection (GFP; green, marked by arrows) of CD169+ macrophages (red) (see also Videos S8 and S9) (representative image of three independent experiments). (C) IVM of the splenic red pulp demonstrates interactions between B220+ cells (green) and VSV-AF647 (blue, marked by arrow) 10 min after i.v. injection. Red, F4/80; representative image of three independent experiments. (D) IVM of B220+ cells (blue) 8 hpi demonstrates VSV infection (GFP; green, marked by arrow) of B cells (see also Video S10). Red, Gr-1; cyan, F4/80; representative image of two independent experiments. (E) IVM of splenic CD11c+ cells (red) interacting with virus (blue, marked by arrows) 10 min after i.v. injection of VSV-AF647. Green, B220; cyan, Ly6g; representative image of three independent experiments. (F) IVM image of VSV-infected DCs (green, marked by arrow) 8 hpi (see also Video S11). Red, F4/80; blue, B220; representative image of three independent experiments. (G and H) IVM of virus bound (arrows) by splenic F4/80+ macrophages (G) and Ly6g+ neutrophils (H) 10 min after i.v. injection of VSV-AF647. Red, F4/80; green, Ly6g; blue, VSV; representative image of three independent experiments; scale bars, 25 μm. (I) Flow cytometry analysis of splenic leukocyte populations 10 min following i.v. injection of VSV-AF647. Cells positive for viral binding are colored blue, and cells negative for viral binding are colored gray; representative plot of two independent experiments. (J) Assessment of viral binding by CD169+, F4/80+, CD11c+, Ly6g+, or B220+ splenocyte populations. Mice were i.v. injected with either PBS or VSV-AF647 and spleens were harvested 10 min post-injection. Numbers indicate percentage of cells interacting with virus; representative plot of two independent experiments. (K) Infected (GFP, green) splenocytes harvested and immunotyped by flow cytometry 8 hpi with VSVGFP delivered i.v. (representative image of three independent experiments). (L) Confocal microscopy of the liver revealed interactions between Kupffer cells (F4/80, red) and virus (blue, arrows) 10 min after i.v. injection of VSV-AF647. Red, F4/80; green, CD11b; scale bars, 50 μm. (M) Higher-magnification image of area denoted in (L). Red, F4/80; green, CD11b; scale bars, 25 μm; representative image of two independent experiments (see also Video S12). (N) Confocal microscopy of the lung revealed interactions between leukocytes (CD45, red) and virus (blue, arrows) 10 min after i.v. injection of VSV-AF647. Red, CD45; green, Ly6g; scale bars, 25 μm; representative image of two independent experiments (see also Video S12).
8 hours following i.v. injection. Cyan, Ly6g.
8 hours following i.v. injection. Cyan, Ly6g/second harmonic.
8 hpi. Z-stack (22 images, 1 μm step) is captured by at a rate of 1 frame/6s for a 360s interval. Red, Gr-1; cyan, F4/80.
Red, F4/80; Blue, B220.
Arrow indicates multiple viruses forming aggregate on cell surface.
Discussion
We describe here the development and validation of an IVM technique that allows for simultaneous visualization of bioactive OV particles and their interactions with, and infections of, host cells in their natural, therapeutic environment. The technique offers two main advantages over existing methods used to study OVT.
-
(1)
Real-time, kinetic visualization in vivo. Our IVM technique allows for monitoring of OV particles, host cells, and their interactions and infections in real time throughout the course of treatment in mouse models of cancer. Dynamic events can be witnessed and studied, such as virus delivery to tumors and secondary lymphoid organs via the bloodstream, virus binding to and surfing upon cells, virus transfer between cells, the expression of viral genes or engineered transgenes within infected cells, the host immune response to the virus infection, and the spread of a virus infection throughout a tumor.
-
(2)
Sensitivity and spatial resolution. Our IVM technique is sufficiently sensitive to allow visualization of single virus particles and individual cells within blood, tumor, and visceral organs of live mice. This allows for the characterization and interrogation of virus interactions (e.g., capture by leukocytes in tumor vessels), virus behaviors (e.g., surfing on leukocyte membranes), and host responses (e.g., leukocyte trafficking in infected tissues). Moreover, our IVM technique allows for high-resolution imaging through all three spatial planes, which enables detailed 3D reconstruction of cells and tissues. This adds significantly to the study of OV-host interactions, for example, in the identification of infected cells using morphology and relationship to tissue landmarks to complement co-localization with molecular markers (many of which lack specificity). For example, OV infection of metallophilic macrophages in the spleen can be identified by a combination of (1) macrophage morphology, (2) anatomical location (surrounding a splenic follicle), and (3) expression of surface antigen CD169.
An important consideration in the development of our technique was the choice of fluorescent labeling strategies. Studying OV-host interactions by IVM requires simultaneous visualization and discernment of non-infected cells (cancer and normal), virally infected cells, and individual bioactive OV particles. When developing our technique, we drew from our own experience12, 24 and published IVM studies19, 49 of pathogenic virus-host interactions that used a combination of genetically engineered FP reporters, fluorophore-conjugated antibodies, and chemical dyes to label and track virus-cell contacts, behaviors, and infections in vivo. Each labeling approach has its unique strengths and limitations, and the choices we made were guided by factors such as specificity, sensitivity, innocuousness, flexibility, ease of use, and cost. For example, genetic strategies were chosen for labeling cancer cells and viruses because, with a few exceptions, cell lines and OVs can be easily and cost-effectively engineered to express a wide range of FPs in a specific, bright, and generally innocuous fashion. In contrast, genetic strategies for labeling normal cells and small virus particles require conditional transgenic reporter mice and chimeric virus-FP fusion proteins, respectively, which can be difficult; expensive; time consuming; and, in the case of fusion proteins, generally attenuating on viral bioactivity.28, 33 We therefore opted for antibody- and chemical labeling approaches for visualizing normal cells and virus particles, respectively. Antibody labeling offers the advantage of ready commercial availability and easy, custom conjugation with a wide range of fluorophores. Moreover, antibody-bound cells are bright and easily imaged by IVM, and multiple antigens on the same or different cell types can be simultaneously labeled and visualized. A major drawback, however, is that antibody binding can sometimes affect the target cell’s biology, for example, by blocking its normal interactions with other cells, causing spurious interactions, or triggering intracellular signaling pathways. Efforts must, therefore, be taken to confirm antibody-based findings using complementary approaches, which we did using flow cytometry but could also be done using transgenic reporter mice.
We chose chemical labeling to visualize OV particles, and indeed a major objective of our study was the identification of the optimal chemical dye. Three commercially available strategies were evaluated with the following objectives in mind: (1) labels individual OV particles with a bright signal that is easily visualized by IVM; (2) does not interfere with viral bioactivity; (3) numerous dyes available to ensure compatibility with different imaging platforms, filter sets, and multiplexing strategies; and (4) labeling is simple, reliable, and compatible with both enveloped and non-enveloped viruses. Of the strategies evaluated, only AF labeling met all four criteria. While FSL-fluorescein labeled VSV for easy visualization in blood vessels, it tended to reduce virus infectivity at increasing concentrations. Moreover, it was not bright enough to discern single particles in tissues with high background auto-fluorescence (i.e., liver and kidney; data not shown), is only available as a single excitation-emission spectra dye, and does not label non-enveloped virions. Likewise, quantum dot labeling by carbodiimide-coupling reaction provided bright fluorescence detection of single VSV particles in vivo, but it dramatically reduced virus infectivity at all doses tested, likely because of its large size. In contrast, AF labeling was bright and easily visualized in vivo. It did not reduce OV bioactivity by any measures tested, both in vitro and in vivo, although it did disrupt reovirus infectivity of cultured cells at higher doses, possibly because reovirus is non-enveloped. AF efficiently labeled all OVs tested, was photostable, and is available in a broad spectrum of colors suitable for both confocal and multiphoton imaging, including those with excitation-emission spectra very distinct from tissue auto-fluorescence. Collectively, these results suggested to us that AF dyes were ideal for labeling virus particles for the study of OV-host interactions by IVM.
While our study was designed to develop and validate the IVM method, several observations were made that may provide mechanistic insight into OV biology. Most striking was that i.v. delivered virus particles commonly interact with leukocytes in tumor blood vessels in seemingly non-random and dynamic ways. For example, VSV was frequently observed being captured by some, but not all, monocytes within an imaged vessel. Because the VSV receptors (low-density lipoprotein receptor [LDLR] and related family members)50 are ubiquitously expressed on mouse cells (except for naive T cells), this observation suggests that the mere presence of the VSV receptor is not sufficient to mediate its capture from the bloodstream. Indeed, it is well established that not all cell surface receptors are capable of capturing particles under the shear stresses associated with blood flow.51 Because VSV is known to be coated in complement and natural immunoglobulin M (IgM) soon after encountering serum,52 it may be that only specialized subpopulations of leukocytes have the appropriate receptors to directly capture coated VSV from the bloodstream, a possibility we are currently exploring.
We also noted several behaviors of cell-bound VSV not previously reported in vivo. These ranged from relatively restricted movement on the cell surface to considerable motion sometimes culminating in co-localization/aggregation with other virions to the occasional transfer from one leukocyte to another. While the significance of these behaviors is not understood, their identification would be essentially impossible in the absence of IVM and our ability to see virions in vivo in real time. It has been reported that some leukocytes can facilitate OV delivery to tumors. Indeed, CD11b+ cells have been reported to carry oncolytic reovirus into the tumor microenvironment, even in the face of a strong neutralizing antibody response.9 The predominance of VSV-leukocyte interactions in the minutes following virus injection, followed hours later by tumor cell infection, is consistent with a potential role for leukocytes in delivering VSV to cancer cells.
Another prominent observation was that of significant particle size variation in vivo. This was not measured in vitro, using either flow cytometry or dynamic light scattering. Observations of virion co-localization on the cell surface (as discussed above) provide a potential explanation for some particle size variation. However, large particles were sometimes observed floating freely in the bloodstream, apparently unbound by cells. While speculative, it may be that these particles represent immune complexes formed between VSV and antibody and/or complement within mouse blood. This possibility and its potential biological relevance is currently under investigation.
Finally, we show that IVM can be used to study the biodistribution, pharmacokinetics, and pharmacodynamics of OV therapies in visceral organs within the mouse. Previous studies have demonstrated that immediately after i.v. VSV administration, the highest viral titers are detected in the liver, spleen, blood, and lung.53 In contrast to the liver, where VSV titers rise rapidly (in minutes) and then gradually decrease, VSV titers in spleen continue to rise until 12 hpi, and virus glycoprotein can be detected in splenic CD169+ macrophages and DCs.41, 53 Consistent with these reports, we found that labeled VSV binds directly to F4/80+ (Kupffer) cells in the liver without infecting them (as determined by GFP expression), whereas VSV binds to and subsequently infects CD169+ macrophages, DCs, and B cells in the spleen. Transient virus replication in CD169+ macrophage is crucial for an effective immune response to VSV and overall animal survival.41 The role of DC and B cell infection is much less studied, but presumably it also plays an important role in eliciting an efficient anti-viral response. In addition, Bridle and colleagues46 recently showed a critical role for B cells and DCs in boosting antigen-specific T cell responses in the spleen after vaccination with a rhabdovirus vaccine. A previous study from the same group showed that B cells loaded ex vivo with a VSV vaccine and adoptively transferred into mice efficiently boosted antigen-specific T cell responses in the spleen.54 The B cell appears, therefore, to be at an interesting position within the OV immunotherapy paradigm, whereby it not only binds and traffics virions throughout both the red and white pulp, potentially delivering virus to DC for T cell priming, but also becomes infected itself, possibly serving the direct role of antigen-presenting cell (APC). Ongoing IVM studies to comprehensively map OV-host interactions in the spleen should allow the precise mechanisms by which OV vaccines boost anticancer T cell responses to be revealed.
There are several limitations that must be considered when using our technique, some of which we are currently working toward addressing: (1) imaging depth is limited to ∼150 μm for confocal IVM and ∼300–500 μm for multiphoton IVM, precluding visualization of deeper regions within the tumor (e.g., hypoxic regions); (2) the experiments are endpoint, with imaging duration being limited to ∼4–6 hours; the availability of imaging window chambers, however, should allow for longer-term, kinetic imaging over days, an approach we are currently exploring; and (3) strategies that further limit the impact of labeling on virus biology would be advantageous for the study of some viruses (i.e., reovirus) that have proven to be more sensitive to the labeling approaches outlined in this study; engineering viruses with small genetic tags, such as iLOV, UnaG, or tetracysteine-tag, is one possible strategy to preserve virus functionality.
Overall, the IVM technique described opens new pathways for studying the complex interactions between individual OVs and host cells in the live, tumor-bearing animal. The method is straightforward and could easily be adopted by other laboratories. Future studies using IVM should aid in the interrogation of OV biology and the subsequent development of more effective OV strains, dosing regimens, and combination therapies.
Materials and Methods
Animals
The 6- to 8-week-old female BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA) and maintained in specific-pathogen-free facilities at the University of Calgary. At the time of use, animals were between 7 and 10 weeks of age and weighed 20–22 g. All animal experiments were approved by the University of Calgary Animal Care Committee and conformed to the guidelines established by the Canadian Council for Animal Care.
Antibodies and Stains
BV421-conjugated rat anti-mouse Ly6g (clone 1A8), BV421-conjugated rat anti-mouse F4/80 (clone T45-2342), FITC-conjugated rat anti-mouse CD11b (clone M1/70), FITC-conjugated rat anti-mouse CD45R/B220 (clone RA3-6B2), phycoerythrin (PE)-conjugated rat anti-mouse CD11b (clone M1/70), PE-conjugated rat anti-mouse CD45 (clone 30-F11), PE-conjugated rat anti-mouse Ly6G and Ly6C (clone R6B-8CP), PE-conjugated rat anti-mouse CD8a (clone 53-6.7), and anti-CD16/CD32 (Fc block, clone 2.4G2) were purchased from BD Biosciences Pharmingen (San Diego, CA). FITC-conjugated rat anti-mouse Ly6g (clone 1A8), FITC-conjugated rat anti-mouse CD45 (clone 2D1), FITC-conjugated rat anti-mouse F4/80 (clone BM8), FITC-conjugated rat anti-mouse IgG2α,κ (clone RTK2758), FITC-conjugated rat anti-mouse IgG2β,κ (clone RTK4530), PE-conjugated rat anti-mouse CD169 (clone 3D6.112), PE-conjugated rat anti-mouse CD31 (clone 390), PE-conjugated rat anti-mouse F4/80 (clone BM8), PE-conjugated Armenian hamster anti-mouse CD11c (clone N418), PE-conjugated rat anti-mouse IgG2α,κ (clone RTK2758), PE-conjugated Armenian hamster anti-mouse IgG (clone HTK888), and PE/Cy7-conjugated rat anti-mouse IgG2β,κ (clone RTK4530) were purchased from BioLegend (San Diego, CA). eFluor 660-conjugated rat anti-mouse CD45R/B220 (clone RA3-6B2), PE/Cy7-conjugated rat anti-mouse CD11b (clone M1/70), and eFluor 660-conjugated rat anti-mouse IgG2α,κ (clone eBR2a) were purchased from eBioscience (San Diego, CA). FITC-conjugated rabbit anti-VSV was purchased from Biorbyt (San Francisco, CA). Qtracker 655 Vascular Labels were purchased from Invitrogen (Eugene, OR).
Viruses and Cells
VSVΔ51 (Indiana strain) engineered to express Fluc and GFP (VSVΔ51-Fluc/GFP, referred to throughout as VSV, VSVFluc, or VSVGFP) was originally generated by John Bell and kindly provided by Dr. Xuiquing Lun (University of Calgary, Calgary, AB, Canada), MarabaMG1 engineered to express GFP (MarabaGFP, referred to throughout as Maraba) was kindly provided by Dr. David Stojdl (CHEO Research Institute, Ottawa, ON, Canada), and Reovirus was kindly provided by Dr. Don Morris (University of Calgary, Calgary, AB, Canada). The VSV and Maraba viruses were propagated in monolayer cultures of Vero cells, purified, and titered using standard protocols.55 Reovirus was propagated in monolayer cultures of L929 cells, purified, and titered using plaque assay.
CT-26 (murine colon adenocarcinoma) cells stably expressing LacZ were kindly provided by Dr. John Bell (University of Ottawa, Ottawa, ON, Canada) and cultured in RPMI supplemented with 10% fetal calf serum (FCS). Vero cells (CCL81) cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in DMEM supplemented with 10% FCS. L929 cells were kindly provided by Dr. Don Morris (University of Calgary) and cultured in DMEM supplemented with 10% FCS. All cell lines routinely tested negative for mycoplasma.
Virus Labeling
Qdot 655 ITK Carboxyl Quantum Dots was purchased from Invitrogen. 5 μL Qdots (0.8 or 8 μM prepared in PBS) was added to 45 μL VSV (6 × 1010 plaque-forming units [PFU]/mL) for a final concentration of 80 or 800 nM and mixed well by stirring. 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC, Invitrogen) was then used to crosslink carboxyl groups of Qdots with viral primary amines. For this, 50 μL freshly prepared EDAC stock solution (10 mg/mL) was added to the mixture of virus and Qdots, following incubation with slow shaking for 1 hr at room temperature.
FSL-fluorescein was obtained from Sigma-Aldrich. 2 μl FSL-fluorescein (20 or 200 μg/mL prepared in PBS) was added to 18 μL VSV (5 × 1010 PFU/mL) for final concentrations of 2 and 20 μg/mL. Virus and FSL-fluorescein were in turn incubated for 1 hr at 37°C or overnight at 4°C.
AF647 or AF555 succinimidyl esters (AF594SE, Molecular Probes, Invitrogen) were reconstituted in DMSO. Five microliter serial dilutions (10, 3, and 1 mg/mL and 300, 200, 100, 30, 20, and 10 μg/mL prepared in PBS) were added to 45 μL VSV (5 × 1010 PFU/mL), 45 μL Maraba (1 × 1010 PFU/mL), or 45 μL reovirus (5 × 1010 PFU/mL) for final concentrations of 1,000, 300, 100, 30, 20, 10, 3, 2, and 1 μg/mL dye, while stirring gently. Virus and dye were incubated 20, 40, or 120 min at room temperature, with gentle inversions every 5–15 min.
For all three approaches, unbound dye was removed by transferring the labeling mixture to Amicon Ultra-4 Centrifugal Filter Units (100-kDa membrane; EMD Millipore) and washing twice in 1 mL PBS by centrifugation (4,000 × g, 10 min, 4°C). Control virus samples were incubated with PBS.
TCID50 Assay
Vero cells were seeded 104/well in 96-well plates and cultured in 10% FCS DMEM until ∼90% confluence. VSV or Maraba was serially diluted (log 10) in serum-free DMEM. Viral dilutions were added to Vero cells and incubated for 1 hr. Infected cultures were washed three times with warm PBS and cultured in fresh 10% FCS DMEM at 37°C. GFP-positive cells were visualized 3 days later by fluorescence microscopy, and TCID50 was calculated using the Reed and Muench method.56
Plaque Assay
L929 cells were seeded 5 × 105/well in 6-well plates, and 24 hr later serial reovirus dilutions in serum-free DMEM were added and incubated for 45 min at 4°C with gentle shaking every 15 min. Infected cells were covered with DMEM (2 times with 10% FCS) and agar (2%) and incubated for 3 days at 37°C. Cells were stained with 1% Neutral Red Solution (Sigma), and virus titer was calculated by counting the plaques.
Virus Growth Kinetics
Vero cells were seeded 2 × 105 per well in 24-well plates, and 24 hr later they were infected with labeled or unlabeled virus (MOI = 1) for 1 hr at 37°C, with gentle shaking every 15 min. Infected cells were washed twice with warm PBS and grown in fresh media (DMEM supplemented with 2% FCS). Virus-containing supernatants were harvested at 6, 12, and 24 hpi and stored at −80°C until being titered by plaque assay.
Flow Virometry and Cytometry
For flow virometry, virus samples were labeled in vitro, diluted 1:10 in PBS, and analyzed immediately using an Attune Acoustic Focusing Cytometer (Life Technologies). PBS was used to set the gating for virus particles. MFI and percentage of labeled events were analyzed, after doublet exclusion, using Attune Cytometer software.
For flow cytometry, blood was collected through cardiac puncture into syringes containing 100 U heparin, and spleens were collected from euthanized animals, placed in ice-cold PBS, and passed through a 70-μm nylon mesh. Red blood cells were lysed using Ammonium-Chloride-Potassium (ACK). Cells were washed 3 times in cold PBS, blocked with anti-CD16/CD32 mAbs in flow cytometry wash buffer (FWB; PBS, 2% FCS, and 5 mM EDTA) for 30 min at 4°C, and stained with fluorophore-conjugated antibodies in FWB for 30 min at 4°C. Cells were washed again 3 times in FWB and quantified using an Attune Acoustic Focusing Cytometer (Life Technologies). Data were analyzed using either FlowJo (Tree Star) or Attune Cytometer software.
Dynamic Light Scattering
The diameter of virus particles was estimated using DLS (Malvern Zetasizer Nano Series). Unlabeled or AF555-labeled VSV was diluted in filtered ultrapure water (18.2 MΩ.cm) in a 1:20 ratio. DLS measurements were performed in 3 cycles of 10 runs (10 s each) at 25°C. Malvern DTS 5 software was used for data processing. Polydispersity index, correlation function, and cumulant fit error values were checked to ensure data quality. Particle sizes are reported as light-scattering intensity averages.
Tumor Model and Evaluation of OV Efficacy
CT-26 tumors were established by injecting 1 × 106 cells (in 50 μL PBS) subcutaneously into the right hind flank. When tumors reached ∼25 mm2 (∼10 days), mice were treated with labeled (30 μg/mL) or unlabeled VSV (5 × 108 PFU; or PBS control) by tail vein injection. At the indicated time points, mice were injected intraperitoneally (i.p.) with 150 mg/kg firefly D-luciferin (Gold Biotechnology) in PBS, and 10 min later they were imaged using an IVIS system series 100 (Xenogen, Alameda, CA). Photon emission values were calculated using Living Image version (v.)2.5 software (Xenogen). Flank tumor diameters were measured every other day using skin calipers, and mice were euthanized when tumors reached 100 mm2.
In Vitro Microscopy
Vero cells were seeded 2 × 105 in 30-mm dishes (Fluorodish) and 24 hr later treated with labeled VSV (10 μg/mL AF647, MOI = 50). Real-time imaging of virus-cell interaction was performed using a Leica SP8 inverted microscope (Leica Microsystems, Concord, ON, Canada) immediately after dispensing virus into the dish.
IVM
The microscopes, surgical preparation for subcutaneous tumor imaging, and intravital imaging were recently described in detail.12
Microscopes
Spinning-disk confocal microscopy was performed using an Olympus IX81 inverted microscope (Olympus, Center Valley, PA), equipped with an Olympus focus drive and a motorized stage (Applied Scientific Instrumentation, Eugene, OR). This microscope is fitted with UPLANSAPO 10×/0.40 and UPLANSAPO 20×/0.70 objective lenses and is mounted to an optical table (Newport, Irvine, CA) to minimize vibration when imaging. This microscope is coupled to a confocal light path (WaveFx; Quorum Technologies, Guelph, ON, Canada) based on a modified Yokogawa CSU-10 head (Yokogawa Electric, Tokyo, Japan). Each of 491-, 561-, and 642-nm excitation laser wavelengths (Cobalt, Stockholm, Sweden) was sequentially controlled and merged into a single optic cable using an LMM5 laser merge module (Spectral Applied Research, Richmond Hill, ON, Canada). Fluorescence was visualized through one of ET 525/50M (green channel), FF 593/40 (red channel), or ET 700/75M (far red channel) band pass emission filters (Semrock, Rochester, NY) and detected with a 512 × 512-pixel back-thinned electron-multiplying charge-coupled device (EMCCD) camera (C9100-13, Hamamatsu, Bridgewater, NJ).
Resonant-scanning confocal and multiphoton microscopies were performed using a Leica SP8 inverted microscope (Leica Microsystems, Concord, ON, Canada), equipped with 405-, 488-, 552-, and 638-nm excitation lasers, 8-kHz tandem scan head, and spectral detectors (conventional photomultiplier tubes [PMTs] and hybrid detectors [HyDs]) for superficial imaging (up to 150 μm). This platform is also equipped with a tunable multiphoton excitation laser (700–1,040 nm; Newport, Irvine, CA) and external PMT detectors (Leica) for imaging deeper inside tumors (up to 300–500 μm).
Surgical Preparations
Mice were anesthetized by intraperitoneal injection of 200 mg/kg ketamine (Bayer Animal Health, Toronto, ON, Canada) and 10 mg/kg xylazine (Bimeda-MTC, Cambridge, ON, Canada), and the tail vein was cannulated with polyethelene tubing (0.28 × 0.60 mm, InStech Laboratories, Plymouth Meeting, PA) for delivering fluorescently labeled antibodies (5–10 μg) and maintaining the anesthetic. Body temperature was maintained using a heated stage.
Skin and tumor preparations were made as described.12 Briefly, a midline incision along the spine was made and the skin reflected. The thin connective tissue membrane overlaying the inside surface of the skin was removed, and edges of this skin flap were secured using sutures to expose and stabilize the tumor/vessels for imaging.
Spleen preparations were made by making a 1-cm incision in the skin and musculature at the left dorsal side of the animal and gently tethering out the spleen using 3-0 sutures tied to its associated connective tissue. The mouse was then laid on a stage with the spleen positioned over a coverslip.
Liver preparations were made by making a midline incision followed by a lateral incision along the costal margin to the midaxillary line, which was performed to expose the liver. The mouse was placed in a right lateral position, and the ligaments attaching the liver to the diaphragm and the stomach were cut, allowing the liver to be externalized onto a glass coverslip on the inverted microscope stage. Exposed abdominal tissues were covered with saline-soaked gauze to prevent dehydration. The liver was draped with a saline soaked tissue to avoid tissue dehydration and to help restrict movement of the tissue on the slide.
For lung preparations, a tracheotomy was performed and mice were mechanically ventilated at 150 breaths/min with 120 μL tidal volume using Inspira ASV (Harvard Apparatus, Holliston, MA). Mice were placed in the right lateral decubitus position, and the lung left lobe was exposed through removal of the overlying skin, fat, and portions of three to four anterior ribs. A thoracic suction window attached to a micromanipulator on the microscope stage was then placed into position, and 20–25 mm Hg of suction was applied (Amvex) to gently immobilize the lung.
In some experiments, following i.v. injection of virus and labeling antibodies, the animal was euthanized and tissues removed (spleen and LN). Tissues were then fixed for 10 min in 1% paraformaldehyde (PFA), sectioned, and ex vivo imaging of these whole mounts was performed.
Image Analysis
Still images were exported from the microscope acquisition software as .tif images and imported directly into ImageJ for analysis of cell number, associations, and behaviors. Display items were processed using Photoshop (Adobe, San Jose, CA) to adjust the minimum threshold values for each of the fluorescence channels. The same threshold values were applied to images from all treatment groups within a single experiment. Videos underwent contrast enhancement for each fluorescence channel. Again, the same settings were applied to the videos of all treatment groups within a given experiment. Videos were exported as .avi files, and they were converted to an appropriate format, size, resolution, and frame rate using Microsoft Movie Maker (Microsoft Canada, Mississauga, ON, Canada).
Statistical Analysis
Statistical analyses were performed using unpaired Student’s t test, ANOVA, and Kaplan-Meier log-rank assessment (GraphPad Prism 5.0). p < 0.05 was considered statistically significant.
Author Contributions
V.N., C.N.J., and D.J.M. designed the study. V.N., S.-J.K, Z.Z., J.D., and A.L. performed intravital imaging. V.N., S.V., and H.D. performed in vitro and animal experiments. C.Z. generated virus. D.-S.K. and N.M. performed dynamic light-scattering experiments in B.H.’s lab. V.N., C.N.J., and D.J.M. analyzed and interpreted data and wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
The authors thank David Stojdl (CHEO), John Bell (OHRI), and Don Morris (University of Calgary) for providing OV strains, cell lines, and access to the Xenogen 100 imager. We also thank Paul Kubes (University of Calgary) for access to the spinning-disk confocal microscope and Mandy Tse (Snyder Translational Lab) for tissue preparation for fluorescence-activated cell sorting (FACS) analysis. The work was supported by grants from the Canadian Foundation for Innovation (D.J.M. and C.N.J.), Canadian Cancer Society Research Institute (D.J.M. and C.N.J.), the Canada Research Chairs program (C.N.J.), the Snyder Institute for Chronic Disease (C.N.J.), and the Alberta Children’s Hospital Foundation (D.J.M.).
Footnotes
Supplemental Information includes five figures and twelve videos and can be found with this article online at https://doi.org/10.1016/j.omto.2018.06.001.
Contributor Information
Craig N. Jenne, Email: cnjenne@ucalgary.ca.
Douglas J. Mahoney, Email: djmahone@ucalgary.ca.
Supplemental Information
References
- 1.Bell J., McFadden G. Viruses for tumor therapy. Cell Host Microbe. 2014;15:260–265. doi: 10.1016/j.chom.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichty B.D., Breitbach C.J., Stojdl D.F., Bell J.C. Going viral with cancer immunotherapy. Nat. Rev. Cancer. 2014;14:559–567. doi: 10.1038/nrc3770. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt C. Oncolytic Virus Approved To Treat Melanoma. J. Natl. Cancer Inst. 2016;108:djw140. doi: 10.1093/jnci/djw140. [DOI] [PubMed] [Google Scholar]
- 4.Inman, S. (2016). Oncolytic poliovirus receives breakthrough designation for glioblastoma. https://www.onclive.com/web-exclusives/oncolytic-poliovirus-receives-breakthrough-designation-for-glioblastoma.
- 5.Panjwani, L. (2015). FDA grants reolysin orphan drug designation for ovarian cancer. https://www.onclive.com/web-exclusives/fda-grants-reolysin-orphan-drug-designation-for-ovarian-cancer#sthash.SNDHB7Kc.dpuf.
- 6.Andtbacka R.H.I., Kaufman H.L., Collichio F., Amatruda T., Senzer N., Chesney J., Delman K.A., Spitler L.E., Puzanov I., Agarwala S.S. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 7.Galanis E., Markovic S.N., Suman V.J., Nuovo G.J., Vile R.G., Kottke T.J., Nevala W.K., Thompson M.A., Lewis J.E., Rumilla K.M. Phase II trial of intravenous administration of Reolysin(®) (Reovirus Serotype-3-dearing Strain) in patients with metastatic melanoma. Mol. Ther. 2012;20:1998–2003. doi: 10.1038/mt.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heo J., Reid T., Ruo L., Breitbach C.J., Rose S., Bloomston M., Cho M., Lim H.Y., Chung H.C., Kim C.W. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilett E., Kottke T., Donnelly O., Thompson J., Willmon C., Diaz R., Zaidi S., Coffey M., Selby P., Harrington K. Cytokine conditioning enhances systemic delivery and therapy of an oncolytic virus. Mol. Ther. 2014;22:1851–1863. doi: 10.1038/mt.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridle B.W., Hanson S., Lichty B.D. Combining oncolytic virotherapy and tumour vaccination. Cytokine Growth Factor Rev. 2010;21:143–148. doi: 10.1016/j.cytogfr.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Schießl I.M., Castrop H. Deep insights: intravital imaging with two-photon microscopy. Pflugers Arch. 2016;468:1505–1516. doi: 10.1007/s00424-016-1832-7. [DOI] [PubMed] [Google Scholar]
- 12.Naumenko V., Jenne C., Mahoney D.J. Intravital Microscopy for Imaging the Tumor Microenvironment in Live Mice. Methods Mol. Biol. 2016;1458:217–230. doi: 10.1007/978-1-4939-3801-8_16. [DOI] [PubMed] [Google Scholar]
- 13.Peti-Peterdi J., Kidokoro K., Riquier-Brison A. Novel in vivo techniques to visualize kidney anatomy and function. Kidney Int. 2015;88:44–51. doi: 10.1038/ki.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Goldstein D.R., Kreisel D. Intravital 2-photon imaging, leukocyte trafficking, and the beating heart. Trends Cardiovasc. Med. 2013;23:287–293. doi: 10.1016/j.tcm.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K., Toiyama Y., Okugawa Y., Okigami M., Inoue Y., Uchida K., Araki T., Mohri Y., Mizoguchi A., Kusunoki M. In vivo optical imaging of cancer metastasis using multiphoton microscopy: a short review. Am. J. Transl. Res. 2014;6:179–187. [PMC free article] [PubMed] [Google Scholar]
- 16.Ellenbroek S.I.J., van Rheenen J. Imaging hallmarks of cancer in living mice. Nat. Rev. Cancer. 2014;14:406–418. doi: 10.1038/nrc3742. [DOI] [PubMed] [Google Scholar]
- 17.Breart B., Lemaître F., Celli S., Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J. Clin. Invest. 2008;118:1390–1397. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigelin B., Bolaños E., Teijeira A., Martinez-Forero I., Labiano S., Azpilikueta A., Morales-Kastresana A., Quetglas J.I., Wagena E., Sánchez-Paulete A.R. Focusing and sustaining the antitumor CTL effector killer response by agonist anti-CD137 mAb. Proc. Natl. Acad. Sci. USA. 2015;112:7551–7556. doi: 10.1073/pnas.1506357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickman H.D., Bennink J.R., Yewdell J.W. Caught in the act: intravital multiphoton microscopy of host-pathogen interactions. Cell Host Microbe. 2009;5:13–21. doi: 10.1016/j.chom.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickman H.D., Bennink J.R., Yewdell J.W. From optical bench to cageside: intravital microscopy on the long road to rational vaccine design. Immunol. Rev. 2011;239:209–220. doi: 10.1111/j.1600-065X.2010.00973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murooka T.T., Deruaz M., Marangoni F., Vrbanac V.D., Seung E., von Andrian U.H., Tager A.M., Luster A.D., Mempel T.R. HIV-infected T cells are migratory vehicles for viral dissemination. Nature. 2012;490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez S.F., Lukacs-Kornek V., Kuligowski M.P., Pitcher L.A., Degn S.E., Kim Y.-A., Cloninger M.J., Martinez-Pomares L., Gordon S., Turley S.J., Carroll M.C. Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for humoral immunity in draining lymph nodes. Nat. Immunol. 2010;11:427–434. doi: 10.1038/ni.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richner J.M., Gmyrek G.B., Govero J., Tu Y., van der Windt G.J.W., Metcalf T.U., Haddad E.K., Textor J., Miller M.J., Diamond M.S. Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection. PLoS Pathog. 2015;11:e1005027. doi: 10.1371/journal.ppat.1005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenne C.N., Wong C.H.Y., Zemp F.J., McDonald B., Rahman M.M., Forsyth P.A., McFadden G., Kubes P. Neutrophils recruited to sites of infection protect from virus challenge by releasing neutrophil extracellular traps. Cell Host Microbe. 2013;13:169–180. doi: 10.1016/j.chom.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Hickman H.D., Reynoso G.V., Ngudiankama B.F., Rubin E.J., Magadán J.G., Cush S.S., Gibbs J., Molon B., Bronte V., Bennink J.R., Yewdell J.W. Anatomically restricted synergistic antiviral activities of innate and adaptive immune cells in the skin. Cell Host Microbe. 2013;13:155–168. doi: 10.1016/j.chom.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada T., Takahashi S., Ishida A., Ishigame H. In vivo multiphoton imaging of immune cell dynamics. Pflugers Arch. 2016;468:1793–1801. doi: 10.1007/s00424-016-1882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mire C.E., White J.M., Whitt M.A. A spatio-temporal analysis of matrix protein and nucleocapsid trafficking during vesicular stomatitis virus uncoating. PLoS Pathog. 2010;6:e1000994. doi: 10.1371/journal.ppat.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S.C., Panda D., Nayak D., Pattnaik A.K. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged m protein allows dynamic imaging of m protein and virus uncoating in infected cells. J. Virol. 2009;83:2611–2622. doi: 10.1128/JVI.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S.C., Nayak D., Zhou Y., Pattnaik A.K. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 2006;80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen L., Lin Y., Zheng Z.-H., Zhang Z.-L., Zhang L.-J., Wang L.-Y., Wang H.Z., Pang D.W. Labeling the nucleocapsid of enveloped baculovirus with quantum dots for single-virus tracking. Biomaterials. 2014;35:2295–2301. doi: 10.1016/j.biomaterials.2013.11.069. [DOI] [PubMed] [Google Scholar]
- 31.Hadac E.M., Federspiel M.J., Chernyy E., Tuzikov A., Korchagina E., Bovin N.V., Russell S., Henry S.M. Fluorescein and radiolabeled Function-Spacer-Lipid constructs allow for simple in vitro and in vivo bioimaging of enveloped virions. J. Virol. Methods. 2011;176:78–84. doi: 10.1016/j.jviromet.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Gonçalves M.S.T. Fluorescent labeling of biomolecules with organic probes. Chem. Rev. 2009;109:190–212. doi: 10.1021/cr0783840. [DOI] [PubMed] [Google Scholar]
- 33.Mire C.E., Dube D., Delos S.E., White J.M., Whitt M.A. Glycoprotein-dependent acidification of vesicular stomatitis virus enhances release of matrix protein. J. Virol. 2009;83:12139–12150. doi: 10.1128/JVI.00955-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B.-H., Lin Y., Zhang Z.-L., Zhuan F., Liu A.-A., Xie M., Tian Z.Q., Zhang Z., Wang H., Pang D.W. Surface labeling of enveloped viruses assisted by host cells. ACS Chem. Biol. 2012;7:683–688. doi: 10.1021/cb2001878. [DOI] [PubMed] [Google Scholar]
- 35.Stojdl D.F., Lichty B.D., tenOever B.R., Paterson J.M., Power A.T., Knowles S., Marius R., Reynard J., Poliquin L., Atkins H. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann M.J., Sherer N.M., Marks C.B., Pypaert M., Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCombs R.M., Melnick M.B., Brunschwig J.P. Biophysical studies of vesicular stomatitis virus. J. Bacteriol. 1966;91:803–812. doi: 10.1128/jb.91.2.803-812.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brun J., McManus D., Lefebvre C., Hu K., Falls T., Atkins H., Bell J.C., McCart J.A., Mahoney D., Stojdl D.F. Identification of genetically modified Maraba virus as an oncolytic rhabdovirus. Mol. Ther. 2010;18:1440–1449. doi: 10.1038/mt.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahoney D.J., Lefebvre C., Allan K., Brun J., Sanaei C.A., Baird S., Pearce N., Grönberg S., Wilson B., Prakesh M. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell. 2011;20:443–456. doi: 10.1016/j.ccr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Coffey M.C., Strong J.E., Forsyth P.A., Lee P.W. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282:1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 41.Honke N., Shaabani N., Cadeddu G., Sorg U.R., Zhang D.-E., Trilling M., Klingel K., Sauter M., Kandolf R., Gailus N. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat. Immunol. 2011;13:51–57. doi: 10.1038/ni.2169. [DOI] [PubMed] [Google Scholar]
- 42.Russell S.J., Peng K.-W., Bell J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burckhardt C.J., Greber U.F. Virus movements on the plasma membrane support infection and transmission between cells. PLoS Pathog. 2009;5:e1000621. doi: 10.1371/journal.ppat.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandenburg B., Zhuang X. Virus trafficking - learning from single-virus tracking. Nat. Rev. Microbiol. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bridle B.W., Stephenson K.B., Boudreau J.E., Koshy S., Kazdhan N., Pullenayegum E., Brunellière J., Bramson J.L., Lichty B.D., Wan Y. Potentiating cancer immunotherapy using an oncolytic virus. Mol. Ther. 2010;18:1430–1439. doi: 10.1038/mt.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bridle B.W., Nguyen A., Salem O., Zhang L., Koshy S., Clouthier D., Chen L., Pol J., Swift S.L., Bowdish D.M. Privileged Antigen Presentation in Splenic B Cell Follicles Maximizes T Cell Responses in Prime-Boost Vaccination. J. Immunol. 2016;196:4587–4595. doi: 10.4049/jimmunol.1600106. [DOI] [PubMed] [Google Scholar]
- 47.Bridle B.W., Boudreau J.E., Lichty B.D., Brunellière J., Stephenson K., Koshy S., Bramson J.L., Wan Y. Vesicular stomatitis virus as a novel cancer vaccine vector to prime antitumor immunity amenable to rapid boosting with adenovirus. Mol. Ther. 2009;17:1814–1821. doi: 10.1038/mt.2009.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pol J.G., Zhang L., Bridle B.W., Stephenson K.B., Rességuier J., Hanson S., Chen L., Kazdhan N., Bramson J.L., Stojdl D.F. Maraba virus as a potent oncolytic vaccine vector. Mol. Ther. 2014;22:420–429. doi: 10.1038/mt.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Junt T., Moseman E.A., Iannacone M., Massberg S., Lang P.A., Boes M., Fink K., Henrickson S.E., Shayakhmetov D.M., Di Paolo N.C. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 50.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helmy K.Y., Katschke K.J., Jr., Gorgani N.N., Kljavin N.M., Elliott J.M., Diehl L., Scales S.J., Ghilardi N., van Lookeren Campagne M. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124:915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 52.Tesfay M.Z., Ammayappan A., Federspiel M.J., Barber G.N., Stojdl D., Peng K.-W., Russell S.J. Vesiculovirus neutralization by natural IgM and complement. J. Virol. 2014;88:6148–6157. doi: 10.1128/JVI.00074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breitbach C.J., Paterson J.M., Lemay C.G., Falls T.J., McGuire A., Parato K.A., Stojdl D.F., Daneshmand M., Speth K., Kirn D. Targeted inflammation during oncolytic virus therapy severely compromises tumor blood flow. Mol. Ther. 2007;15:1686–1693. doi: 10.1038/sj.mt.6300215. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L., Bridle B.W., Chen L., Pol J., Spaner D., Boudreau J.E., Rosen A., Bassett J.D., Lichty B.D., Bramson J.L., Wan Y. Delivery of viral-vectored vaccines by B cells represents a novel strategy to accelerate CD8(+) T-cell recall responses. Blood. 2013;121:2432–2439. doi: 10.1182/blood-2012-06-438481. [DOI] [PubMed] [Google Scholar]
- 55.Diallo J.-S., Vähä-Koskela M., Le Boeuf F., Bell J. Propagation, purification, and in vivo testing of oncolytic vesicular stomatitis virus strains. Methods Mol. Biol. 2012;797:127–140. doi: 10.1007/978-1-61779-340-0_10. [DOI] [PubMed] [Google Scholar]
- 56.Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arrows indicate co-localization of multiple virus particles on the cell surface. Images are captured at 1 frame/s for a 420s interval.
Using resonant scanning confocal microscopy followed by imaging the same vessel with multiphoton microscopy. Blue, CD11b; green, Ly6g; cyan, collagen.
Arrows indicate examples of leukocytes bound by many virus particles, forming a “halo” around the cell surface. Images are captured at 1 frame/6 s for a 420 s interval.
Arrow indicates multiple virus particles co-localizing on the cell surface. Images are captured at 1 frame/6s for a 420 s interval. Green, Ly6g.
Using IVM 5 minutes after i.v. injection of virus.
Prominent examples are indicated by arrows. Red, CD11b; Blue, VSV-AF647; Green, Ly6g. Each of 3 videos were captured with 1 frame/6s acquisition rate for a 300 s interval.
30 minutes after virus injection.
8 hours following i.v. injection. Cyan, Ly6g.
8 hours following i.v. injection. Cyan, Ly6g/second harmonic.
8 hpi. Z-stack (22 images, 1 μm step) is captured by at a rate of 1 frame/6s for a 360s interval. Red, Gr-1; cyan, F4/80.
Red, F4/80; Blue, B220.
Arrow indicates multiple viruses forming aggregate on cell surface.