Abstract
Background/Aim: Microsatellite instability (MSI) results from genetic alterations involving the mismatch repair (MMR) genes MLH1, PSM2, MSH2, and MSH6. MSI has been implicated in both sporadic CRC and Lynch syndrome. The aim of the study was to assess the frequency of alterations in MMR protein expression in both primary colorectal cancer and precursor lesions among Puerto Rican patients. Patients and Methods: A retrospective study of 84 Puerto Rican patients was performed to assess the frequency of MMR protein expression alterations in both primary CRC and precursor lesions using tissue microarray and immunohistochemistry. Results: The loss of expression of both MLH1 and PMS2 proteins was present in 6.3% of adenomas, 9.1% of adenomas with high-grade dysplasia and 9.4% of colon adenocarcinomas. Negative nuclear staining for both MSH2 and MSH6 proteins was found in 2.4% of colon adenocarcinomas. Conclusion: When compared to prior reports, this study suggests a lower frequency of MSI among the Puerto Rican population. The higher prevalence of MLH1 mutations correlates with previous studies of protein expression among the Hispanic community including Colombian, Uruguay and Brazilian populations.
Keywords: DNA mismatch repair, proteins, colon cancer, PuertoRicans
Colorectal cancer (CRC) is the third most common cancer in the United States and the fourth most common cause of death worldwide. Studies suggest that approximately 20% of colorectal cancers arise due to microsatellite instability, a mechanism characterized by deletions or insertions within repetitive sequences of a distinct cluster of genes known as the mismatch repair genes (MMR). The MMR system is responsible for correcting mismatches generated during DNA replication. Novel studies have unveiled new roles of these genes defects in carcinogenesis involving DNA damage signaling caused by exogenous carcinogens through synergistic interaction with p53-homologous proteins, meiotic crossover promotion, reparative recombination prevention, somatic hypermutation process of immunoglobulin diversification, trinucleotides expansion associated with multiple neurodegenerative conditions and modulation of microRNA (1).
Microsatellite instability is the hallmark of hereditary nonpolyposis colorectal cancer (HNPCC) which accounts for 12-15% of sporadic colon cancer. This diagnosis harbors distinct clinical features such as an autosomal-dominant inheritance pattern with up to 90% of colorectal cancer penetrance with an onset during 4th or 5th decades of life, tumor location at the ascending colon, lymphocytes infiltration, mucinous histology and higher risk of developing extracolonic malignancies especially gastric and endometrial cancers (1,2). These microsatellite genetic alterations also account for 85% of hereditary colorectal cancer cases as well (3). These tumors are distinguished by multiple molecular mechanisms leading to the common denominator of MMR impairment including truncation of protein and subsequent loss of expression or hypermethylation of MLH1 promoter region and additional mutation of the proto-oncogene BRAF which plays a major role in growth factor signaling process (4).
The most frequently observed genetic alterations among MMR proteins involve the MLH1, MSH2, PMS2 and MSH6 proteins. The MSH2 and MSH6 bind together to form a complex known as MutSalpha that predominantly identifies base pairs mismatch. MLH1 and PMS2 also form a complex known as MutLalpha whose main role consists in interacting with the MutSalpha and proliferating cellular nuclear antigen (PCNA) in order to conduct excision and DNA re-synthesis. Therefore, mechanisms leading to loss of expression of any of these proteins cause an inability to repair DNA mismatches and consequently accumulation of mutations that favor oncogenesis (5-7). Researchers have stated that the clinical phenotype depends on the specific gene mutations. It has been described that colorectal cancer incidence is higher among individuals with MLH1 mutation (8,9). The most frequently observed genetic alteration among sporadic CRC with microsatellite instability was hypermethylation of the 5-end CpG region of the MLH1 gene, therefore affecting the promoter function and consequently protein transcription and expression (1,10). However, individuals with MSH2 genetic alterations have higher risk of developing extracolonic malignancies (11). Genetic alterations of secondary proteins such as PMS2 and MSH6 are associated with early tumor development expressing high microsatellite instability and late tumor development during the 6th decade of life expressing low microsatellite instability respectively (12-14).
The mutation spectrum has been shown to vary among the Hispanic populations. It has been described that MLH1 loss-of-expression mutations are more prevalent among non-Hispanic populations with colorectal cancer than other MMR genes mutations (15,16). Similarly, higher prevalence of MLH1 mutations has been described among several Hispanic populations from Colombia, Uruguay, Brazil, Portugal and Spain. (17). However, Caribbean Puerto Ricans MMR studies suggest a higher prevalence of MSH2 mutations among Puerto Rican patients compared to MLH1 mutations among Portuguese and Spanish patients despite of shared Spanish ancestry (18). The aim of the study is to assess the frequency of alterations in MMR protein expression in both primary colorectal cancer and precursor lesions of Puerto Rican patients.
Patients and Methods
Study design. We conducted a retrospective study including 84 CRC cancer patients among the Puerto Rican population from St. Luke’s Episcopal Hospital at Ponce, Puerto Rico between 2010 and 2013. The pathology reports and H&E slides of each case were evaluated by pathologists (SG and DC) with interest in Gastro-intestinal pathology, at both Institutions, to confirm the diagnosis and mark the regions of interest (normal colon, adenoma, HGD, and carcinoma). All tissue samples were collected, to construct tissue microarrays (TMA). When present, the tissue types included normal colonic mucosa (NM), adenoma (AD), high grade dysplasia adenoma (HGD) and colon adenocarcinoma (CA). Study criteria included individuals older than 18 years old with colorectal cancer. Those patients with inflammatory bowel disease or family history of it, were excluded from the study. To construct the TMAs, three core tissue cylindrical punches (diameter 1 mm) were taken from each “donor” paraffin-embedded tissue block and precisely arrayed into a new “recipient” paraffin block using Beecher Instruments (Manual tissue arrayer Model MTA-1). The constructed TMAs contained 84 cases of colonic adenocarcinoma, 11 cases of adenoma with high grade dysplasia, 16 cases of adenoma, and 30 cases of non-neoplastic colonic mucosa.
Mismatch repair protein expression. All samples included in the TMAs consisted of formalin fixed paraffin embedded tissues. Four micrometer sections were taken from each TMA and stained with hematoxylin and eosin. The mismatch repair protein expression of the tissues was assessed by immunohistochemistry using the following antibodies: MLH 1 (Clone ES05, Dako, Carpinteria, CA, USA), PMS2 (Clone EP51, Dako), MSH2 (Clone FE11, Dako) and MSH6 (Clone EP49, Dako). The immunostains were performed using the automated Ventana Ultra instrument. The immunostains were interpreted as positive if >10% of the available tissue for each sample was staining.
Ancestry analysis of Colon TMA tumors. A set of 106 single nucleotide polymorphisms (SNPs) that can discriminate indigenous American, African, and European ancestry was used to estimate the proportion of genetic ancestry in all cases tested. This panel has been described previously (30). SNPs included were selected for their large difference in allelic frequencies between ancestral populations and have a well-balanced distribution across the 22 autosomal chromosomes. Genotyping of the ancestry informative markers was done using a multiplex PCR coupled with single base extension methodology with allele calls using a Sequenom analyzer. All of the 84 CRCs, the 11 AD with HGD and the 16 AD present in the TMAs were subjected to ancestry analysis. Out of the 106 AIMS genotyped, 5 were excluded due to a genotyping rate <80%. Genotype frequencies at all remaining markers did not significantly deviate from those expected under the Hardy-Weinberg equilibrium law. Individuals with a call rate of <80% were excluded from the analysis. A total of 6 duplicates were also genotyped and concordance was >90%. For each sample, global genetic ancestry proportions were estimated using ADMIXTURE (31) under a supervised model with k of 3.
Statistical analysis. In order to evaluate the distinct expression of mismatch proteins expression of MLH1, PMS2, MSH2, and MSH6 in Puerto Rican population, a paired T-test was performed. The determined p-value measured as statistically significant was less than 0.05.
Results
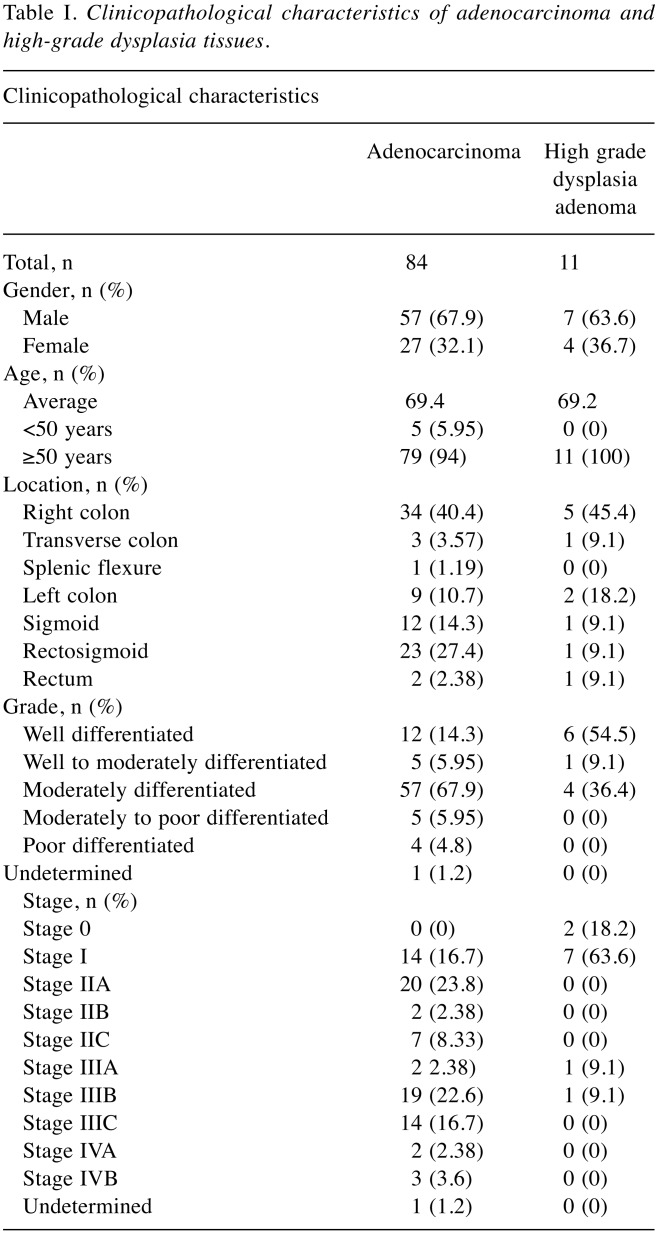
Clinicopathological features. A total of 95 samples were collected from 64 male and 31 female CRC/HGD patients ranging from 26 to 93 years old. Eighty-four tissues were colon adenocarcinoma from the right colon (40.4%), transverse colon (3.57%), splenic flexure (1.19%), left colon (10.7%), sigmoid colon (14.3%), rectosigmoid (27.4%) and rectum (2.38%). Eleven tissues were high-grade dysplasia (HGD) adenoma from the right colon (45.4%), transverse colon (9.1%), left colon (18.2%), sigmoid colon (9.1%), rectosigmoid (9.1%) and rectum (9.1%). Most tissues of colon adenocarcinoma were moderately differentiated (67.9%) and stage IIA or extending into serosa (23.8%). The average adenocarcinoma size was 4.27 centimeters, with the median being 4 centimeters and ranging from 0.6 to 12.5 centimeters. Clinicopathological characteristics of colon adenocarcinoma and HGD-adenoma respectively are detailed in Table I.
Table I. Clinicopathological characteristics of adenocarcinoma and high-grade dysplasia tissues.
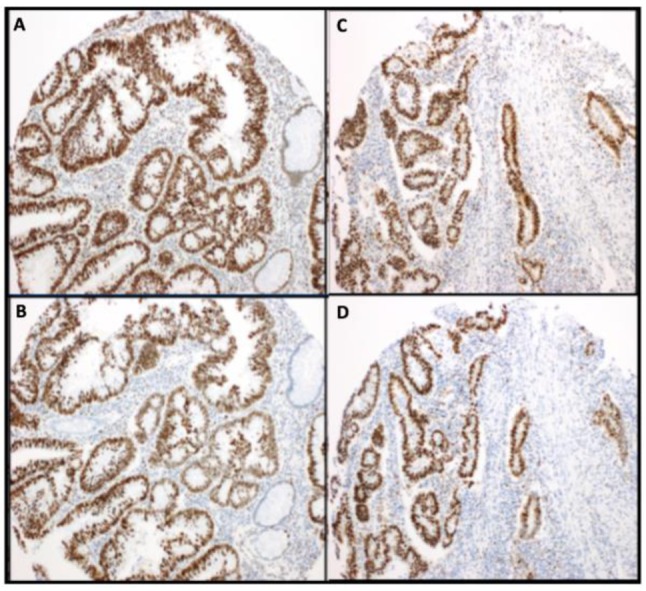
Frequency of mismatch repair protein expression. Among all of the analyzed samples the MMR stains were localized to the nucleus. The loss of expression of both MLH1 and PMS2 proteins was present in 6.3% of AD, 9.1% of AD with HGD and 9.4% of CRCs (Figure 1). However, no loss of expression of both MLH1 and PMS2 proteins was observed in tissue samples corresponding to non-neoplastic colonic mucosa, as it was expected. Negative nuclear staining for both MSH2 and MSH6 proteins was found in 2.4% of CRCs (Figure 2). Nonetheless, no loss of expression of both MSH2 and MSH6 proteins was observed in tissue samples of ADs, ADs with HGD and non-neoplastic colonic mucosa.
Figure 1. MLH1 and PMS2 proteins are expressed normally in microsatellite stable tumors (Image A and Image B respectively). Loss of MLH1 and PMS2 from tumor tissue in microsatellite instable colonic adenocarcinoma tumor (Image C and Image D respectively).
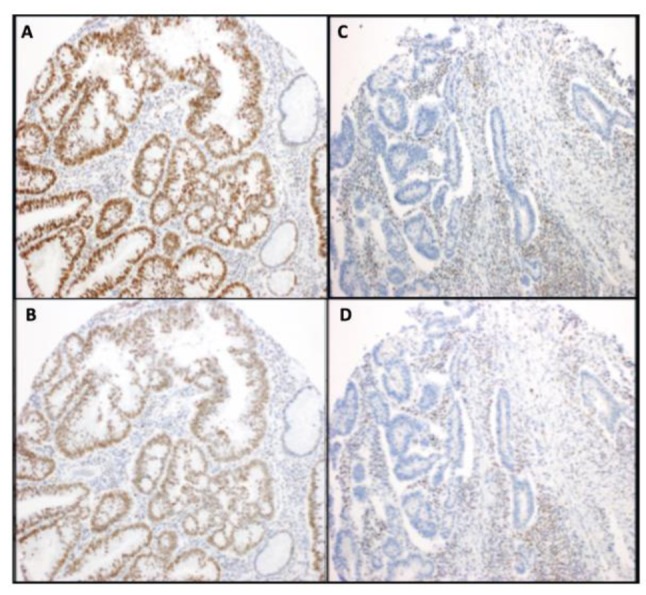
Figure 2. MSH2 and MSH6 proteins are expressed normally in microsatellite stable tumors (Image A and Image B respectively). Loss of MSH2 and MSH6 from tumor tissue in microsatellite instable colonic adenocarcinoma tumor (Image C and Image D respectively).
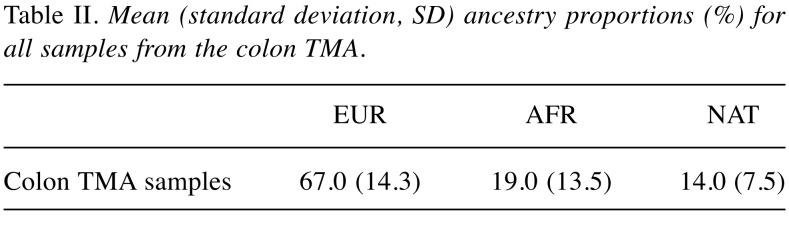
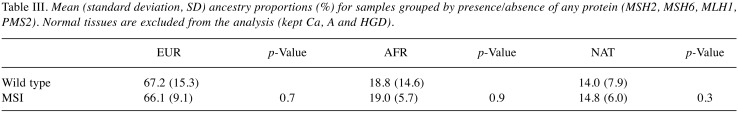
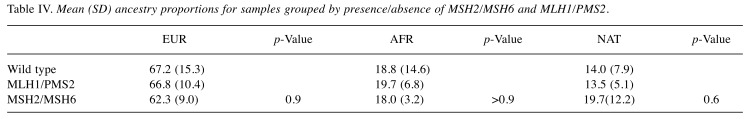
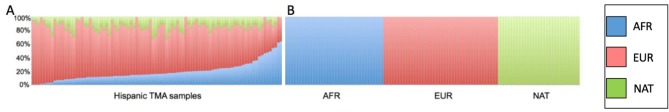
Ancestry analysis of colon TMA samples. The respective contribution of each ancestral population to the TMA samples was European 67.0% (SD 14.3%), African 19.0% (SD 13.5) and Native American 14.0% (SD 7.5%) (Table II). There were no significant differences in global ancestry proportions after grouping the TMA samples by microsatellite instability status (Table III), or according to the presence or absence of MSH1/PMS2 or MSH2/MSH6 (Table IV). [There was a marked reduction in European ancestry and a consequent increase in the Native American component when compared to wild type tumors. Although statistical significance was not reached, this observation warrants further investigation in a larger cohort (Tables III and IV)].
Table II. Mean (standard deviation, SD) ancestry proportions (%) for all samples from the colon TMA.
Table III. Mean (standard deviation, SD) ancestry proportions (%) for samples grouped by presence/absence of any protein (MSH2, MSH6, MLH1, PMS2). Normal tissues are excluded from the analysis (kept Ca, A and HGD).
Table IV. Mean (SD) ancestry proportions for samples grouped by presence/absence of MSH2/MSH6 and MLH1/PMS2.
Discussion
In this study we assessed the MMR protein expression in both primary colorectal cancer and precursor lesions using the tissue microarray technology and immunohistochemistry. We used samples from colon resections from Puerto Rican patients with no family history of colorectal cancer or inflammatory bowel disease. Family history is the main component of the Amsterdam II criteria for Identifying individuals with Hereditary Non-Polyposis Colorectal Cancer (Lynch syndrome), thus individuals with Lynch syndrome are not likely included in our study (19,20). We found that 6.3% of AD, 9.1% of HGDs and 9.4% of CAs showed loss of expression of both MLH1 and PMS2 proteins. Negative nuclear staining for both MSH6 and MSH2 proteins was found in 2.4% of CAs, while the expression of these two proteins was not lost in either ADs or HGDs. The increase of loss of expression according to different stages of disease suggest the accumulation of mutations leading to gene silencing as the disease progresses. The data also suggest a higher prevalence of MLH1 and PMS2 mutations compared to MSH2 and MSH6.
Previous studies of MMR genetics in the Hispanic population including individuals from Puerto Rico and Dominican Republic with diagnosis of HPNCC demonstrated that MSH2 was the most commonly affected gene compared to MLH1 in non-Hispanic populations. Also, higher prevalence of MSH2 mutations has been reported among other Hispanic populations such as Argentinian while higher prevalence of MLH1 mutations has been reported among Colombian, Uruguay and Brazilian populations. Therefore, our study correlated with the latter evidence. Regarding previous studies of mismatch repair protein among Caucasian populations, the study suggests a lower frequency among Puerto Rican population.
Ancestry analysis of the tissue microarray samples demonstrated that the majority of the patients had European ancestry (67%) followed by African (19%) and Native Puerto Rican (14%) (Figure 3).
Figure 3. Genetic ancestry proportions of TMA samples from colonic adenoma and adenocarcinoma depicted by percentages of African (blue), European (red) and Native American ancestry (green) (A). Reference populations from African (AFR), European (EUR) and Native American (NAT) ancestry (B).
CRC can arise from two main mechanisms, the first involving the chromosomal instability pathway, and second involving the microsatellite instability (MSI) pathway. The purpose of this paper is focused on assessing the presence of MSI pathway alterations in CRCs of Puerto Ricans. MSI occurs when a cell loses the ability to properly fix DNA mismatch errors that occur during cell replication via the DNA mismatch repair (MMR) mechanism. The MMR system is a DNA repair mechanism that prevents mutations from occurring in the cell. Without MMR, an increase in the frequency of mutations occurs and thus increases the potential for a cell to convert into a neoplastic process. The genes involved in the MMR system mainly include MLH1, PMS2, MSH2, and MSH6. MLH1 and PMS2 form dimers and loss of MLH1 on immunohistochemistry will manifest as loss of both MLH1 and PMS2 because PMS2 is not stable by itself. MSH2 and MSH6 form dimers and loss of MSH2 and MSH6 on immunohistochemistry will manifest as loss of both MSH2 and MSH6 because MSH6 is also not stable by itself (21).
CRCs with MSI as a result of sporadic or inherited MMR mutations have distinct clinicopathologic characteristics and therapeutic implications. DNA MMR deficient tumors are characteristically located in the proximal colon and usually exhibit prominent T-lymphocytes infiltration. MMR-deficient neoplasms have a correlation to a morphologic phenotype depending on the extent of neoplasm proliferation, for example, adenomas are usually sessile serrated and adenocarcinomas are usually of the mucinous phenotype (22). The tumors with MMR deficiency are said to be Microsatellite instability high (MSI-H) (22).
Management of colorectal adenocarcinomas depends on the stage of the disease. With local disease, surgery is the definitive treatment with adjuvant chemotherapy varying between stage. Adjuvant chemotherapy for resected stage II (node-negative) disease is not the standard of care for all patients, however, patients with stage III (node-positive) disease can be considered for FOLFOX (folic acid, leucovorin, Fluro-Uracil, oxaliplatin) or XELOX (oxaliplatin plus capecitabine), with the latter combination being more toxic (23). Patients with MMR-deficient colorectal carcinomas have been documented to have resistance against fluorouracil-only treatment. However, patients with MMR-deficient colorectal carcinomas have a better prognosis compared to those with a proficient MMR system unless if the MMR deficiency was caused by a BRAF V600E mutation (24,25). In the case of metastatic (stage 4) disease, immune checkpoint inhibitors are used as second-line treatment. Specifically, pembrolizumab was approved by the FDA in May of 2017 for the treatment of metastatic CRCs with MSI-high that is refractory to other conventional treatments. Pembrolizumab is a monoclonal antibody targeted against PD-1, and acts to activate the “shut down” immune response against the neoplasm. The programmed death-1 pathway is a negative feedback system that down-regulates T-helper cell activation via the programmed death-1 ligand (PDL-1) (26-29).
The ancestry analysis data presented herein describe similar ancestry proportions previously reported in genetic studies among the Puerto Rican population. MSI was considered high if any of the four proteins (MSH2, MSH6, MLH1 or PMS2) was absent in any of the cores. Since the study had a limited number of samples, the results may not be generalized to the entire Puerto Rican population. However, this is the first MMR study that reports frequency of protein expression in Puerto Ricans among the spectrum of colon cancer progression from non-neoplastic colon to pre-malignant ADs and to CRC. Furthermore, this is the first study done within a Puerto Rican region (Ponce) where the population was found to have a high percentage of Native Puerto Ricans as proven by the ancestry analysis. This study may be of interest when contemplating the application of the newer therapeutic approaches for CRC Puerto Rican patients and to decrease any cancer disparity it may exist for the diagnosis, therapy and surveillance of Puerto Ricans affected by this type of tumor.
References
- 1.Liccardo R, De Rosa M, Izzo P, Duraturo F. Novel Implications in Molecular Diagnosis of Lynch Syndrome. Gastroenterol Res Pract. 2017;2017:2595098. doi: 10.1155/2017/2595098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karahan B, Argon A, Yildirim M, Vardar E. Relationship between MLH-1, MSH-2, PMS-2, MSH-6 expression and clinicopathological features in colorectal cancer. Int J Clin Exp Pathol. 2015;8(4):4044–4053. [PMC free article] [PubMed] [Google Scholar]
- 3.De Jesús-Monge W, González-Keelan C, Zhao R, Hamilton S, Rodríguez-Bigas M, Cruz-Correa M. Mismatch repair protein expression and colorectal cancer in Hispanics from Puerto Rico. Fam Cancer. 2010;9(2):155–166. doi: 10.1007/s10689-009-9310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gausachs M, Mur P, Corral J, Pineda M, González S, Benito L, Menéndez M, Alfons J, Brunet J, Iniesta M, Gruber S, Lázaro C, Blanco I, Capellá G. MLH1 promoter hypermethylation in the analytical algorithm of Lynch syndrome: a cost-effectiveness study. Eur J Hum Genet. 2012;20(7):762–768. doi: 10.1038/ejhg.2011.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Martin A. Mismatch repair and colon cancer: mechanisms and therapies explored. Trends Mol Med. 2016;22(4):274–289. doi: 10.1016/j.molmed.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Ling G. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 7.Poulogiannis G, Frayling I, Arends M. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology. 2010;56:167–179. doi: 10.1111/j.1365-2559.2009.03392.x. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129(7-8):391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Møller P, Seppälä T, Bernstein I, Holinski-Feder E, Sala P, Gareth D, Lindblom A, Macrae F, Blanco I, Sijmons R, Jeffries J, Vasen H, Burn J, Nakken S, Hovig E, Andreas E, Tharmaratnam K, H de Vos tot Nederveen Cappel W, Hill J, Wijnen J, Green K, Lalloo F, Sunde L, Mints M, Bertario L, Pineda M, Navarro M, Morak M, Renkonen-Sinisalo L, Frayling I, Plazzer J, Pylvanainen K, Sampson J, Capella G, Mecklin J, Möslein G. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2015;66(3):464–472. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carethers J. Hereditary, sporadic and metastatic colorectal cancer are commonly driven by specific spectrums of defective DNA mismatch repair components. Trans Am Clin Climatol Assoc. 2016;127:81–97. [PMC free article] [PubMed] [Google Scholar]
- 11.Dowty J, Win A, Buchanan D, Lindor N, Macrae F, Clendenning M, Antill Y, Thibodeau S, Casey G, Gallinger S, Le Marchand L, Newcomb P, Haile R, Young G, James P, Giles G, Gunawardena S, Leggett B, Gattas M, Boussioutas A, Ahnen D, Baron J, Parry S, Goldblatt J, Young J, Hopper J, Jenkins M. Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat. 2013;34(3):490–497. doi: 10.1002/humu.22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houlleberghs H, Goverde A, Lusseveld J, Dekker M, Bruno MJ, Menko FH, Mensenkamp AR, Spaander M, Wagner A, Hofstra R, Te Riele H. Suspected Lynch syndrome associated MSH6 variants: A functional assay to determine their pathogenicity. PLoS Genet. 2017;13(5):e1006765. doi: 10.1371/journal.pgen.1006765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plaschke J, Engel C, Kruger S, Holinski-Feder E, Pagenstecher C, Mangold E, Moeslein G, Schulmann K, Gebert J, Knebel Doeberitz M von, Ruschoff J, Loeffler M, Schackert H. Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: The German hereditary nonpolyposis colorectal cancer consortium. J Clin Oncol. 2004;22:4486–4494. doi: 10.1200/JCO.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers J, Bannwart F, Yurtsever H, Neuweiler J, Riehle H, Cattaruzza M, Heinimann K, Schar P, Jiricny J, Marra G. Immunohistochemical Analysis Reveals High Frequency of PMS2 Defects in Colorectal Cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 15.Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, Al-Mawaly K, Al-Mjeni R, Al-Sayegh A, Raeburn S, Lee E, Giardiello F, Smoot D, Vilkin A, Richard Boland C, Goel A, Hafezi M, Nouraie M, Ashktorab H. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Xiao Z, Braciak T, Ou Q, Chen G, Oduncu F. Systematic immunohistochemical screening for mismatch repair and ERCC1 gene expression from colorectal cancers in China: Clinopathological characteristics and effects on survival. PLoS ONE. 2017;12(8):e0181615. doi: 10.1371/journal.pone.0181615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz-Correa M, Díaz-Algorri Y, Pérez-Mayoral J, Suleiman-Suleiman W, González-Pons M, Bertrán C, Casellas N, Rodríguez N, Pardo S, Rivera K, Mosquera R, Rodríguez-Quillchini S. Clinical characterization and mutation spectrum in Caribbean Hispanic families with Lynch syndrome. Fam Cancer. 2015;14(3):415–425. doi: 10.1007/s10689-015-9795-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Correa M, Pérez-Mayoral J, Dutil J, Echenique M, Mosquera R, Rivera-Román K, Umpierre S, Rodríguez-Quilichini S, González-Pons M, Olivera M, Pardo S. Hereditary cancer syndromes in Latino populations: genetic characterization and surveillance guidelines. Hered Cancer Clin Pract. 2017;15:3. doi: 10.1186/s13053-017-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Da Silva F, Wernhoff P, Dominguez-barrera C, Dominguez-valentin M. Update on hereditary colorectal cancer. Anticancer Res. 2016;36:4399–4406. doi: 10.21873/anticanres.10983. [DOI] [PubMed] [Google Scholar]
- 20.Sideris M, Papagrigoriadis S. Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer. Anticancer Res. 2014;34(5):2061–2068. [PubMed] [Google Scholar]
- 21.Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res. 2016;22(4):813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 22.Robbins S, Cotran R, Kumar V, Abbas A, Aster J. Pathologic basis of disease. Saunders Elsevier. 2015 [Google Scholar]
- 23.Sargent D, Shi Q, Yothers G, Van Cutsem E, Cassidy J, Saltz L, Wolmark N, Bot B, Grothey A, Buyse M, de Gramont A, Adjuvant Colon Cancer End-points (ACCENT) Group Two or three year disease free survival (DFS) as a primary endpoint in stage III adjuvant colon cancer trials with fluoropyrimidines with or without oxaliplatin or irinotecan: Data from 12,676 patients from MOSAIC, X-ACT, PETACC-3, C-06, C-07, and C89803. Eur J Cancer. 2011;47(7):990–996. doi: 10.1016/j.ejca.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, Gray R, Quirke P. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29(10):1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 25.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF-V600E in patients with colon cancer. Clin Cancer Res. 2008;14(11):3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedoeem A, Azoulay-Alfaguter I, Strazza M, Silverman GJ, Mor A. Programmed death-1 pathway in cancer and autoimmunity. Clin Immunol. 2014;153(1):145–152. doi: 10.1016/j.clim.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev. 2010;236:219–242. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubo Terufumi, Hirohashi Y, Matsuo K, Sonoda T, Sakatomo H, Furumura K, Tsukahara T, Kanaseki T, Nakatsuga M, Hirano H, Furuhata T, Takemasa I, Hasegawa T, Torigoe T. Mismatch repair protein deficiency is a risk factor for aberrant expression of HLA class I molecules: A putative "adaptive immune escape" phenomenon. Anticancer Res. 2017;37:1289–1296. doi: 10.21873/anticanres.11446. [DOI] [PubMed] [Google Scholar]
- 30.Fejerman L, John EM, Huntsman S, Beckman K, Choudhry S, Perez-Stable E, Burchard EG, Ziv E. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res. 2008;68(23):9723–9728. doi: 10.1158/0008-5472.CAN-08-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]