Abstract
Background: Peritoneal dissemination (PD) is one of the most common causes of cancer-related mortality in gastric cancer (GC). We aimed to identify PD-associated genes and investigate their role in GC. Materials and Methods: We identified FGFR1 as a putative PD-associated gene using a bioinformatics approach. The biological significance of FGFR1 in epithelial-to-mesenchymal transition (EMT) was evaluated according to the correlation with genes that participated in EMT and FGFR1 knockdown experiments. The associations between FGFR1 expression and the clinicopathological features were examined. Results: FGFR1 expression positively correlated with SNAI1, VIM and ZEB1 expression, and negatively correlated with CDH1 expression. Knockdown of FGFR1 suppressed the malignant phenotype of GC cells. High FGFR1 expression significantly correlated with the peritoneal lavage cytology and synchronous PD positivity as well as poor prognosis. Conclusion: High FGFR1 expression was associated with PD via promotion of EMT and led to a poor prognosis of GC patients.
Keywords: Gastric cancer, FGFR1, peritoneal dissemination, epithelial to mesenchymal transition
Gastric cancer (GC) is the third leading cause of cancer-related mortality in both sexes worldwide, and the prognosis of patients with GC is dismal (1,2). In most patients with advanced GC, GC-related mortality is caused by distant metastasis, and PD is the most frequent metastasis from GC (3). Once GC metastasizes to the peritoneal cavity, there is no therapeutic strategy to cure GC radically. Understanding the mechanisms of PD and the consequent development of sensitive biomarkers and therapeutic targets is necessary to conquer PD and improve the prognoses of patients.
Fibroblast growth factor receptor 1 (FGFR1) is a member of the FGFR family, and it is a receptor tyrosine kinase that activates mitogen activated protein kinase signaling and phosphoinositide-3-kinase/AKT signaling (4,5). Aberrant expression and somatic mutation of FGFR1 have been reported in several cancers, and FGFR inhibitors have been focused on as anticancer agents (6-8). In GC tissue, FGFR1 has been reported to be overexpressed and to be associated with poor prognosis (9,10). However, the molecular mechanisms of FGFR1 contributing to metastasis and poor prognosis in GC are not fully known. Establishment of a metastatic focus requires a multistep process, and epithelial to mesenchymal transition (EMT) plays a pivotal role in several steps, including vessel invasion, detachment from primary lesion and anoikis resistance (11). To the best of our knowledge, there are currently no studies on the association between FGFR1 and EMT in GC. Therefore, the aim of the present study was to assess the clinical significance of FGFR1 expression in GC focusing on EMT and PD.
Materials and Methods
Identification of putative PD-associated genes. To identify genes that mediate PD, we performed extraction-of-expression modules (EEM) analysis as previously described (12,13). EEM indicated that the gene set that was up-regulated in the 58As9 cell line, that was established from the HSC-58 cell line and is considered to be a highly peritoneal-metastatic cell line compared to the HSC-58 cell line, was most significantly associated with poor overall survival (OS), and FGFR1 was included in the upregulated gene set.
Cell lines and knockdown (KD) of FGFR1. The human scirrhous GC cell lines HSC-58 and 58As9Luc and the culture conditions have been previously described (13-16). The pcDNA6.2-GW/EmGFP-miR plasmid from the Block-iT Pol II miR RNAi Expression Vector Kit (Invitrogen, Carlsbad, CA, USA) was obtained for KD of FGFR1. Three double-stranded shRNAs specific for FGFR1 (sh#1, #2 and #3) were designed using an online software (http://www.invitrogen.com/rnai) (Table I). shRNAs specific for FGFR1 and negative control shRNA (shNC) were transfected into 58As9Luc cells using Lipofectamine 2000 (Invitrogen). Stable transfectants were selected by 10 μg/mL blasticidin and fluorescence activated cell sorting for GFP.
Table I. Sequences of shRNAs and primers.
Clinical GC samples. Primary GC samples of the Singapore dataset were obtained from 198 patients who underwent gastric resection at the Singapore Health Services and deposited in National University Hospital System tissue repositories. Primary GC samples of the Beppu dataset were obtained from 197 patients who underwent gastric resection at the Oita Prefectural Hospital and the Kyushu University Beppu Hospital. All patients provided written informed consent, and the study protocol was approved by the appropriate ethics committee. Experiments with these samples were performed in accordance with the approved guidelines.
RNA extraction, gene expression array and quantitative reverse transcription polymerase chain reaction (qRT-PCR). Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Chatsworth, CA, USA). The gene expression array of the Singapore dataset has been previously described (13). Using the Beppu dataset and GC cell lines, qRT-PCR was performed as previously described, and glyceraldehyde-3-phosphate dehydrogenase mRNA expression was quantified for standardization (13). The specific primers are listed in Table I.
Protein extraction and western blotting. Cells were lysed in lysis buffer, and protein expression was evaluated by WB as previously described (17). A primary rabbit monoclonal antibody against FGFR1 (#9740; Cell Signaling Technology, Beverly, MA, USA) was used at a dilution of 1:1,000 at 4˚C overnight. A primary mouse monoclonal antibody against β-actin (sc-47778; Santa Cruz Biotechnology, Dallas, TX, USA) was used at a dilution of 1:1,000 at room temperature for 1 h.
Cell proliferation, migration and invasion assay. The cell proliferation capacity was evaluated by the MTT assay using the Cell Proliferation Kit 1 (Roche Applied Science, Penzberg, Germany) according to the manufacturer’s protocol in biological sextuplicates. The cell migration capacity and invasion capacity were assessed using Corning BioCoat Control Inserts (#354578; BD Biosciences, Bedford, MA, USA) and Matrigel Invasion Chambers (#354480; BD Biosciences) according to manufacturers’ protocol in biological quadruplicates. After an appropriate incubation time (migration, 24 h; invasion, 96 h), we used a light microscope to count the cells present on the surface of the membrane.
Gene Set Enrichment Analysis (GSEA). The associations between FGFR1 expression and previously defined gene sets were analyzed by GSEA using gene expression profiles from The Cancer Genome Atlas (TCGA) dataset as previously described (18).
Statistical analysis. For continuous variables, statistical analyses were performed using Student’s t-tests. Categorical variables were compared using Pearson’s correlation coefficients and χ2 tests or Fisher’s exact tests. Survival time was evaluated using the Kaplan-Meier method, and survival curves were compared using log-rank tests. Multivariate analysis for survival time was estimated by the Cox proportional hazard model. All statistical analyses were performed using R version 3.4.1 (Vienna, Austria. URL: http://www.R-project.org/).
Results
Identification of FGFR1 as a putative PD-associated gene. EEM analysis detected module genes for PD in a gene set obtained from comparison between HSC-58 and 58As9 cells. In this study, we focused on FGFR1, which was overexpressed in 58As9 cells by 4.03 log2-fold compared to HSC-58 cells, and its high expression was significantly associated with poor OS in the Singapore dataset.
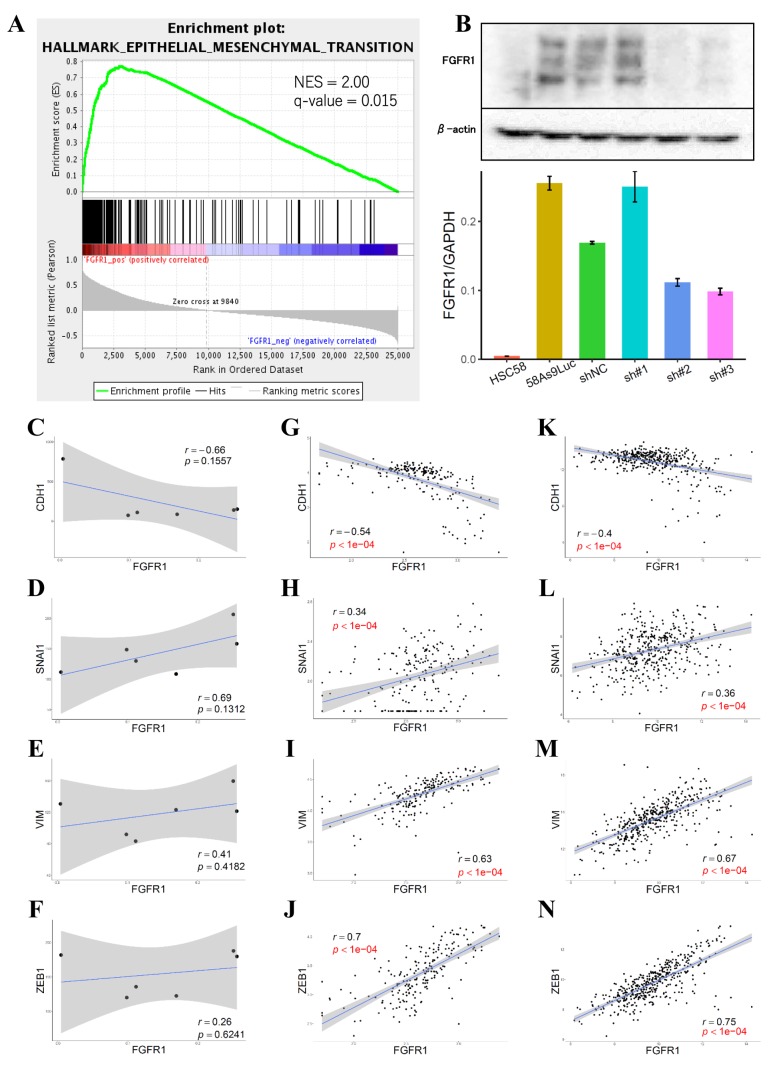
High FGFR1 expression is positively correlated with EMT markers. GSEA revealed that high FGFR1 expression was positively correlated with the gene set associated with EMT (Figure 1A). We established stable FGFR1 KD cell lines, and inhibition of FGFR1 expression was confirmed by qRT-PCR and WB (Figure 1B). sh#2 and sh#3 worked well, and sh#1 did not have a KD effect. FGFR1 in the GC cell lines negatively correlated with cadherin 1 (CDH1) and positively correlated with snail family transcriptional repressor 1 (SNAI1), vimentin (VIM) and zinc finger E-box binding homeobox 1 (ZEB1) (Figure 1C-F). A significant negative correlation with CDH1 and significant positive correlations with SNAI1, VIM and ZEB1 were observed in both the Singapore dataset (Figure 1G-J) and TCGA dataset (Figure 1K-N) with statistical significance. In subsequent experiments, sh#1-transfected cells were excluded due to an insufficient KD effect.
Figure 1. FGFR1 expression is associated with EMT-associated genes. A: GSEA revealed that high FGFR1 expression positively correlated with a gene set associated with EMT. B: KD of FGFR1 was confirmed by qRT-PCR and western blotting. C-F: Correlation analysis between FGFR1 and EMT-associated genes in the GC cell line. G-J: Correlation analysis between FGFR1 and EMT-associated genes in the Singapore dataset. K-N: Correlation analysis between FGFR1 and EMT-associated genes in the TCGA dataset.
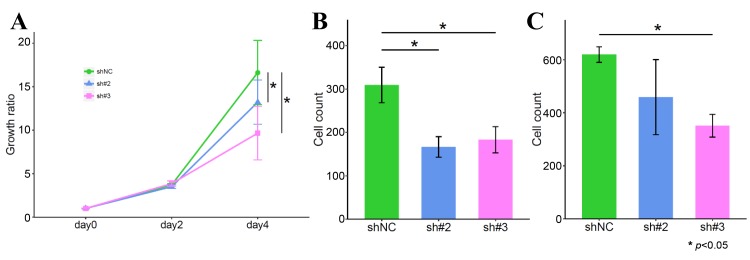
KD of FGFR1 suppresses the malignant phenotype of GC cells. To determine the influence of FGFR1 on the cell phenotype, we evaluated the proliferation, migration and invasion capacities using FGFR1 KD cells. KD of FGFR1 significantly decreased the cell proliferation capacity (Figure 2A). The number of GC cells migrating across the micropore membrane decreased in FGFR1 KD cells (Figure 2B). Moreover, there was significant change in the invasion capacity in sh#3-transfected cells (Figure 2C).
Figure 2. Effect of KD of FGFR1 on the cell phenotype. A; MTT assay. B; Migration assay. C; Invasion assay.
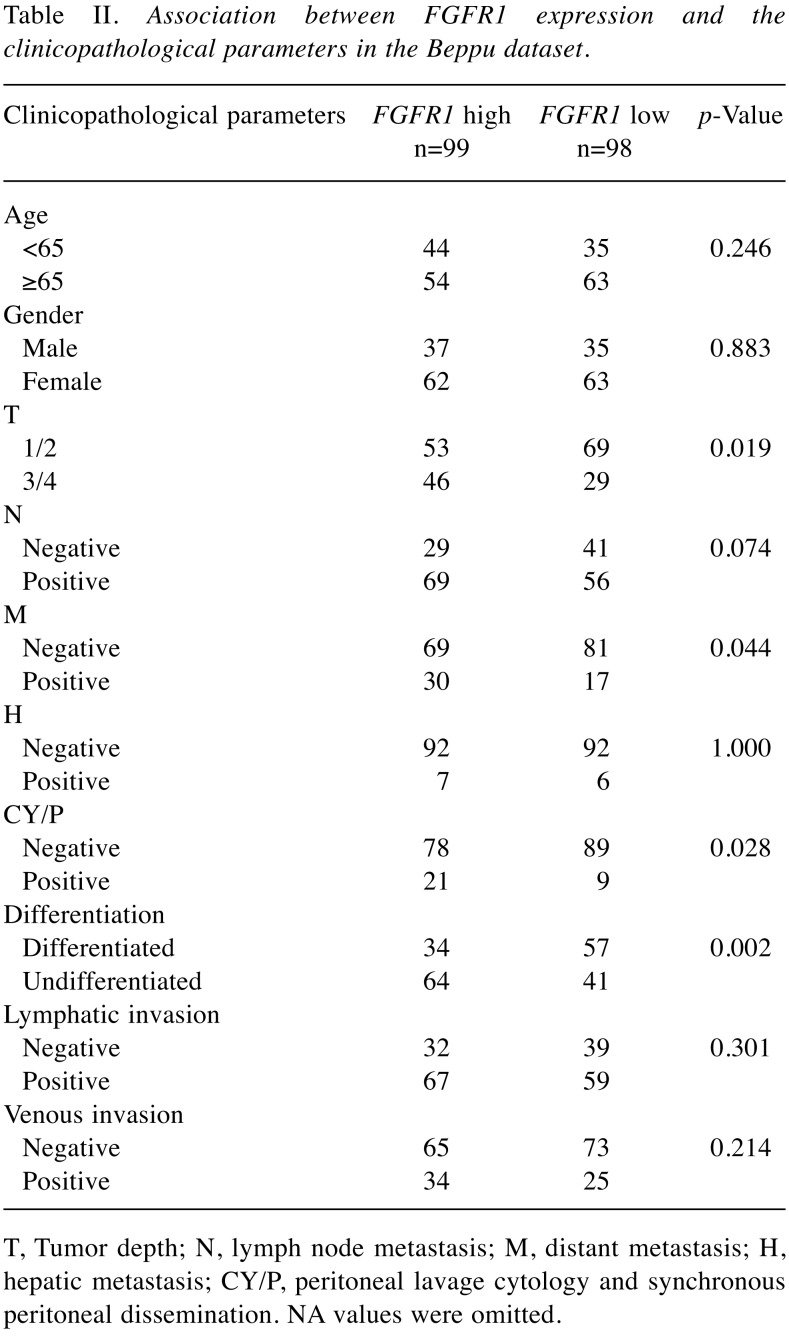
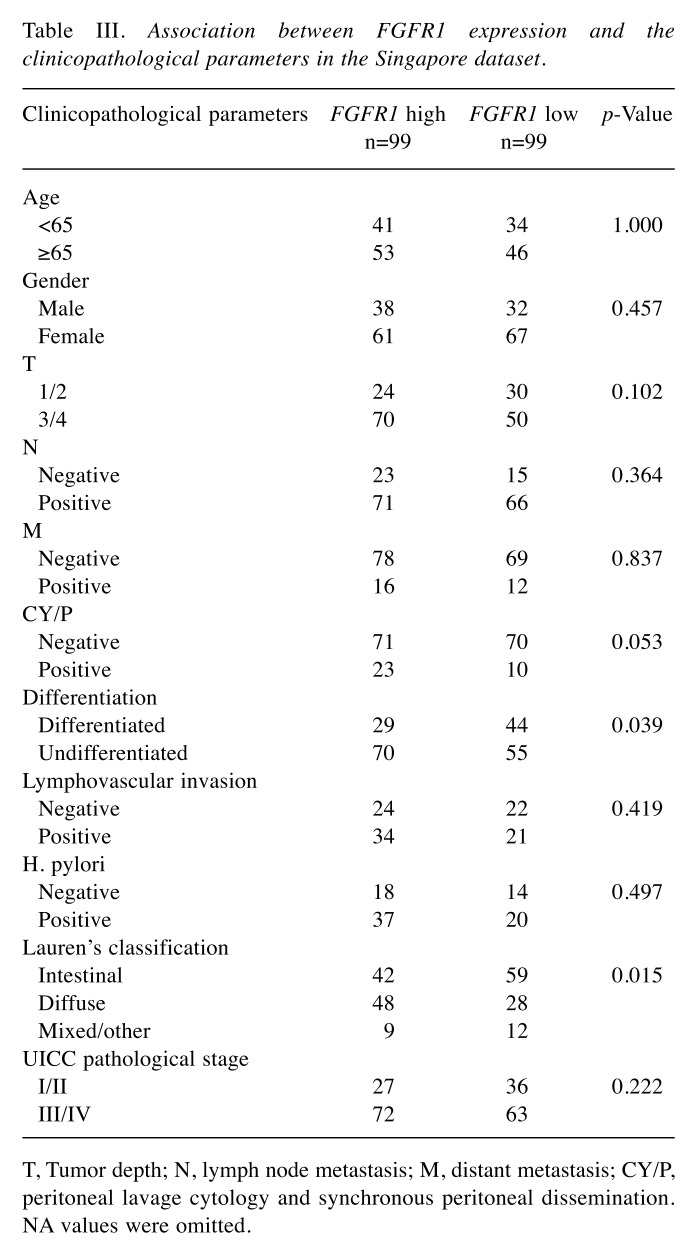
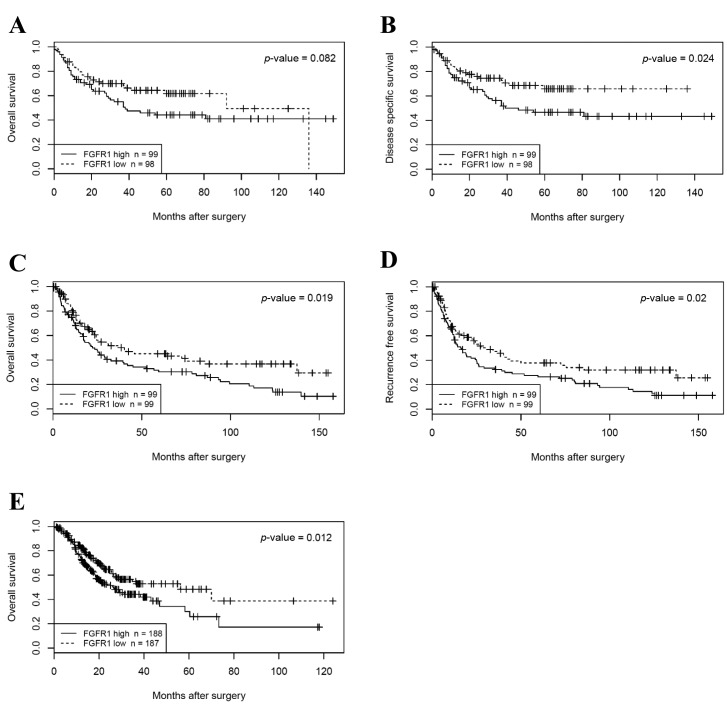
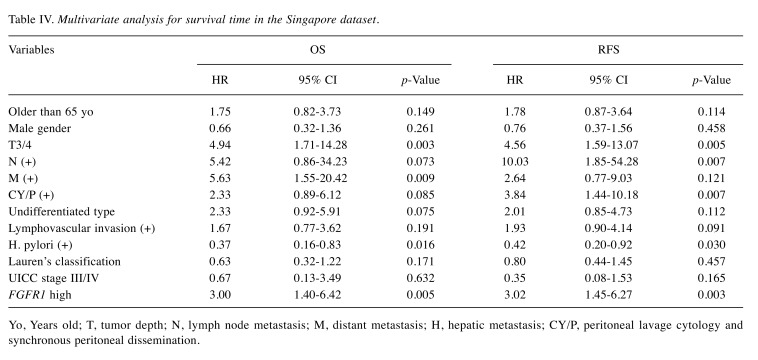
Clinical significance of FGFR1 expression. Patients were divided into two groups according to the median value of FGFR1 expression in the Beppu and Singapore datasets. High FGFR1 expression significantly correlated with tumor depth, distant metastasis, undifferentiated type and the peritoneal lavage cytology and synchronous PD (CY/P) positivity in the Beppu dataset (Table II), and high FGFR1 expression significantly correlated with undifferentiated type and Lauren’s classification in the Singapore dataset (Table III). Patients with high FGFR1 expression had the following results for each dataset: significantly short disease-specific survival time in the Beppu dataset, significantly short OS and recurrence free survival (RFS) in the Singapore dataset, and short OS in the TCGA dataset (Figure 3). High FGFR1 expression was an independent prognostic factor for OS and RFS in the Singapore dataset (Table IV).
Table II. Association between FGFR1 expression and the clinicopathological parameters in the Beppu dataset.
T, Tumor depth; N, lymph node metastasis; M, distant metastasis; H, hepatic metastasis; CY/P, peritoneal lavage cytology and synchronous peritoneal dissemination. NA values were omitted.
Table III. Association between FGFR1 expression and the clinicopathological parameters in the Singapore dataset.
T, Tumor depth; N, lymph node metastasis; M, distant metastasis; CY/P, peritoneal lavage cytology and synchronous peritoneal dissemination. NA values were omitted.
Figure 3. Survival analysis according to FGFR1 expression. A, B: OS and DSS in the Beppu dataset. C-D: OS and RFS in the Singapore dataset. E: OS in the TCGA dataset.
Table IV. Multivariate analysis for survival time in the Singapore dataset.
Yo, Years old; T, tumor depth; N, lymph node metastasis; M, distant metastasis; H, hepatic metastasis; CY/P, peritoneal lavage cytology and synchronous peritoneal dissemination.
Discussion
PD from GC greatly affects the quality of life of patients and is the main cause of cancer-related mortality. Understanding the mechanism and management of PD should contribute to the improvement of patient prognosis. We aimed to identify the PD-associated genes and investigate how those genes are associated with GC progression. Some studies have reported that high expression of FGFR1 is associated with poor prognosis in GC patients, and the utility of FGFR1 inhibitors has been reported in GC in vitro and in vivo (9,10,19-22). However, the underlying mechanism of the exacerbation of GC patient prognosis via high FGFR1 expression is not fully understood. We performed GSEA to explore the FGFR1 contribution to poor prognosis in GC and revealed that high FGFR1 expression was positively correlated with a gene set associated with EMT. The association between FGFR1 and EMT has been reported in prostate cancer, bladder cancer, head and neck squamous cell carcinomas and chordoma, but not in digestive cancers, including GC (23-27). We demonstrated that FGFR1 negatively correlated with CDH1 and positively correlated with SNAI1, VIM and ZEB1 in GC cells and clinical GC samples. KD of FGFR1 significantly suppressed the malignant phenotype of GC cells. These results supported the relevance of FGFR1 to EMT. In the Beppu dataset, high FGFR1 expression significantly correlated with CY/P positivity, although there was no correlation with hepatic metastasis. In the Singapore dataset, high FGFR1 expression correlated with CY/P positivity. Therefore, these data suggest that FGFR1 specifically contributes to the establishment of PD via promoting EMT. Moreover, high FGFR1 expression correlated with the undifferentiated type in both datasets. The comprehensive analyses of large cohorts indicated that FGFR1 specifically associates with PD because the undifferentiated type has been reported to be a risk factor for PD in GC (28). Furthermore, patients with high FGFR1 expression had poor prognosis, and high FGFR1 expression was an independent prognostic factor for OS and RFS in the Singapore dataset. Thus, FGFR1 might be a putative prognostic biomarker for GC patients.
One limitation of our study was that the molecular mechanism through which FGFR1 promoted EMT remains unknown. Receptor tyrosine kinase has been reported to activate the PI3K/AKT signaling pathway, which is essential for EMT (5,29,30). Additionally, it has been reported that AKT phosphorylates and suppresses GSK3β, which leads to the degradation of β-catenin via promoting ubiquitination (31-34). Another limitation was the lack of an in vivo study. Further investigation into the effect of FGFR1 on downstream pathways and in vivo using an appropriate orthotopic xenograft model would lead to a better understanding of the association between FGFR1 and PD. In conclusion, our study demonstrated that FGFR1 associates with PD via promoting EMT in GC and that FGFR1 might be a putative prognostic biomarker.
Conflicts of Interest
The Authors declare no competing financial interests
Acknowledgements
The GC cell lines, HSC-58 and 58As9, were gratuitously given to us by Kazuyoshi Yanagihara, Division of Translational Research, Exploratory Oncology and Clinical Trial Center, National Cancer Center Hospital East, Japan. This work was supported, in part, by the following grants and foundations: Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (Grant No.: 15H05912, 15H05707, hp160219, hp170227, and hp170227).
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss L. Metastasis of cancer: a conceptual history from antiquity to the 1990s. Cancer Metastasis Rev. 2000;19:I–xi, 193-383. [PubMed] [Google Scholar]
- 4.Zwick E, Bange J, Ullrich A. Receptor tyrosine kinase signalling as a target for cancer intervention strategies. Endocr Relat Cancer. 2001;8:161–173. doi: 10.1677/erc.0.0080161. [DOI] [PubMed] [Google Scholar]
- 5.Wheler JJ, Atkins JT, Janku F, Moulder SL, Stephens PJ, Yelensky R, Valero V, Miller V, Kurzrock R, Meric-Bernstam F. Presence of both alterations in FGFR/FGF and PI3K/AKT/mTOR confer improved outcomes for patients with metastatic breast cancer treated with PI3K/AKT/mTOR inhibitors. Oncoscience. 2016;3:164–172. doi: 10.18632/oncoscience.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelleher FC, O’Sullivan H, Smyth E, McDermott R, Viterbo A. Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis. 2013;34:2198–2205. doi: 10.1093/carcin/bgt254. [DOI] [PubMed] [Google Scholar]
- 7.Jiang XF, Dai Y, Peng X, Shen YY, Su Y, Wei MM, Liu WR, Ding ZB, Zhang A, Shi YH, Ai J. SOMCL-085, a novel multi-targeted FGFR inhibitor, displays potent anticancer activity in FGFR-addicted human cancer models. Acta Pharmacol Sin. 2018;39:243–250. doi: 10.1038/aps.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 9.Murase H, Inokuchi M, Takagi Y, Kato K, Kojima K, Sugihara K. Prognostic significance of the co-overexpression of fibroblast growth factor receptors 1, 2 and 4 in gastric cancer. Mol Clin Oncol. 2014;2:509–517. doi: 10.3892/mco.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inokuchi M, Murase H, Otsuki S, Kawano T, Kojima K. Different clinical significance of FGFR1-4 expression between diffuse-type and intestinal-type gastric cancer. World J Surg Oncol. 2017;15:2. doi: 10.1186/s12957-016-1081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanda M, Kodera Y. Molecular mechanisms of peritoneal dissemination in gastric cancer. World J Gastroenterol. 2016;22:6829–6840. doi: 10.3748/wjg.v22.i30.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niida A, Smith AD, Imoto S, Aburatani H, Zhang MQ, Akiyama T. Gene set-based module discovery in the breast cancer transcriptome. BMC Bioinformatics. 2009;10:71. doi: 10.1186/1471-2105-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurashige J, Hasegawa T, Niida A, Sugimachi K, Deng N, Mima K, Uchi R, Sawada G, Takahashi Y, Eguchi H, Inomata M, Kitano S, Fukagawa T, Sasako M, Sasaki H, Sasaki S, Mori M, Yanagihara K, Baba H, Miyano S, Tan P, Mimori K. Integrated Molecular Profiling of Human Gastric Cancer Identifies DDR2 as a Potential Regulator of Peritoneal Dissemination. Sci Rep. 2016;6:22371. doi: 10.1038/srep22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanagihara K, Takigahira M, Tanaka H, Komatsu T, Fukumoto H, Koizumi F, Nishio K, Ochiya T, Ino Y, Hirohashi S. Development and biological analysis of peritoneal metastasis mouse models for human scirrhous stomach cancer. Cancer Sci. 2005;96:323–332. doi: 10.1111/j.1349-7006.2005.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagihara K, Takigahira M, Takeshita F, Komatsu T, Nishio K, Hasegawa F, Ochiya T. A photon counting technique for quantitatively evaluating progression of peritoneal tumor dissemination. Cancer Res. 2006;66:7532–7539. doi: 10.1158/0008-5472.CAN-05-3259. [DOI] [PubMed] [Google Scholar]
- 16.Yanagihara K, Tanaka H, Takigahira M, Ino Y, Yamaguchi Y, Toge T, Sugano K, Hirohashi S. Establishment of two cell lines from human gastric scirrhous carcinoma that possess the potential to metastasize spontaneously in nude mice. Cancer Sci. 2004;95:575–582. doi: 10.1111/j.1349-7006.2004.tb02489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nambara S, Masuda T, Nishio M, Kuramitsu S, Tobo T, Ogawa Y, Hu Q, Iguchi T, Kuroda Y, Ito S, Eguchi H, Sugimachi K, Saeki H, Oki E, Maehara Y, Suzuki A, Mimori K. Antitumor effects of the antiparasitic agent ivermectin via inhibition of Yes-associated protein 1 expression in gastric cancer. Oncotarget. 2017;8:107666–107677. doi: 10.18632/oncotarget.22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Q, Masuda T, Sato K, Tobo T, Nambara S, Kidogami S, Hayashi N, Kuroda Y, Ito S, Eguchi H, Saeki H, Oki E, Maehara Y, Mimori K. Identification of ARL4C as a Peritoneal Dissemination-Associated Gene and Its Clinical Significance in Gastric Cancer. Ann Surg Oncol. 2018;25:745–753. doi: 10.1245/s10434-017-6292-6. [DOI] [PubMed] [Google Scholar]
- 19.Schafer MH, Lingohr P, Strasser A, Lehnen NC, Braun M, Perner S, Holler T, Kristiansen G, Kalff JC, Gutgemann I. Fibroblast growth factor receptor 1 gene amplification in gastric adenocarcinoma. Hum Pathol. 2015;46:1488–1495. doi: 10.1016/j.humpath.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt K, Moser C, Hellerbrand C, Zieker D, Wagner C, Redekopf J, Schlitt HJ, Geissler EK, Lang SA. Targeting Fibroblast Growth Factor Receptor (FGFR) with BGJ398 in a Gastric Cancer Model. Anticancer Res. 2015;35:6655–6665. [PubMed] [Google Scholar]
- 21.Ying S, Du X, Fu W, Yun D, Chen L, Cai Y, Xu Q, Wu J, Li W, Liang G. Synthesis, biological evaluation, QSAR and molecular dynamics simulation studies of potential fibroblast growth factor receptor 1 inhibitors for the treatment of gastric cancer. Eur J Med Chem. 2017;127:885–899. doi: 10.1016/j.ejmech.2016.10.066. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Du X, Li W, Zhou Y, Bai E, Kang Y, Chen Q, Fu W, Yun D, Xu Q, Qiu P, Jin R, Cai Y, Liang G. A novel non-ATP competitive FGFR1 inhibitor with therapeutic potential on gastric cancer through inhibition of cell proliferation, survival and migration. Apoptosis. 2017;22:852–864. doi: 10.1007/s10495-017-1361-7. [DOI] [PubMed] [Google Scholar]
- 23.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–571. doi: 10.1016/j.ccr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Tomlinson DC, Baxter EW, Loadman PM, Hull MA, Knowles MA. FGFR1-induced epithelial to mesenchymal transition through MAPK/PLCgamma/COX-2-mediated mechanisms. PLoS One. 2012;7:e38972. doi: 10.1371/journal.pone.0038972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen PT, Tsunematsu T, Yanagisawa S, Kudo Y, Miyauchi M, Kamata N, Takata T. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br J Cancer. 2013;109:2248–2258. doi: 10.1038/bjc.2013.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Mintz A, Shah SR, Quinones-Hinojosa A, Hsu W. The FGFR/MEK/ERK/brachyury pathway is critical for chordoma cell growth and survival. Carcinogenesis. 2014;35:1491–1499. doi: 10.1093/carcin/bgu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao J, Zhao X, Liang Y, Tang D, Pan C. FGF1-FGFR1 axis promotes tongue squamous cell carcinoma (TSCC) metastasis through epithelial-mesenchymal transition (EMT) Biochem Biophys Res Commun. 2015;466:327–332. doi: 10.1016/j.bbrc.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge of gastric cancer. Histol Histopathol. 2018;33:11–26. doi: 10.14670/HH-11-898. [DOI] [PubMed] [Google Scholar]
- 29.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, Lee EY, Weiss HL, O’Connor KL, Gao T, Evers BM. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y, Yang Z, Xu A, Dong P, Huang Y, Liu H, Li F, Wang H, Xu Q, Wang Y, Sun D, Zou Y, Zou X, Wang Y, Zhang D, Liu H, Wu X, Zhang M, Fu Y, Cai Z, Liu C, Wu S. PIK3R1 negatively regulates the epithelial-mesenchymal transition and stem-like phenotype of renal cancer cells through the AKT/GSK3beta/ CTNNB1 signaling pathway. Sci Rep. 2015;5:8997. doi: 10.1038/srep08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh M, Kim H, Yang I, Park JH, Cong WT, Baek MC, Bareiss S, Ki H, Lu Q, No J, Kwon I, Choi JK, Kim K. GSK-3 phosphorylates delta-catenin and negatively regulates its stability via ubiquitination/proteosome-mediated proteolysis. J Biol Chem. 2009;284:28579–28589. doi: 10.1074/jbc.M109.002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C, Kim NG, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8:4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]