Abstract
Blood plasma microRNAs (miRNAs) are emerging as a clinically useful tool for non-invasive detection and prognosis estimation in various cancer types including pancreatic ductal adenocarcinoma (PDAC). The aim of the present study was to provide an independent validation of circulating miRNAs identified in previous studies as diagnostic and/or prognostic biomarkers in PDAC. Based on the literature search, 6 miRNAs were chosen as candidates for independent validation; miR-21-5p, miR-375, miR-155, miR-17-5p, miR-126-5p and miR-1290. Validation of these miRNAs was performed in a cohort of 25 patients with PDAC undergoing surgical resection and 24 healthy donors. Plasma levels of miRNAs were determined using quantitative real-time PCR. We confirmed significantly higher levels of all tested miRNA in blood plasma of PDAC patients in comparison to healthy controls with miR-21-5p showing the highest analytical performance (p<0.001; AUC>0.99). Increased levels of miR-21-5p (p=0.045) and miR-375 (p=0.013) were significantly associated with overall survival. Multivariate analysis demonstrated that miR-21-5p is a significant unfavorable prognostic factor independent on other clinical variables including adjuvant chemotherapy (hazard ratio 2.95; 95% CI 1.06-8.18; p=0.038). Our preliminary data indicate promising diagnostic and prognostic utility of plasma miR-21-5p in PDAC patients.
Keywords: Pancreatic ductal adenocarcinoma, microRNAs, plasma, miR-21-5p, diagnosis, prognosis
Pancreatic ductal adenocarcinoma cancer (PDAC) is one of the most lethal and aggressive cancers, with most patients dying within one year after diagnosis and a less than 6% five-year survival rate (1,2). Currently, radical surgical resection is the only chance for cure and the five-year survival rate for PDAC patients increases significantly with curative resection of early-stage disease (3). However, PDAC develops no symptoms, local invasiveness, and metastases to distant organs in the early stage of clinical course (4). Therefore, most patients are diagnosed with locally advanced or metastatic disease without the possibility of radical resection. In addition, even patients with operable PDAC (approx. 20%) have only a small chance to live longer than five years after surgery (3). Therefore, there is a tremendous medical need for novel diagnostic, and prognostic biomarkers that would contribute to an early diagnosis, and accurate pre-operative determination of prognosis as there are high morbidity rates associated with surgery.
In the clinical setting, only few molecules have been validated as diagnostic biomarkers for PDAC. Conventional serum tumour markers, such as carcinoembryonic antigen and mainly carbohydrate antigen 19-9 (CA19-9), are used as diagnostic assays for early detection and monitoring of the disease (5). Unfortunately, CA19-9 is not reliable enough on its own because its detection can be significantly influenced by high levels of bilirubine (6), it lacks sufficient sensitivity and specificity and often lead to incorrect diagnosis for PDAC and other non-cancer pancreatic diseases (e.g., chronic pancreatitis) (5,6).
Therefore, novel and accurate biomarkers are required to enable detection of PDAC at an early stage and pre-operative estimation of the prognosis. Accurate prognosis prediction may prevent patients from surgery-related morbidity in situation of aggressive disease where surgical intervention is not expected to significantly improve survival. Among candidate prognostic biomarkers, in last years, microRNAs (miRNAs) became an attractive option.
MiRNAs are small, non-coding, single-stranded RNAs, 20~25 nucleotides in length, that post-transcriptionally regulate gene expression (7,8). MiRNAs have potential to regulate more than half of the human genes, among them also significant number of important oncogenes and tumour suppressor genes (9,10), and their deregulation is tightly associated with various cancers, including PDAC (11,12). Circulating miRNAs in blood plasma/serum were also evaluated as potential non-invasive biomarkers in PDAC (13). Based on these previous studies, we selected 6 miRNA candidates (miR-21-5p, miR-375, miR-155, miR-17-5p, miR-126-5p and miR-1290), compared the plasma levels of each miRNA between PDAC patients and healthy controls, and evaluated prognostic potential of these miRNAs in PDAC patients undergoing surgical resection.
Materials and Methods
Study population. Blood plasma samples from PDAC patients were collected at the Masaryk Memorial Cancer Institute (MMCI; Brno, Czech Republic) and University Hospital Brno (UHB; Brno, Czech Republic). The study group included 25 patients diagnosed for PDAC who underwent surgical resection at Department of Surgery of UHB (characteristics of patients in Table I) and 24 cancer-free controls from Department of Preventive Oncology at MMCI (male: 17, female: 17, age median 64/45-79/) with no history of any type of cancer. Both cancer patients and healthy controls were of the same ethnicity (Caucasian). Clinical and pathological characteristics including sex, age, localization, TNM stage, CA19-9, and adjuvant chemotherapy information are summarized in Table I. The study has been approved by the local Ethical Committees at MMCI and UHB and all participants provided written informed consent before entering the study.
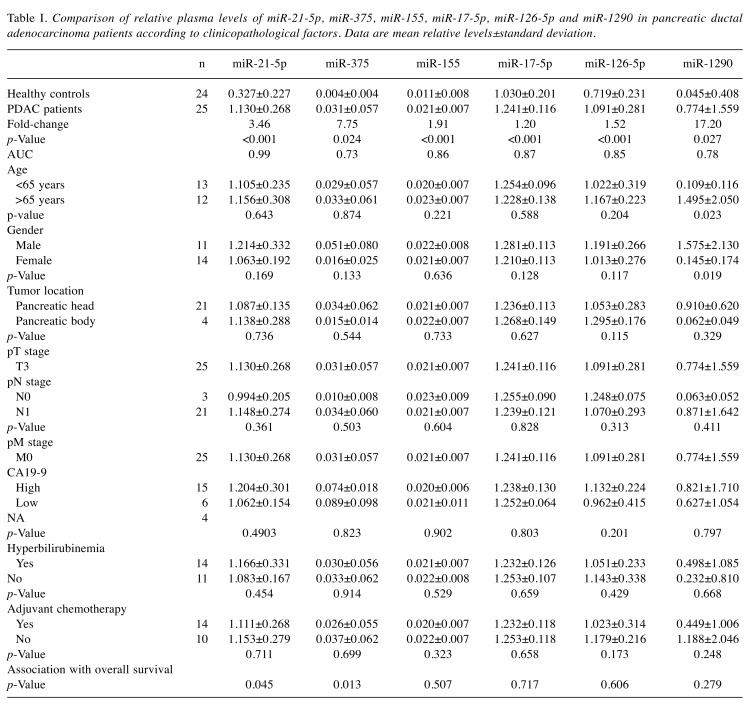
Table I. Comparison of relative plasma levels of miR-21-5p, miR-375, miR-155, miR-17-5p, miR-126-5p and miR-1290 in pancreatic ductal adenocarcinoma patients according to clinicopathological factors. Data are mean relative levels±standard deviation.
RNA isolation. Blood plasma samples were collected just before surgery and initiation of any treatment. Blood was processed for plasma within one hour after extraction. Plasma was stored at –80˚C and median of storage time to endpoint analysis was 20 months. Plasma was obtained by centrifugation at 1,200 × g for 10 min at 4˚C. To complete the removal of residual cellular components, plasma samples were re-centrifuged at 12,000 × g for a further 10 min at 4˚C. Total RNA enriched for small RNAs was isolated using Qiagen miRNeasy Serum/Plasma Kit (Qiagen, GmbH, Germany) from 250 μL of blood plasma according to manufacturers’ protocol. Concentration and purity of RNA were determined using NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The samples were either stored at –80˚C or immediately processed.
qRT-PCR quantification of miRNA expression in serum. MiRNAs were quantified by use of TaqMan MicroRNA Assays (miR-21-5p:ID000397, miR-375:ID000564, miR-155:ID002623, miR-17-5p:ID002308, miR-126-5p:ID000451 and miR-1290:ID002863). TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used for reverse transcription of 3 μl of RNA sample. Real-time PCR was performed using the QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems). The 20-μl PCR reaction mixture included 1.33 μl of RT product, 1 X TaqMan (NoUmpErase UNG) Universal PCR Master Mix (Applied Biosystems) and 1 μl of primer and probe mix of the TaqMan MicroRNA Assay kit (Applied Biosystems). Reactions were incubated in a 96-well optical plate at 95˚C for 10 min, followed by 40 cycles at 95˚C for 15 s and 60˚C for 1 min. The threshold cycle data were calculated by QuantStudio 12K Flex software (Applied Biosystems). All real-time PCR reactions were run in triplicates.
Statistical analysis. The average levels of all measured miRNAs were normalized by use of miR-106a (ID:002169; Applied Biosystems) and subsequently analyzed by the 2−ΔCt method. Statistical analyses were performed using GraphPad Prism version 6 (GraphPad software, USA). Sensitivity, specificity and area under curve (AUC) for miRNA levels were determined using Receiver Operator Characteristic (ROC) analysis. Clinical-pathological parameters and miRNA levels were evaluated using the Mann-Whitney-U-test. Kaplan-Meier analysis, long-rank test and multivariate Cox regression analysis were used for survival analysis.
Results
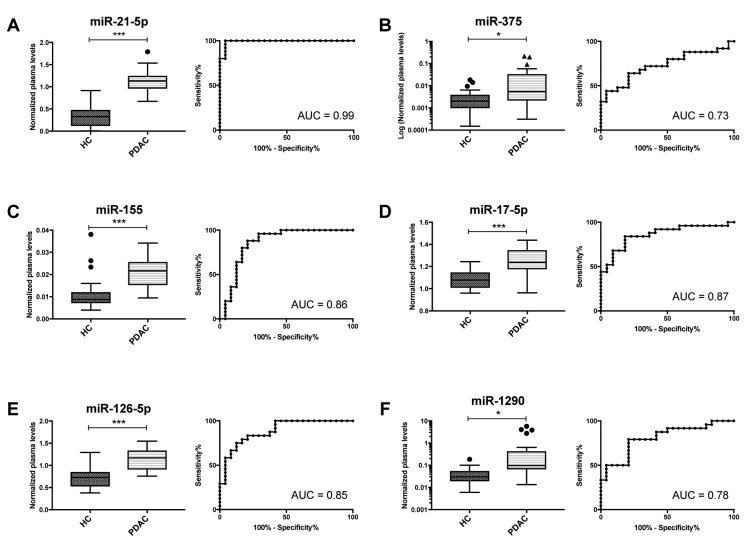
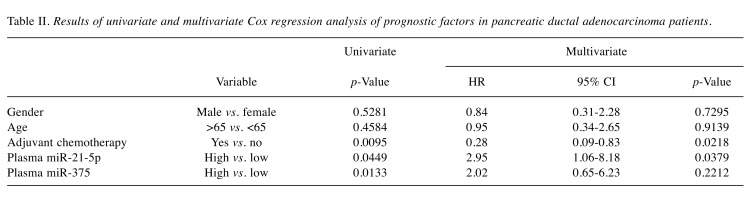
Expression levels of 6 candidate miRNAs chosen from the literature were determined by qRT-PCR in blood plasma specimens of 25 PDAC patients and 24 matched healthy controls. All miRNA tested had significantly higher levels in blood plasma of PDAC patients in comparison to healthy donors (Figure 1), whereas miR-21-5p, miR-155, miR-17-5p and miR-126-5p were the most significant (p<0.001). In addition, miR-1290 had higher levels in blood plasma of older (p=0.023) and male patients (p=0.019). Higher levels of miR-21-5p (p=0.045) and miR-375 (p=0.013) were significantly associated with worse overall survival of PDAC patients (Figure 2). We have not observed any other significant associations of miRNA levels and clinic/pathological features of PDAC patients (pTNM stage, tumour location or CA19-9 levels). Results from statistical analysis are summarized in Table I. When analysed by use of multivariate Cox regression model, the only significant prognostic factors were application of adjuvant chemotherapy (p=0.0218) and plasma levels of miR-21-5p (p=0.0379) (Table II).
Figure 1. MicroRNAs with significantly different levels in blood plasma of pancreatic ductal adenocarcinoma (PDAC) patients and healthy controls (HC). Results of Mann-Whitney U test and ROC analysis for miR-21-5p (A), miR-375 (B), miR-155 (C), miR-17-5p (D), miR-126-5p (E) and miR-1290 (F). AUC, Area under curve. *p<0.05, **p<0.01, ***p<0.001.
Figure 2. Kaplan–Meier analysis of overall survival (OS) based on the plasma levels of miR-21-5p and miR-375 in pancreatic ductal adenocarcinoma patients.

Table II. Results of univariate and multivariate Cox regression analysis of prognostic factors in pancreatic ductal adenocarcinoma patients.
Discussion
At present, the only clinically used blood-based biomarker for PDAC is CA19-9. Unfortunately, it is not reliable enough on its own since its diagnostic utility is limited by poor sensitivity, false negative results in Lewis negative phenotype and increased false positivity in case of non-pancreatic neoplastic conditions and numerous benign and malignant biliary conditions (14). Moreover, CA19-9 serum levels alone cannot differentiate between benign, precursor lesions and malignant pancreatic conditions (6,14). Therefore, first aim of our study, was to validate whether candidate plasma miRNAs selected from the previous research enable non-invasive and sensitive detection of PDAC from blood plasma specimen.
Consistently with the published data (for review (13)), significantly increased levels of miR-21-5p, miR-375, miR-155, miR-17-5p, miR-126-5p and miR-1290 have been observed in blood plasma specimens of PDAC patients when compared to healthy donors. Out of these miRNAs, miR-21-5p, miR-155, miR-17-5p and miR-126-5p indicated promising analytical performance (p<0.001, AUC>0.85). MiR-21-5p indicated the strongest performance with AUC being 0.99, which is in accordance with previous research showing miR-21-5p to be up-regulated in PDAC tumour tissue (15-17). Using in situ hybridization increased miR-21-5p expression was found in 79% of PDAC; however, only 8% of benign pancreas and 27% of chronic pancreatitis tissue expressed this miRNA suggesting its important role in the development of PDAC (18). Very recently, serum level of miR-21-5p was confirmed to be sensitive and specific diagnostic biomarker of PDAC also in meta-analysis of 9 previous studies (19). MiR-155 was previously shown to be up-regulated in PDAC tumour tissue (20). Regarding its extra-cellular form, increased levels of miR-155 were observed in blood plasma (21) and pancreatic juice (20). In addition, this miRNA controls exosome synthesis and promotes gemcitabine resistance (22) and it is biologically linked to conversion of normal fibroblasts to cancer-associated fibroblasts in PDAC (23). MiR-17-5p is up-regulated in PDAC tumour tissue (24) and has increased levels in exosomes from blood serum of PDAC patients (25). MiR-126-5p was shown to act as tumour suppressor in PDAC (26), but interestingly, only miR-126-3p (passenger strand) was detected in blood plasma and indicated higher levels in PDAC patients compared to healthy controls, which is in accordance with increased levels of miR-126-5p in PDAC patients observed in our study (27).
In PDAC, identification of reliable prognostic biomarkers still present important unmet medical need. Despite the fact, that surgical resection is the only chance for cure, it is still necessary to estimate whether individual patient will benefit from a surgery, which is associated with high rates of morbidity. Therefore, the second aim of our study was evaluation whether candidate plasma miRNAs enable identification of patients who will not profit benefit from surgical resection, e.g. patients with same or shorter overall survival as non-surgically treated patients. We proved that high levels of miR 375 and miR-21-5p are significantly associated with poor prognosis of PDAC patients. Tumour tissue level of miR-375 was demonstrated to be significant prognostic biomarkers in PDAC (28). In our study, we have shown, for the first time, that pre-operative plasma level of miR-375 is associated with overall survival of PDAC patients. However, using multivariate Cox regression analysis, miR-375 was not confirmed to be independent prognostic factor. Concerning miR-21-5p, several studies have been previously published demonstrating prognostic function of this miRNA in PDAC (15,16,29). High level of tumour tissue miR 21-5p was associated also with poor response to gemcitabine and its levels were increased after the exposure to this drug (29-31). Serum levels of miR-21-5p have been also associated to overall survival, however, PDAC patients of all clinical stages were included in the study (32). In our study, we evaluated prognostic functioning of pre-operative plasma miR-21-5p levels in uniform clearly defined cohort of patients who underwent radical surgery for PDAC. We confirmed plasma level of miR-21-5p to be significantly associated with overall survival of PDAC patients and further, by use of Cox proportional hazards regression, confirmed that miR-21-5p level could serve as an indicator for predicting the survival rate of PDAC patients independent of other clinic/pathological factors (age, gender, adjuvant chemotherapy).
This prognostic role of miR-21-5p is biologically plausible. MiR-21-5p in an oncogenic miRNA, which participates on the number of important hallmarks of cancer including sustained proliferation through PTEN (phosphatase and tensin homolog), Sprouty, PI3K (phosphoinositide 3-kinase) and PDCD4 (tumor suppressor gene tropomyosin 4); impaired apoptosis through BTG2 (B-cell translocation gene 2), FasL (pro-apoptotic FAS ligand), FBXO11 (a member of the F-box subfamily 1), and TIMP3 (inhibitor of metalloproteinases 3); and angiogenesis and invasion through PTEN, TIMP3, and TPM1 (tropomyosin 1), as well as some other pathways related to inflammatory processes and genome instability (for review 33).
In conclusion, we validated previous research describing higher levels of miR-21-5p, miR-375, miR-155, miR-17-5p, miR-126-5p and miR-1290 in blood plasma of PDAC patients in comparison to healthy controls, with miR-21-5p being the most promising candidate for further independent evaluation. Further, we showed that pre-operative plasma levels of miR-375 and miR-21-5p are associated with survival of PDAC patients undergoing radical surgery and miR-21-5p is independent of other clinic/pathological factors.
Conflicts of Interest
The Authors declare no conflict of interest.
Acknowledgements
Supported by Ministry of Health of the Czech Republic, grant No. 16-31314A. All rights reserved.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144(6):1252–1261. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffin JF, Poruk KE, Wolfgang CL. Pancreatic cancer surgery: past, present, and future. Chin J Cancer Res. 2015;27(4):332–348. doi: 10.3978/j.issn.1000-9604.2015.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma J, Jemal A. The rise and fall of cancer mortality in the USA: why does pancreatic cancer not follow the trend. Future Oncol. 2013;9(7):917–919. doi: 10.2217/fon.13.76. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int J Clin Exp Med. 2015;8(7):11683–11691. [PMC free article] [PubMed] [Google Scholar]
- 6.Scara S, Bottoni P, Scatena R. CA 19-9: Biochemical and Clinical Aspects. Adv Exp Med Biol. 2015;867:247–260. doi: 10.1007/978-94-017-7215-0_15. [DOI] [PubMed] [Google Scholar]
- 7.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 8.Winter J, Diederichs S. MicroRNA biogenesis and cancer. Methods Mol Biol. 2011;676:3–22. doi: 10.1007/978-1-60761-863-8_1. [DOI] [PubMed] [Google Scholar]
- 9.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62(1):33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 10.Ercan G, Karlitepe A, Ozpolat B. Pancreatic Cancer Stem Cells and Therapeutic Approaches. Anticancer Res. 2017;37(6):2761–2775. doi: 10.21873/anticanres.11628. [DOI] [PubMed] [Google Scholar]
- 11.Diab M, Muqbil I, Mohammad RM, Azmi AS, Philip PA. The role of microRNAs in the diagnosis and treatment of pancreatic adenocarcinoma. J Clin Med. 2016;5(6) doi: 10.3390/jcm5060059. doi:10.3390/jcm5060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Liu J, Chen-Xiao K, Zhang X, Lee WN, Go VL, Xiao GG. Advance in microRNA as a potential biomarker for early detection of pancreatic cancer. Biomark Res. 2016;4:20. doi: 10.1186/s40364-016-0074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebrahimi S, Hosseini M, Ghasemi F, Shahidsales S, Maftouh M, Akbarzade H, Parizadeh SA, Hassanian SM, Avan A. Circulating microRNAs as potential diagnostic, prognostic and therapeutic targets in pancreatic cancer. Curr Pharm Des. 2016;22(42):6444–6450. doi: 10.2174/1381612822666160817095047. [DOI] [PubMed] [Google Scholar]
- 14.Ballehaninna UK, Chamberlain RS. Serum CA 19-9 as a Biomarker for Pancreatic Cancer-A Comprehensive Review. Indian J Surg Oncol. 2011;2(2):88–100. doi: 10.1007/s13193-011-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vychytilova-Faltejskova P, Kiss I, Klusova S, Hlavsa J, Prochazka V, Kala Z, Mazanec J, Hausnerova J, Kren L, Hermanova M, Lenz J, Karasek P, Vyzula R, Slaby O. MiR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:38. doi: 10.1186/s13000-015-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavano F, di Mola FF, Piepoli A, Panza A, Copetti M, Burbaci FP, Latiano T, Pellegrini F, Maiello E, Andriulli A, di Sebastiano P. Changes in miR-143 and miR-21 expression and clinicopathological correlations in pancreatic cancers. Pancreas. 2012;41(8):1280–1284. doi: 10.1097/MPA.0b013e31824c11f4. [DOI] [PubMed] [Google Scholar]
- 17.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, Danesi R, Campani D, Verheul HM, Boggi U. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70(11):4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 18.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12(12):2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S, Zhou L, Wei J, Chang H, Li K, Wang Z, Liu C, Wu Z. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7(1):1692. doi: 10.1038/s41598-017-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8(4):340–346. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131(3):683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 22.Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K, Gotoh K, Takeda Y, Tanemura M, Mori M, Doki Y. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:42339. doi: 10.1038/srep42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang W, Su J, Wang Y, Feng H, Dai X, Yuan Y, Chen X, Yao W. Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Sci. 2015;106(10):1362–1369. doi: 10.1111/cas.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. doi: 10.4161/cbt.10.8.13083. [DOI] [PubMed] [Google Scholar]
- 25.Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. doi: 10.1186/1477-7819-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10(1):3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 27.Cao Z, Liu C, Xu J, You L, Wang C, Lou W, Sun B, Miao Y, Liu X, Wang X, Zhang T, Zhao Y. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016;7(27):41575–41583. doi: 10.18632/oncotarget.9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KH, Lee JK, Choi DW, Do IG, Sohn I, Jang KT, Jung SH, Heo JS, Choi SH, Lee KT. Postoperative prognosis prediction of pancreatic cancer with seven microRNAs. Pancreas. 2015;44(5):764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 29.Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, Kim SW, Del Chiaro M, Peters GJ, Giaccone G. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5(5):e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morinaga S, Nakamura Y, Atsumi Y, Murakawa M, Yamaoku K, Aoyama T, Kobayashi S, Ueno M, Morimoto M, Yokose T, Miyagi Y. Locked nucleic acid in situ hybridization analysis of microRNA-21 predicts clinical outcome in patients after resection for pancreatic cancer treated with adjuvant gemcitabine monotherapy. Anticancer Res. 2016;36(3):1083–1088. [PubMed] [Google Scholar]
- 31.Paik WH, Kim HR, Park JK, Song BJ, Lee SH, Hwang JH. Chemosensitivity induced by down-regulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. 2013;33(4):1473–1481. [PubMed] [Google Scholar]
- 32.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y, Zhang CY. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 33.Shi J. Considering exosomal miR-21 as a biomarker for cancer. J Clin Med. 2016;5(4):42. doi: 10.3390/jcm5040042. [DOI] [PMC free article] [PubMed] [Google Scholar]