Abstract
Background/Aim: Glioblastoma multiforme (GBM) is the most common primary tumor of the central nervous system. The serum and glucocorticoid-regulated kinase SGK1 gene is required for the growth and survival of GBM stem-like cells under both normoxic and hypoxic conditions. It has been reported that oxygenation significantly affects cellular genetic expression; 30% of the genes required in hypoxia were not required under normoxic conditions. Therefore, we examined SGK1 expression to determine if it may be a novel potential drug target for GBM. Materials and Methods: We assessed the association between SGK1 and glioblastoma patient overall survival using the GBM cohort in TCGA (The Cancer Genome Atlas) database (TCGA-GBM). To access and analyze the data we used the UCSC Xena browser (https://xenabrowser.net). Survival data of the GBM subgroup were extracted for analysis and generation of Kaplan–Meier curves for overall survival. The best cut-off was identified by methods described in the R2 web-based application (http://r2.amc.nl). Results: We analyzed patient survival by tumor SGK1 copy number segments after removal of common germ-line copy-number variants (CNVs). Copy number segments (log2 tumor/normal) ≤0.009700 were associated with significantly poorer survival (p=0.016). Conclusion: Increased median overall survival associated with increased SGK1 copy number segments may be a reflection of better tumor oxygenation. Therefore, besides being a drug target, SGK1 may also be a prognostic marker. Among molecular tumor markers, only the methylation status of the O-6-methylguanine-DNA methyltransferase (MGMT) gene has shown a significant association with survival in patients with GBM.
Keywords: Glioblastoma, survival, cancer genome atlas
Glioblastoma multiforme (GBM) is the most common primary tumor of the central nervous system. A subpopulation of cells known as GBM stem-like cells (GBM-SC) initiate and nurture tumor growth and are molecularly similar to the tumor from which they arise. GBM-SCs are known to thrive in hypoxic regions and may contribute to therapeutic resistance (1-3).
Hypoxia is a hallmark of glioblastoma, with larger hypoxic, necrotic regions a poor prognostic factor. Hypoxia is an impediment to the treatment of many malignant tumors with radiation and chemotherapy. But attempts to increase oxygenation with hyperbaric oxygen have not been encouraging (4).
More recently, efforts have been made to understand the molecular mechanisms that govern glioblastoma and cancer stem cells, in order to develop novel targeted therapeutics for GBM and other brain cancers. Cancer stem cells are not independent entities; rather, they function within an ecological system, actively changing the microenvironment and receiving critical maintenance cues from surrounding tissue.
Kulkarni et al. have reported that the serum and glucocorticoid-regulated kinase (SGK1) gene is required for the growth and survival of GBM stem-like cells under both normoxic and hypoxic conditions (5). In this analysis, we examined the role of the SGK1 gene in glioblastoma survival to determine if it might be a potential drug target. At present, only temozolomide has been conclusively shown to prolong survival of GBM patients (6).
Materials and Methods
We assessed the association between SGK1 and glioblastoma overall survival using the GBM cohort in TCGA (The Cancer Genome Atlas) database (TCGA-GBM). To access and analyze the data we used the UCSC Xena browser (https://xenabrowser.net). Survival data of the GBM subgroup were extracted for analysis and generation of Kaplan–Meier curves for overall survival. The best cut-off was identified by methods described in the R2 web-based application (http://r2.amc.nl).
Results
Data from 665 patients were analyzed. The mean age at diagnosis was 57.8±14.5 (mean±SD). The longest tumor dimension was 1.1±0.43 cm; shortest dimension 0.45±0.24 cm. Karnofsky performance score was 77.2±15.8. The patients were 38% women and 61% men; in 1% of cases gender was not specified. 64% of patients received chemotherapy; 71% received radiation therapy. 11% received hormonal treatment (glucocorticoids).
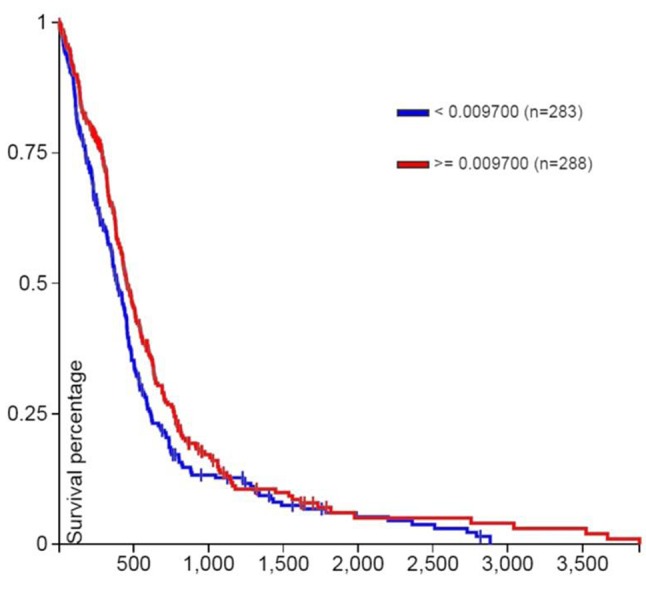
Patient overall survival (days) by tumor SGK1 copy number segments (log2 tumor/normal), after removal of common germ-line copy-number variants (CNVs), is shown in (Figure 1). Note that copy number segments ≤0.009700 are associated with significantly poorer survival (p=0.016, log rank test). 283 patients with copy number segments ≤0.009700 had median survival 382 days; 288 patients with copy number segments ≥0.009700 had median survival 455 days.
Figure 1. Patient survival (days) by SGK1 copy number segments (log2 tumor/normal) after removal of common germ-line copy-number variants (CNVs). Removing the common germline copy-number variants can make it easier to focus on the copy variation that is just in the tumor. Note that copy number segments ≤0.009700 are associated with significantly poorer survival (p=0.016, log rank test).
TCGA studies have identified four subtypes of GBM. These subtypes are defined by genomic characteristics, survival length, patient age and treatment response. The four groups were named Proneural, Neural, Classical and Mesenchymal (7).
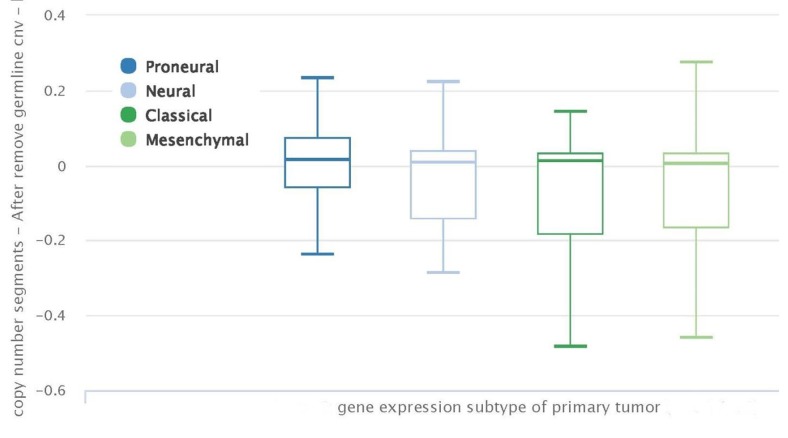
Figure 2 shows Factor SGK1 copy number segments (log2 tumor/normal) in the four subtypes after removal of common germ-line copy-number variants (CNVs). There is no significant variation in SGK1 copy number segments among the 4 subtypes.
Figure 2. Serum and glucocorticoid-regulated kinase (SGK1) gene copy number segments (log2 tumor/normal) in the four primary tumor subtypes after removal of common germ-line copy-number variants (CNVs). There is no significant difference in SKG1 copy number segments among the 4 subtypes.
Figure 3 shows patient overall survival (days) of the four primary tumor subtypes. The proneural subtype has the best survival.
Figure 3. Patient survival (days) by primary tumor subtype.
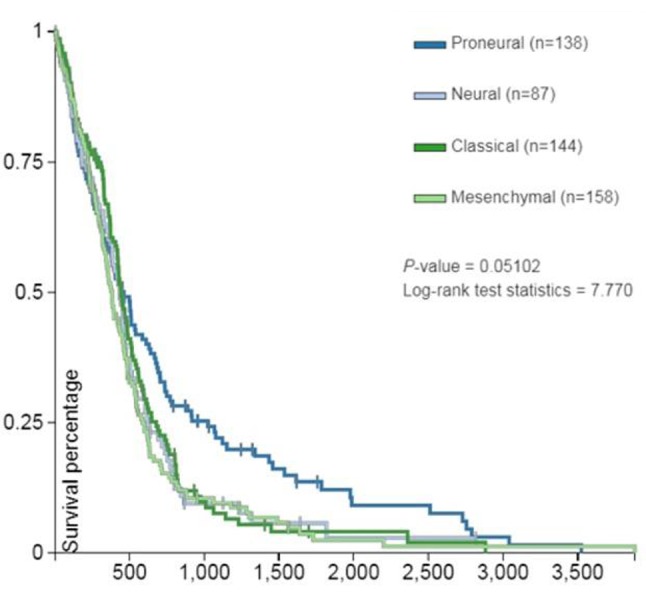
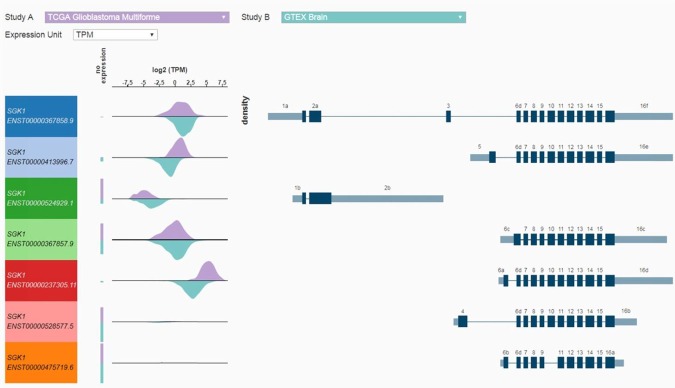
Figure 4 shows SGK1 transcript-specific expression (8), log2 transcripts per million (TPM) in glioblastoma TCGA data and normal brain tissue from the Genotype-Tissue Expression (GTEx) project. Expression of transcripts is different in tumor as compared to normal brain tissue. Xena uses transcript models directly from Ensembl/Gencode (hence the Ensembl IDs in the left most column).
Figure 4. SGK1 Transcript-specific expression (7 transcripts) for ‘tumor’ TCGA data (lavender) and ‘normal’ (blue-green) GTEX data (TPM transcripts per million). All RNAseq data were generated by the Toil pipeline, recompute done by the UCSC Computational Core using the RSEM package. All seven transcripts are from Gencode V23 comprehensive annotation. The exons are numbered using a UCSC Computational Core method which may not line up with exon numbering in the literature. Regions that are intronic in all transcripts are removed. The remaining exonic regions are numbered 1-16. For example, exon 4 is the unique exon in the 4th exonic region. Different exons within a given region are labeled starting with ‘a’ for the left-most exon (in transcript direction). Exons 6a, 6b, 6c, and 6d are four different exons in the sixth exonic region. In other words, different exons across all transcripts which overlap transitively, that is, partially across the adjoining exon, are assigned the same integer. So if one transcript has exons 6a and 6c, there must be exons in other transcripts that overlap them, and each other. Note that it is fairly common for a gene to have transcripts with non-overlapping exons, depending on the gene, its function, the location of the protein coding domains, and where that particular transcript was observed.
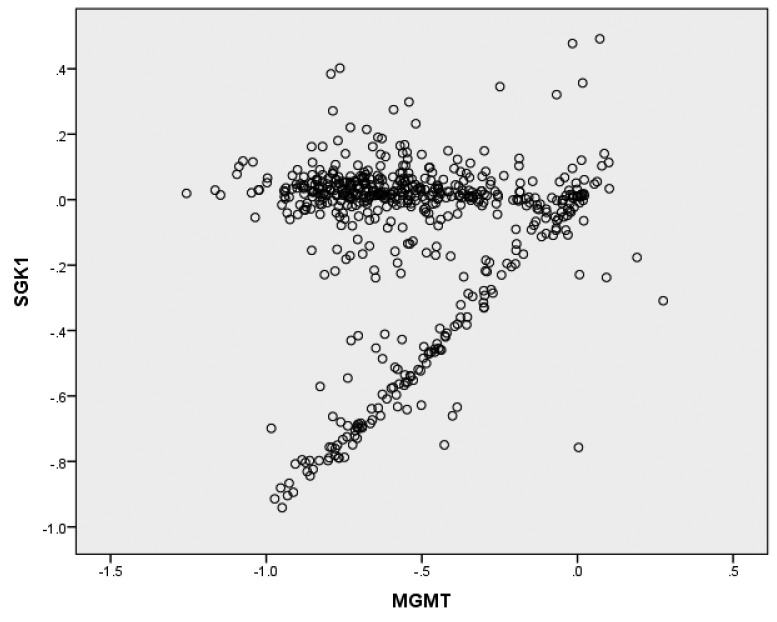
O-6-methylguanine-DNA methyltransferase (MGMT) copy number segments (log2 tumor/normal) are plotted against SGK1 copy number segments (log2 tumor/normal) in Figure 5. SGK1 copy number segments correlate significantly overall with MGMT copy number segments (r=0.139, p=0.001) (Figure 5). The data appear to define two distinct groups: one in which MGMT values close to zero do not relate to SGK1; and a second group in which SGK1 and MGMT are linearly correlated.
Figure 5. O-6-methylguanine-DNA methyltransferase (MGMT) copy number segments (log2 tumor/normal) plotted against SGK1 copy number segments (log2 tumor/normal). SGK1 copy number segments correlate significantly overall with MGMT copy number segments (r=0.139, p=0.001). The data appear to define two distinct groups: one in which MGMT values close to zero do not relate to SGK1; and a second group in which SGK1 and MGMT are linearly correlated.
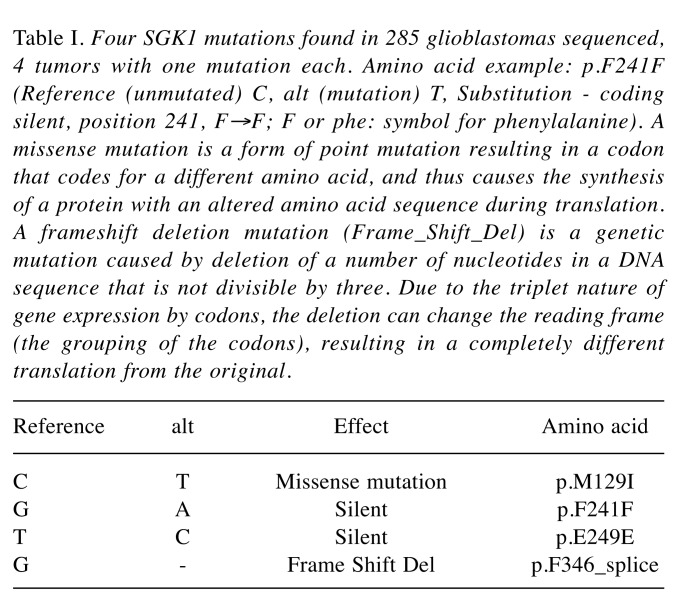
Mutations of SGK1 are rare in GBM. Table I lists the 4 mutations found in 285 tumors sequenced, 4 tumors with one mutation each.
Table I. Four SGK1 mutations found in 285 glioblastomas sequenced, 4 tumors with one mutation each. Amino acid example: p.F241F (Reference (unmutated) C, alt (mutation) T, Substitution - coding silent, position 241, F→F; F or phe: symbol for phenylalanine). A missense mutation is a form of point mutation resulting in a codon that codes for a different amino acid, and thus causes the synthesis of a protein with an altered amino acid sequence during translation. A frameshift deletion mutation (Frame_Shift_Del) is a genetic mutation caused by deletion of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the deletion can change the reading frame (the grouping of the codons), resulting in a completely different translation from the original.
According to the newly revised WHO classification of brain tumors, GBM has been divided into two representative forms, that is, ‘GBM with wildtype IDH’, which used to be called ‘primary GBM’, and ‘GBM with mutant IDH’, which used to be called ‘secondary GBM’ (9)). Whether IDH is mutated or not in GBM is very important since it is an established, powerful, and reliable prognostic biomarker. Therefore, we evaluated survival versus SGK1 copy number segments in the 13 patients
Discussion
Kulkarni et al. have verified the importance of SGK1 for cellular survival; their report is the only one to date that analyzes the relationship of SGK1 and glioblastoma. They studied multiple patient-derived GBM stem-like cell lines. They found that SGK1 depletion and inhibition has little effect on serum grown glioma lines and on differentiated GBM stem-like cells, indicating its specific importance in GBM stem-like cell survival only on glioblastoma cells interacting with multiple other genes. Kulkarni et al. validated SGK1 as critical for GBM stem-like cell proliferation and survival, using shRNA, CRISPR and an SGK1-specific inhibitor (5).
SGK1 belongs to a subfamily of serine/threonine kinases that is under acute transcriptional control by several stimuli, including serum and glucocorticoids. Kinases regulate many important cellular processes and are important drug targets. More than 25 kinase inhibitors are already approved as oncology drugs. SGK1 is responsible for cell survival and resistance to therapy in multiple cancer types (10).
The increased median overall survival associated with increased SGK1 copy number segments (2.4 months) (Figure 1) is comparable to the increased median overall survival after treatment with temozolomide (2.6 months) (6). Although the SGK1 sample size and temozolomide study sample size were about the same and median survival increments appeared similar, the statistical significance of the difference for the increase seen with temozolomide was far greater. This implies that the survival increase seen with the SGK1 difference was more variable than that seen with temozolomide.
We hypothesize that increased median overall survival associated with increased SGK1 copy number segments is a reflection of better tumor oxygenation. Indeed, increased SGK1 expression is a favorable prognostic marker in cervical cancer (https://www.proteinatlas.org/ENSG00000118515-SGK1/pathology), where tumor oxygenation predicts radiation response and survival (11).
The histology of GBM is characterized by pseudo-palisading necrosis, which may be the result of ‘scrambling for oxygen’ by rapidly growing tumor cells (12). If it is true that increased SGK1 copy number segments may be a reflection of better tumor oxygenation, increased SGK1 could lead to the longer survival of the tumors by way of better tumor oxygenation; as a result, the patients may have poorer prognosis. But this is not the case here: Increased SGK1 is associated with better survival of patients. This ambiguity might reflect the fact that radiation and chemotherapy are more effective in better oxygenated tumors (13) and outweigh the effect of pseudo-palisading necrosis.
Kulkarni et al. report that oxygenation significantly affects cellular genetic expression; 30% of the genes required in hypoxia were not required under normoxic conditions. Therefore, besides being a drug target, SGK1 might also be a molecular marker for survival. Among molecular markers, only the methylation status of the O-6-methylguanine-DNA methyltransferase (MGMT) gene has shown a significant association with survival in patients with GBM (14). Multiple copy number segments and transcript-specific expression of SGK1 may therefore represent a second molecular marker. Further studies would be worthwhile.
Conflicts of Interest
The Authors declare that they have no conflicts of interest.
References
- 1.Tasaki T, Fujita M, Okuda T, Yoneshige A, Nakata S, Yamashita K, Yoshioka H, Izumoto S, Kato A. MET expressed in glioma stem cells is a potent therapeutic target for glioblastoma multiforme. Anticancer Res. 2016;36:3571–3577. [PubMed] [Google Scholar]
- 2.Ceresa C, Nicolini G, Semperboni S, Requardt H, Le DG, Santini C, Pellei M, Bentivegna A, Dalpra L, Cavaletti G, Bravin A. Synchrotron-based photon activation therapy effect on cisplatin pre-treated human glioma stem cells. Anticancer Res. 2014;34:5351–5355. [PubMed] [Google Scholar]
- 3.Teicher BA, Dupuis NP, Emi Y, Ikebe M, Kakeji Y, Menon K. Increased efficacy of chemo- and radio-therapy by a hemoglobin solution in the 9L gliosarcoma. In Vivo. 1995;9:11–18. [PubMed] [Google Scholar]
- 4.Tibbles PM, Edelsberg JS. Hyperbaric-oxygen therapy. N Engl J Med. 1996;334:1642–1648. doi: 10.1056/NEJM199606203342506. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni S, Goel-Bhattacharya S, Sengupta S, Cochran BH. A Large-Scale RNAi Screen Identifies SGK1 as a Key Survival Kinase for GBM Stem Cells. Mol Cancer Res. 2018;16:103. doi: 10.1158/1541-7786.MCR-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 7.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moll AG, Lindenmeyer MT, Kretzler M, Nelson PJ, Zimmer R, Cohen CD. Transcript-specific expression profiles derived from sequence-based analysis of standard microarrays. PLoS One. 2009;4:e4702. doi: 10.1371/journal.pone.0004702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232:165–177. doi: 10.1002/path.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talarico C, Dattilo V, D’Antona L, Menniti M, Bianco C, Ortuso F, Alcaro S, Schenone S, Perrotti N, Amato R. SGK1, the new player in the game of resistance: Chemo-radio molecular target and strategy for inhibition. Cell Physiol Biochem. 2016;39:1863–1876. doi: 10.1159/000447885. [DOI] [PubMed] [Google Scholar]
- 11.Fyles AW, Milosevic M, Wong R, Kavanagh MC, Pintilie M, Sun A, Chapman W, Levin W, Manchul L, Keane TJ, Hill RP. Oxygenation predicts radiation response and survival in patients with cervix cancer. Radiother Oncol. 1998;48:149–156. doi: 10.1016/s0167-8140(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 12.Rong Y, Durden DL, Van Meir EG, Brat DJ. Pseudopalisading necrosis in glioblastoma: A familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J Neuropathol Exp Neurol. 2006;65:529–539. doi: 10.1097/00005072-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Harrison L, Blackwell K. Hypoxia and anemia: Factors in decreased sensitivity to radiation therapy and chemotherapy. Oncologist. 2004;9:31–40. doi: 10.1634/theoncologist.9-90005-31. [DOI] [PubMed] [Google Scholar]
- 14.Kreth S, Thon N, Eigenbrod S, Lutz J, Ledderose C, Egensperger R, Tonn JC, Kretzschmar HA, Hinske LC, Kreth FW. O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One. 2011;6:e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]