Abstract
Over the past decade, major discoveries in retrotransposon biology have depicted the neural genome as a dynamic structure during life. In particular, the retrotransposon LINE-1 (L1) has been shown to be transcribed and mobilized in the brain. Retrotransposition in the developing brain, as well as during adult neurogenesis, provides a milieu in which neural diversity can arise. Dysregulation of retrotransposon activity may also contribute to neurological disease. Here, we review recent reports of retrotransposon activity in the brain, and discuss the temporal nature of retrotransposition and its regulation in neural cells in response to stimuli. We also put forward hypotheses regarding the significance of retrotransposons for brain development and neurological function, and consider the potential implications of this phenomenon for neuropsychiatric and neurodegenerative conditions.
Keywords: retrotransposon, LINE-1, neuron, neurogenesis, mosaicism
1. Introduction
The mammalian brain is remarkably complex in form and function. Neural cell diversity underpins this complexity, and has classically been defined in terms of the morphological differences between cell types, their diverse connectivity patterns, physiological and functional properties and the expression of various transcription factors (TFs), cell surface and secreted molecules [1,2]. At the same time, it has been assumed that the neural cells of any individual will all carry the same genetic instructions, or genotype, and that cellular diversity is achieved by changing how these instructions are read, for example, through epigenetic modifications [3]. However, recent advances in DNA sequencing and genetic analysis, as well as bioinformatics, have made it possible to identify mutations generating distinct genotypes among the neurons of an individual human brain. These mutations include single-nucleotide variants, copy number variants (CNVs) and retrotransposon insertions [4–10]. Collectively, these genetic variants form a landscape of somatic mosaicism within the brain.
Somatic mosaicism is defined as the existence of two or more cells with different genotypes within one individual. Depending on its developmental timing, mosaicism can encompass mutations that are heritable (germline mosaicism), non-heritable (somatic mosaicism) or a combination of these two outcomes [9,11–14]. Retrotransposons, colloquially referred to as ‘jumping genes’, are DNA fragments that have the ability to copy their sequences from one location in the genome and insert themselves into a new genomic position, causing mutations and changing the genomic landscape at the new integration site. It is now established that retrotransposons are endogenous mediators of somatic mosaicism in the brain [4–7,10,14]. As remnants of mammalian genome evolution, retrotransposons may occupy up to two-thirds of the human genome [15], but most human retrotransposon copies have lost their ability to mobilize further [16]. However, some elements are still capable of mobilization, in the germline and in the soma. Over the past decade, several studies have shown that retrotransposons contribute to intra-individual variation in neuronal genomes, defining a new layer of neural diversity and heterogeneity. Maintenance of this complex heterogeneity may be essential for healthy brain function, while its disruption may play a role in neurological diseases [17,18]. To provide a comprehensive understanding of the roles retrotransposons play in neural function, and their potential contribution to disease, we will first present an overview of the active retrotransposon families in humans, their mobilization mechanism, and experimental strategies for studying neuronal retrotransposition.
2. Types of retrotransposons in the human genome
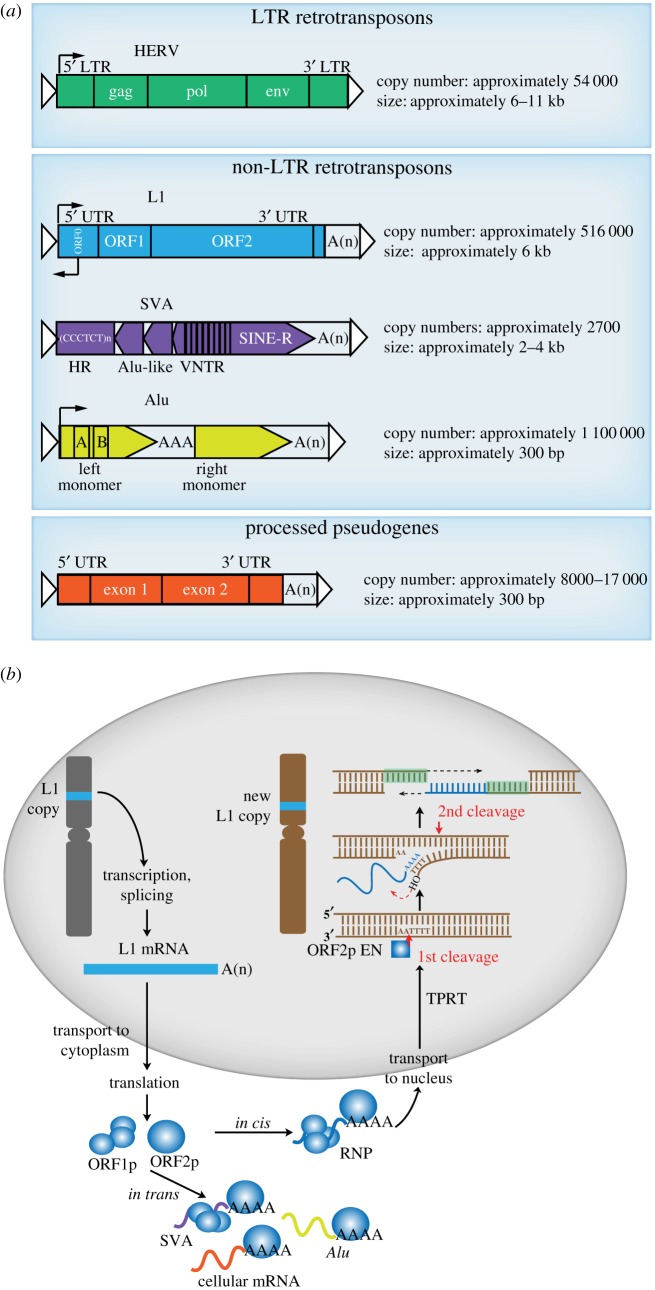
Retrotransposons are the main class of transposable elements found in most mammalian genomes. Based on the presence of long terminal repeats (LTR) in their structure, they are classified into LTR and non-LTR retrotransposons. The LTR group consists of ERVs (endogenous retroviruses), which comprise approximately 8% of the human genome and represent remnant retrovirus sequences incorporated into the host germline after ancient viral infections [19,20]. The non-LTR retrotransposon group is further subdivided based on the ability of elements to mobilize independently or only with the machinery encoded by another retrotransposon, into autonomous and non-autonomous elements, respectively. Long interspersed element 1 (LINE-1 or L1) is the only autonomous non-LTR retrotransposon in humans. In addition to its own mobilization in cis, L1 can mobilize in trans non-autonomous short interspersed element (SINE) retrotransposons. These include 7SL-derived Alu elements, composite SVA (SINE–variable number tandem repeat–Alu) elements, and cellular mRNAs, resulting in processed pseudogenes [21–25]. L1 copies can be over 6 kb in length and occupy the highest proportion of the human genome by sequence of all the transposable elements (approx. 17%) [19]. Alu copies are approximately 300 bp in size and represent the most abundant transposable element by copy number (approx. 11% by sequence) [19,26,27]. SVAs are approximately 2 kb in size, with far fewer copies than L1 or Alu, and comprise only about 0.2% of the genome sequence [19,28–30] (figure 1a).
Figure 1.
Human retrotransposon families and mobilization mechanism. (a) Types of retrotransposons, their mobilization capacity, size and number of copies in the human genome. HERV, human endogenous retroviruses consisting of two LTRs; gag, group-specific antigen; pol, polymerase; env, envelope gene; long interspersed element-1 (L1) structure: 5′ untranslated region (UTR) with open reading frame (ORF)0 and promoter activity; ORF1 and ORF2; 3′ UTR and poly(A) tail; SVA structure: CCCTCT hexamer repeats (HR), Alu sequence in reverse orientation (Alu-like); VNTR, variable number of tandem repeats; SINE-R, a short interspersed element of HERV origin; An, poly(A) tail; Alu structure: a left monomer with internal RNA polymerase III promoter binding sites (A, B boxes); AAA, adenosine-rich linker; right monomer ending in poly(A) tail (An); processed pseudogene: sequence derived from cellular messenger RNA, which has been reverse transcribed into DNA and has no introns; target side duplications are shown as white triangles. (b) L1 retrotransposition mechanism as an example of retrotransposon mobilization. A full-length, retrotransposition-competent L1 is present at one genomic locus (blue box in the left grey chromosome). The L1 encodes two proteins essential for its mobility, ORF1p (with nucleic acid chaperone activity [31,32]) and ORF2p (with endonuclease [33] and reverse transcriptase [34] activity), as well as an unusual antisense ORF0 that may facilitate retrotransposition [35]. L1 transcription results in a bicistronic, polyadenylated mRNA (blue rectangle), which is transported to the cytoplasm for translation. Upon translation, ORF1p and ORF2p (blue spheres) bind to their encoding L1 mRNA in cis and form an RNP complex. Note that the L1 proteins can retrotranspose cellular mRNAs to generate processed pseudogenes, as well as SVA and Alu retrotransposons (purple, red and yellow wavy lines). Once the RNP is formed, it enters the nucleus through a still poorly understood process, where a new L1 insertion occurs by target-site primed reverse transcription (TPRT) [36,37]. During TPRT, the ORF2p endonuclease makes a first and second cleavage (red arrows) in the genomic DNA at the consensus sequence 5'-TTTT/AA [33], and releases a 3' hydroxyl (OH) group from which the ORF2p reverse transcriptase initiates reverse transcription of the attached L1 mRNA (indicated by the red dashed line arrow). The DNA fragment between the two cleavages is highlighted in green to indicate the formation of TSDs. The black dashed arrow indicates completing synthesis across the second strand of cDNA, resulting in a new L1 copy, and the TSDs, which flank the new L1 copy.
Owing to truncation, deletion, internal rearrangement and other mutations, the vast majority of human retrotransposon sequences are incapable of further mobilization and are considered molecular fossils [16]. Computational-, molecular- and genomics-based studies have to date identified and catalogued retrotransposons into evolutionary families and subfamilies. Only a few L1, Alu and SVA subfamilies remain mobile. Moreover, functional studies have identified that most retrotransposition-competent L1s are part of a small group of elements termed Ta (transcribed-active) L1s [38]. All of the L1s reported to be highly active in vitro to date belong to the L1-Ta and L1-preTa families [39–42]. Similarly, particular Alu subfamilies, such as AluYa5 and AluYb8 [43], and SVA E, F and F1 subfamilies [19,24,29] have been identified as the most active in humans. Human ERVs (HERVs) are considered to be immobile in modern humans, although a recent study revealed several polymorphic HERV-K insertions in the global population and a full-length, intact, insertion with potentially functional open reading frames (ORFs) [44]. However, it remains unclear whether this insertion is a recently integrated provirus or has simply been lost in some individuals.
3. An overview of the retrotransposon mobilization mechanism
All retrotransposons mobilize via a replication mechanism involving an RNA intermediate [45]. During this process, termed retrotransposition, the host polymerase starts the synthesis of an RNA copy of the element and, subsequently, the RNA is reverse transcribed into DNA by an element-encoded reverse transcriptase. HERV reverse transcription takes place in the cytoplasm and the viral DNA is then transported into the nucleus and integrated into a viral integrase which catalyses insertion of the HERV into its new target site [46]. L1 produces a bicistronic mRNA that is transported to the cytoplasm and translated. L1 encodes proteins that present a strong cis-preference [47], meaning they tend to bind to their encoding mRNA. This results in the formation of ribonucleoprotein particles (RNPs) that may localize to cytoplasmic stress granules [48,49] or enter the nucleus. In the case of L1, reverse transcription occurs in the nucleus, where the L1-encoded endonuclease [33] makes a single-strand cleavage at the target DNA, exposing a 3' hydroxyl group, which is used as a primer for reverse transcription by the L1-encoded reverse transcriptase [34] (termed target-primed reverse transcription, or TPRT) [36,37]. TPRT typically results in L1 insertions with the following sequence features: insertion at an L1 endonuclease motif (5'-TTTT/AA), target site duplications (TSDs) and a poly(A) tail (figure 1b) [33,50,51].
Retrotransposition does not always result in the faithful duplication of an element, particularly in the case of L1. Owing to errors during reverse transcription, only approximately 40% of L1 copies are identical to their source, or donor, L1 [52]. Also, 5' truncation, which is a common characteristic of recent human L1 insertions [19,53], may occur, likely because of the premature dissociation of the reverse transcriptase from the L1 mRNA strand during TPRT. The precise mechanism through which 5' truncation occurs is not clearly understood, although it has been speculated that DNA repair may play a role as host DNA repair factors, such as ATM (ataxia telangiectasia mutated), may limit the size of L1 insertions [54].
As a result of 5' truncations, internal inversions or deletions [55] and point mutations, only about 80–100 [41,42] L1 copies are estimated to be able to retrotranspose per average human genome, with even fewer copies accounting for most new L1 insertions [41,56–58]. One new L1 insertion is generated per 100–200 human births [42,59]. By contrast, 3000 L1 copies [60] are estimated to be retrotransposition-competent in the mouse genome, with at least one in eight mice harbouring a new L1 insertion [11].
4. Detection of retrotransposon activity in the healthy brain
Although nearly 70 years have passed since Barbara McClintock's seminal studies of transposable elements in maize [61,62], ongoing L1 retrotransposition in modern human genomes was only demonstrated in 1988 [63]. Subsequently, endogenous L1 retrotransposition has been described in early embryonic development, germ cells and various diseases, such as cancer [63–69]. Insertions arising during early development or in cancer cells are likely to undergo clonal expansion, resulting in a significant proportion of cells harbouring a particular insertion. Studying the contribution of L1 insertions to somatic mosaicism in normal tissues, however, presents a particular technical challenge, as these genomic variants may be found only in a limited number of cells.
Over the past decade, three main tools have facilitated the discovery and study of L1 retrotransposition in the brain. Firstly, an engineered L1 reporter construct, previously developed to recapitulate L1 retrotransposition in cultured cells [50,70], has been used as a functional read-out of L1 retrotransposition in human embryonic stem cell (hESC)-derived neural progenitor cells (NPCs), rodent NPC cultures and transgenic animal models [71]. The L1 construct consists of a human L1 sequence and a gene encoding enhanced green fluorescent protein (EGFP) in reverse orientation to the L1 transcript. The EGFP gene is interrupted by an intron in the same transcriptional orientation as the L1. Thus, only cells with successful L1 retrotransposition events can express GFP because the EGFP intron is removed from the RNA intermediate, followed by reverse transcription and integration into a genomic location. When the L1-EGFP system is employed in neuronal cultures, GFP co-localizes with neuronal markers Map2 and β-tubulin, whereas very little to no GFP co-localizes with astrocyte or oligodendrocyte markers [72]. Neurons obtained from transgenic L1-EGFP mouse brain sections were positive for GFP expression, in a variety of different brain areas, such as cortex, hypothalamus, amygdala and hippocampus [72–74]. Although the genomic location of the engineered L1 transgene may result in unknown epigenetic effects that do not necessarily recapitulate the regulation of endogenous L1 copies, this type of assay remains a valid tool to understand the effects of L1 and other retrotransposon activity on neural cell function. With recent advances in genome editing technology [75], the design of site-targeted reporter constructs, as well as conditional inducible constructs for temporal and cell-specificity, is more easily achievable and may be instrumental in dissecting the parameters of retrotransposition in the brain.
Secondly, to estimate the number of L1 copies across different tissues and brain areas, Coufal et al. [76] developed a TaqMan quantitative PCR-based L1 CNV assay. This assay estimated an enrichment of L1 copies in the human brain when compared with other tissues, with more L1 copies also found in the hippocampal dentate gyrus than in other brain areas [4,74,76]. The authors [76] estimated 80 somatic L1 insertions occurred per neuron. Owing to the high copy numbers of L1 sequences already present in the genome [19], this assay offers very limited sensitivity when attempting to quantify variation in the number of L1 insertions in somatic tissues. Moreover, the assay does not discriminate integrated copies generated by TPRT from potential extrachromosomal L1 DNAs or any other source of L1 CNV not arising from retrotransposition [77]. New quantitative PCR approaches, such as digital droplet PCR (ddPCR), may prove a better tool to quantify retrotransposon CNV in a more accurate but still cost-effective manner [78,79].
Thirdly, high-throughput DNA sequencing strategies have been developed to detect, quantify and resolve L1 insertions. Whole-genome sequencing (WGS) or targeted sequencing of retrotransposon–genome junctions has been applied to bulk brain, pooled neurons and whole-genome amplified (WGA) material from single neuronal nuclei from post-mortem human brain [4–7,10,80]. Tissues from other parts of the body, such as liver or blood, have been used to discriminate somatic and polymorphic insertions (i.e. insertions found in neurons and absent in non-brain tissue are annotated as somatic). Databases of known retrotransposon insertion polymorphisms can also be used to discriminate novel and known insertions. To fully characterize most new insertions, thorough PCR validation followed by Sanger sequencing is required [81,82]. One of the advantages of using high-throughput DNA sequencing is that it allows not only for detection and quantification of endogenous L1 variants, but also for resolving the structure and genomic location of these insertions. Resolving the structure of a specific insertion is paramount in understanding the potential functional impact of the insertion on the genome.
Providing a valuable orthogonal approach to single-cell genomic analysis, a recent study used somatic cell nuclear transfer to reprogramme postmitotic olfactory bulb neurons into enucleated oocytes and cloned the neuronal genome to produce enough DNA for WGS without the need for WGA. WGS analysis of the resulting clones revealed several de novo L1 insertions [14]. These studies altogether demonstrate that L1-driven somatic mosaicism can occur in the mammalian brain. However, with single-cell genomic analysis in its infancy, methodological variation, including the use of different cell populations as starting material, has led to estimates ranging from one L1 insertion per 25 neurons [5] to 13.7 L1 insertions per neuron [10]. Therefore, although these studies offer strong evidence of L1 retrotransposition in the brain, the frequency of retrotransposition in neural cells requires further investigation [83].
5. Is there a temporal niche for retrotransposition in the brain?
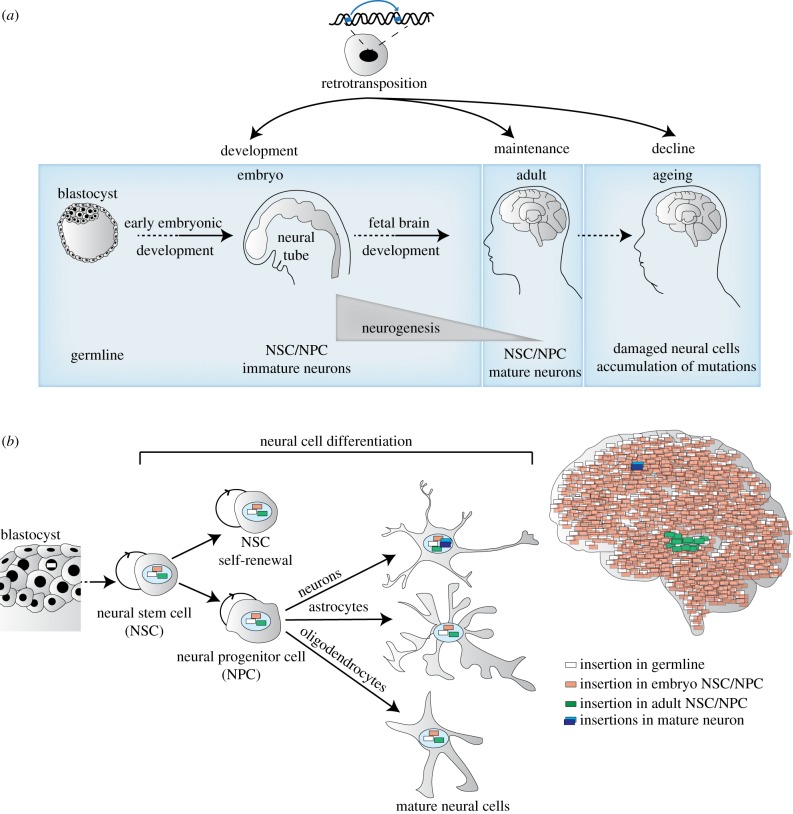
Retrotransposition in the brain could occur at any stage of life, from early brain development to brain maintenance and decline during adulthood, and also influence neural diversity and survival (figure 2a). Neural diversity arises during early embryogenesis through the spatial and temporal patterning of neuroblasts and neuronal progenitor cells. A complex network of gene expression determines a particular cell type-specific progenitor domain and fate [84]. Expression of pan-neuronal genes marks the beginning of neurogenesis and specific TFs are required for neuronal differentiation. Differentiating neurons migrate, grow axonal processes and later form synapses. Various signalling molecules, TFs and cell receptor molecules tightly control this development [85]. Although the bulk of neurons are generated during embryonic neurogenesis, neurons are also born in specific areas of the postnatal brain, such as hippocampus, olfactory system and amygdala [86,87]. An estimated 700 neurons are renewed in the human hippocampus daily [88]. Whether retrotransposition occurs during adult neurogenesis, or during embryonic development, in neuroblasts, committed neural progenitors, or in mature neural cells, determines the number of somatic L1 insertions harboured by a given cell. The spatio-temporal parameters of somatic L1 retrotransposition therefore will likely determine the number of cells that are mosaic for a particular insertion and, most importantly, will govern the overall impact an individual insertion may have on brain activity (figure 2b).
Figure 2.
Timeline of retrotransposition activity in the brain. (a) Retrotransposition can occur in the germline, during embryogenesis, as well as during nervous system development, and can involve neural stem cells (NSCs), NPCs and post-mitotic neurons. Retrotransposition events could accumulate over time and also, potentially, take place later in life as a consequence of the ageing process. (b) Depending on its timing and cell type, a single retrotransposition event can become clonally expanded in the neural cells of the adult brain, with potentially significant effects on brain physiology. When retrotransposition occurs later in brain development, or even in post-mitotic neural cells, new insertions are restricted to one cell, which can in turn be selected against during development or maintained in adults but with a lesser probability of affecting overall brain function.
5.1. Retrotransposition in embryonic and adult neurogenesis
Over a decade ago, Muotri et al. [72] presented the first evidence that L1 retrotransposons can mobilize during neuronal differentiation. Using a human L1-EGFP reporter system, L1 expression was observed both in vitro, in rodent primary hippocampus-derived NPC cultures, and in vivo, in transgenic mice. L1 expression in transgenic mice was observed as early as embryonic day (E) 12.5, when the first waves of immature differentiated neurons appear in the brain [72]. These data were later corroborated and advanced by Coufal et al. [76] in a study reporting L1 reporter activity and endogenous L1 mRNA expression in human NPCs either isolated from fetal brain or derived from hESCs. The preference of retrotransposition for proliferating progenitor cells may be related to the cell cycle. Several studies reported an elevated rate of L1 retrotransposition in dividing cells which suggests that, although non-dividing cells can accommodate L1 retrotransposition [12,89], the cell cycle may promote efficient retrotransposition [90–92]. The mechanism through which the L1 RNP enters the nucleus is not well understood. However, live-cell imaging experiments tracking an L1 reporter construct during cell cycle progression indicate that L1 enters the nucleus as the cell starts to divide, presumably facilitated by nuclear envelope disassembly during mitosis [92].
Another explanation for L1 activity in proliferating NPCs may be a more relaxed chromatin state. A relaxed chromatin state is correlated with pluripotency and proliferation, and differentiated cells have more condensed chromatin than progenitor cells [93]. Open chromatin has been assumed to facilitate L1 activation. By using a transcription activator-like (TAL) effector to target different parts of a mouse L1, which was then fused with a transcriptional activator domain, a recent study demonstrated that L1 transcription directly impacts global chromatin accessibility in the early mouse embryo [94]. L1 transcriptional activity to maintain chromatin openness may be a prerequisite for normal development of the early embryo [94] and it will be interesting to further extend this study into later developmental stages and in the context of neurodevelopment.
Finally, TFs can stimulate L1 to retrotranspose during neuronal differentiation. Sex determining region Y-box 2 (SOX2), which is expressed in stem cells and neural progenitors, and is essential in maintaining a proliferative state, can bind to the 5' UTR of L1 and repress promoter activity [72]. During neuronal differentiation, when SOX2 is downregulated, L1 can become active [72]. Concomitantly, members of the Wnt signalling pathway such as wingless-related integration site family member 3a (WNT3a), which promotes neurogenesis, can also stimulate and increase L1 transcription, providing a window of opportunity for L1 mobilization to occur in differentiating neurons [73]. Hence, the shifting epigenetic and transcriptional landscapes of neurogenesis may provide a unique situation for somatic retrotransposition to occur in the brain.
5.2. Retrotransposition in postmitotic neurons
If we accept that endogenous L1 mobilization can occur during adult neurogenesis, can it also occur in postmitotic neurons? This is a fundamental question because the lifespan of mature neurons can be as long as a human lifespan, and therefore, any potential for somatic retrotransposition in this context may lead to the largest absolute accumulation of new L1 insertions found in the brain. Crucially, Kubo et al. [89] showed in 2006 that engineered L1 retrotransposition can occur in non-dividing human fibroblasts and hepatocytes and, much more recently, Macia et al. [12] confirmed that this was also true for postmitotic neurons. Macia et al. compared the retrotransposition of a hybrid adenoviral L1-EGFP vector in dividing versus non-dividing neural cells by infecting NPC cultures with the L1 reporter and concomitantly adding 5'-bromo-2'-deoxyuridine (BrdU), which labels dividing cells. Immunostainings of differentiated NPC cultures infected with L1-EGFP at 31 days post-NPC differentiation showed the presence of neurons expressing only EGFP but not the cycling marker, BrdU. When comparing the number of integrated L1-EGFP copies in cultures infected with the L1 reporter at day 0 (multi-potent NPC) versus day 31 of differentiation (in mature neurons), a sixfold increase in L1-EGFP copies was observed by qPCR against the EGFP splice junction in mature neurons, even when correcting for differences in proliferation rate [12]. This was an astonishing result, as it supports speculation that not only do mature neurons support retrotransposition, but also retrotransposition occurs at even higher rates in neurons than in proliferating neural progenitors. The study relied heavily on qPCR detection of EGFP copies, and used an in vitro assay to compare dividing versus non-dividing neural cells. Going forward, it will be interesting to address this question in vivo, perhaps by using an inducible knock-in reporter model. Furthermore, it would be very interesting to explore in detail the mechanisms through which retrotransposons escape epigenetic silencing and mobilize in postmitotic neurons.
6. Is retrotransposition necessary for healthy brain function?
It is not established whether retrotransposition in the neural genome is part of normal brain function. It is however plausible that, if retrotransposons did contribute to normal brain function, it would be via altered splicing and DNA methylation of genes, with these being routes to perturb the transcriptional output of cells. For example, pseudogene transcripts can carry microRNA (miRNA) recognition sites and facilitate degradation of their target miRNAs, acting as competitive miRNA targets to the miRNA targets of their parent genes [95]. Such pseudogenes carrying miRNA recognition sites are present in temporal lobe neurons, where they may play an important role in regulating the expression of miRNAs [96]. Somatic pseudogene insertions in the brain, if found, may therefore regulate gene regulation as miRNA ‘sponges’ [97]. In another example, experiments using L1-EGFP transgenic rodents, which were either sedentary or allowed to voluntarily run, demonstrated that exercise increased the total number of newborn hippocampal neurons, which correlated with an increase in engineered L1 retrotransposition [98]. However, it remains unclear whether the increase in GFP was due to a greater number of insertions in the running mice or an activation of GFP expression from previously silenced L1-EGFP insertions, due to changes in the epigenetic landscape. Nevertheless, as the hippocampus is an important area for brain structural plasticity, being involved in learning, memory as well as stress regulation, retrotransposition in hippocampal neurons could potentially contribute to neuronal plasticity. Whether retrotransposon insertions can actively contribute to neuronal physiology, and thus impact behaviour, is still debatable, as to date no functional analyses have been published in this area.
7. Retrotransposition-induced genomic alterations in neuronal genes and their functional consequences
Retrotransposon insertions can significantly alter protein-coding and regulatory regions of the genome, and thereby affect gene expression and other cellular outputs [63,68,99–103]. L1 insertions can occur within neuronal genes, and therefore have the potential to change the structure of genes or how highly they are expressed [4,7,10,72,99]. The consequences of an intragenic L1 insertion for a cell depend on: (i) the characteristics of the insertion (full-length or 5' truncated L1, sense or antisense to the gene; internally inverted and deleted); (ii) the locus where it integrates; and (iii) the ability of the host cell to silence or compensate for the effects of the L1 insertion [104,105]. The various routes by which L1 insertions can lead to gene expression changes have been reviewed previously [106–108]. L1 insertions can delete sequences at the target site [52,109]. In some cases, these deletions can be quite large, such as the 46 kb deletion reported for the PDHX gene encoding pyruvate dehydrogenase, which results in pyruvate dehydrogenase deficiency, a neurodegenerative disorder [110]. Genomic sequences flanking the source L1 5' or 3' end can be transduced along with the L1 to the new integration site, potentially leading to exon shuffling and new genes [111]. Recombination can occur between retrotransposons, causing deletions, duplications or rearrangements in genes [112]. Transcriptional stop sites and polyadenylation signals can be introduced by new retrotransposon sequences, leading to premature transcriptional termination [99,113]. An antisense promoter located in the L1 5' UTR can also create new transcription start sites for genes upstream of the L1 [35,114–116], meaning that both intragenic and intergenic L1 insertions may alter gene expression.
New L1 insertions can lead to aberrant transcriptional splicing [117–121]. Two prominent examples of this are provided by mice bearing spontaneous mutations with neurological phenotypes: the Spastic and Orleans reeler mice. Firstly, the Spastic mouse harbours a germline full-length L1 insertion into a non-coding region of the glycine neurotransmitter receptor β (Glyrβ) gene which leads to aberrant splicing of the pre-mRNA by exon skipping, thus resulting in reduced intact subunit β expression in the brain [118,119]. As the β subunit is required for glycine receptor protein assembly, this insertion leads to a decrease in glycine receptors in the brain and a complex motor deficit phenotype. Secondly, the Orleans reeler mouse incorporates a full-length L1 insertion into a coding region of the Reelin (Reln) gene, which also leads to exon skipping and a frame shift, in this case generating a 220 bp deletion of the Reln mRNA, and leading to inefficient secretion of Reln truncated protein [120,122,123]. As Reln is essential for neuronal migration and cortical lamination, deficiency in its secretion leads to a severe impairment of neuronal migration and, as a consequence, cortical and cerebellar delamination. Subsequent neurological symptoms in Reln mutant mice recapitulate the phenotype seen in patients with lissencephaly caused by other Reln mutations [124]. Hence, de novo L1 insertions arising in the germline can alter genes expressed in the brain and cause a neurological phenotype, suggesting that similar mutations arising in neurons have the potential to generate a functional change.
8. Retrotransposon regulation and environmental factors impacting retrotransposition
8.1. Locus-specific regulation of long interspersed element 1 retrotransposition
Retrotransposition depends on a mobile element's intrinsic ability to ‘jump’, and its host cell's capacity to defeat this mobilization. Despite their significant presence in the genome, almost all human L1 copies are 5' truncated, or contain internal mutations and rearrangements, and are not able to mobilize. A few L1 copies are full length, remain transcriptionally active and have intact ORFs and therefore have the capacity to act as source, or donor, elements in a cycle of retrotransposition. An even smaller subset (approx. 6 L1s per individual genome) retrotranspose efficiently in vitro when tagged with a fluorescent or selectable marker [50,70] and are referred to as ‘hot’ donor L1s [41,42], although retrotransposition efficiency can vary significantly in different cell types. Hot L1s tend to be the most recently acquired L1 copies in the genome, have very low sequence divergence from the L1 consensus sequence and are polymorphic in the population (found in some individuals but absent in others) [41]. If a cell type supports a high L1 retrotransposition rate, is this due to the activity of a few hot donor L1s, or is it the result of L1 mRNAs being widely expressed in line with concerted, genome-wide dysregulation? To address this question, Philippe et al. [105] recently designed an assay that discriminated near-identical donor L1s by mapping the 5' and 3' genome junction sequences of all of the potentially active donor L1s in a set of somatic cell lines and intersecting these data with transcriptional and epigenetic signatures. The authors found that donor L1s were generally inactive, with a limited number active in each of the cell lines examined. These results suggested cell-type-specific activity of retrotransposition-competent L1s, presumably as only some L1s escaped silencing in each cell type. However, a previous study [125] identified a large number of L1s that were actively expressed in multiple hESC lines, suggesting that patterns of donor L1 activation may differ in pluripotent and differentiated cells, albeit with a different approach in each study.
8.2. Retrotransposon silencing mechanisms
As retrotransposition events can cause such large effects on gene expression, cell function and ultimately organism fitness, several mechanisms exist to limit and regulate L1 mobilization in germ and neural cells. These mechanisms have been reviewed comprehensively [126,127]. We will focus here on mechanisms thought to regulate retrotransposition during neural cell differentiation, a process that must be tightly regulated during critical stages of brain development, as differences have been observed in retrotransposition efficiency between proliferating NPCs and postmitotic neurons [12,76]. It is known that different DNA methylation patterns are present across neurodevelopmental stages [128]. Presumably, dynamic methylation during developmental processes, such as neuronal differentiation, can offer windows of opportunity for retrotransposition to occur [76]. Additionally, different neuronal subtypes (such as GABAergic interneurons and glutamatergic projection neurons from the prefrontal cortex (PFC)) are known to present differences in their methylation patterns [129]. A recent study investigating methylation of the L1 5' UTR, which is a critical predictor of L1 promotor activity [76,130], found no differences in the levels of methyl-cytosine and 5-hydroxy-methyl-cytosine across frontal cortex, hippocampus, cerebellum and basal ganglia regions in the adult mouse brain [131]. Nevertheless, subtle differences in L1 methylation state across neuronal types and brain areas could potentially be missed when analysing bulk tissue samples.
Several epigenetic factors are known to repress L1. For example, methyl-CpG-binding protein 2 (MeCP2) is present ubiquitously in the body and in higher abundance in mature neurons. MeCP2 binds 5-methyl-cytosine residues in CpG dinucleotides and interacts with histone deacetylase protein (HDAC) and transcriptional SIN3A corepressor complexes to repress transcription [132–134]. MeCP2 knockout mice show an increase in L1 promoter activity consistent with MeCP2 functioning as an L1 repressor [74,135]. Valproic acid inhibition of HDAC1 enhances the transcriptional activity of L1 [136], indicating that HDAC1 is also involved in L1 repression. Another deacetylase suggested to inhibit L1 is mono-ADP ribosyltransferase enzyme, or Sirtuin 6 (SIRT6), that was shown to localize to the L1 promoter [137]. SIRT6 is expressed in neurons and appears to be displaced in oxidative stress conditions and during ageing [138]. Although L1 is repressed by these epigenetic mechanisms, the brain appears to exhibit lower L1 methylation than found for other tissues, such as skin [76].
In addition to epigenetic regulation, retrotransposon activity can be regulated by specific TFs, such as yin yang 1 (YY1), runt-related transcription factor 3 (RUNX3), SOX2 and tumour protein p53 (p53). YY1 is a zinc finger protein expressed ubiquitously in the brain. It is critical for regional patterning of the brain and neuronal differentiation, as it regulates genes including Otx2 and Engrailed 2 [139–141]. YY1 positively regulates L1 by directing the RNA polymerase II complex to its proper binding site [142,143]. RUNX3, which is involved in neurogenesis, development and survival of proprioceptive neurons, also positively regulates the L1 promoter [144–146]. Conversely, the TF SOX2, involved in the maintenance of the multi-potent state of neural stem cells (NSCs), has been shown to inhibit L1 transcription [72,73,76,147–149]. This is also the case for p53, a master regulator of cell cycling that is involved in neural proliferation and differentiation (reviewed in [150]). p53 may suppress L1 retrotransposition through its involvement in H3K9 trimethylation (H3K9me3), a silencing marker, at the L1 5' UTR [151,152]. As new L1 insertions carry TF binding sites and potentially attract epigenetic suppression [153], the integration of an L1 into an intron or an intergenic region immediately upstream of a protein-coding gene can alter the expression pattern of that gene. L1 insertions are thought to be particularly harmful if oriented in sense to the gene, based on a depletion of these insertions from the human population [99,154], and also the length of the insertion may influence its impact [99]. Host factors, such as apolipoprotein B mRNA editing enzyme catalytic subunit 3A (APOBEC3A) [155], or DNA repair factors such as ATM [54], can therefore counteract the impact of L1 at the insertion site by limiting the length of L1 insertions [156]. Hence, even if the host cell fails to prevent the integration of a new L1 copy, it may mitigate the potential consequences that L1 insertion has upon the genome.
8.3. Environmental factors impacting retrotransposition
Numerous studies have proposed that environmental factors may trigger hyperactivation of L1 and other retrotransposons. However, most of these studies are preliminary and the experiments have been largely performed on cell lines. For instance, heavy metals such as mercury, nickel and cadmium seem to increase L1 retrotransposition in HeLa and neuroblastoma cells [157–159], and so does oxidative stress [160]. Pollutants such as benzo[a]pyrene (BaP), a polycyclic aromatic hydrocarbon ubiquitously found in grilled meats, tobacco smoke, vehicle exhaust, domestic wood and coal fires, have also been found to be involved in L1 retrotransposition [161]. Similarly, an L1-lacZ retrotransposition assay showed that UV light and heat-shock induces an increase in β-galactosidase activity in tumour cell lines [162]. The increase in L1 activity as a result of environmental insults may occur via an indirect pathway, for example, L1 retrotransposon may leverage the DNA double-stranded breaks induced by heavy metal toxicity to mobilize and insert at new genomic locations. Indeed, Morrish et al. [163] described an endonuclease-deficient engineered L1 that could readily retrotranspose in cell lines lacking DNA repair mechanisms. However, Farkash et al. [164] reported no increase in retrotransposition of an endonuclease-deficient L1 after application of gamma radiation, which is known to induce DNA double-strand breaks. It therefore remains unclear whether L1 can easily integrate, through an endonuclease-independent mechanism, at preformed DNA double-strand breaks. Alternatively, the cellular machinery could use L1 to induce apoptosis or repair DNA damage through a compensatory mechanism [161,163,165]. Although these environmental insults are known to impact neurogenesis and brain function, a link between these factors and an increase in retrotransposition in neurons or brain tissue has not been formally demonstrated in vivo.
9. Retrotransposition in neurological disease
Retrotransposition in the early embryo and committed germline can result in heritable mutations that cause genetic disease [11,63]. Several retrotransposon insertions involved in genetic neurologic diseases have been reported to date (table 1). As stress-responsive elements, retrotransposons could be activated by an abnormal neural cell environment, and thereby generate somatic insertions with the potential to influence neurological diseases of developmental and degenerative origin. Early developmental stress factors, as well as environmental insults occurring later in life, are major risk factors for developing neuropsychiatric and neurodegenerative disorders [208]. Although many studies report elevated retrotransposon copy numbers, mRNA levels or retroviral markers in neurological diseases, to date there are essentially no confirmed causative links established between any neurological condition and somatic retrotransposition. Here, we present an overview of the most relevant findings in this area from L1 and Alu data. The implication of HERVs in neurological disease, such as multiple sclerosis, schizophrenia and bipolar disorder, has been recently reviewed [178,209,210]. The involvement of SVA retrotransposons in neurological disease has also been recently reviewed [211,212]. Processed pseudogenes in the context of neuropathology have not been studied extensively, to our knowledge.
Table 1.
Studies associating retrotransposons with neurological disease. Here, we consider association as a study having shown that a retrotransposon insertion, or insertions, may cause a disease, or the study having reported elevation of retrotransposon copy number or mRNA level in the brain tissue of affected individuals when compared with healthy individuals.
| retrotransposon | neurological disease |
|
|---|---|---|
| insertions | activity (mRNA levels, CNV, biomarkers) | |
| HERV | multiple sclerosis [166–168] 3q13.31 microdeletion syndrome [169] |
amyotrophic lateral sclerosis [170,171] multiple sclerosis [166–168,172] schizophrenia [173–176] bipolar disorder [173,175,177] HIV-associated dementia [178] major depression [177] autism [179] ADHD [180] |
| L1 | pyruvate dehydrogenase complex deficiency [110] Fukuyama-type congenital muscular dystrophy [181] neurofibromatosis type I [182] ataxia with oculomotor apraxia 2 [183] glioblastoma [184,185] schizophrenia [176] ataxia telangiectasia [54] Coffin Lowry syndrome [186] |
major depression [187,188] schizophrenia [187–189] Rett syndrome [74] autism [190] cocaine addiction [191] post-traumatic stress disorder [192] |
| Alu | autosomal dominant optic atrophy [193] adrenoleukodystrophy [194] neurofibromatosis type I [182,195] lipoprotein lipase deficiency [196] spastic paraplegia [197,198] Hunter disease [199] Menkes disease [200] Walker–Warburg syndrome [201] Lesch–Nyhan disease [202] glycogen storage disease type II [203] |
post-traumatic stress disorder [192] |
| SVA | neurofibromatosis type I [204] X-linked dystonia-parkinsonism [101–103,205] Fukuyama muscular dystrophy [206] |
n.a. |
| processed pseudogenes | spinal muscular atrophy [207] | n.a. |
9.1. Neurodevelopmental and psychiatric disease
A prominent example of a neurodevelopmental disorder that has been linked to retrotransposon activity is Rett syndrome (RTT). RTT is a progressive, severe neurodevelopmental disease associated with a mutation in the MeCP2 gene [213]. MeCP2 globally regulates methylated DNA and, as mentioned above, has been shown to repress L1 transcription and retrotransposition [74,135]. Mice lacking functional MeCP2 exhibit elevated L1 mRNA expression and L1 copy number in brain [74]. Increased retrotransposition of the L1-EGFP reporter has been observed in MeCP2 mutant rat NSC cultures and transgenic mice [74]. Elevated L1 mRNA abundance and L1-EGFP reporter activity were also found in NPCs differentiated from induced pluripotent stem cells (iPSCs) derived from RTT patient fibroblasts [74]. However, it remains unclear whether susceptibility to increased L1 retrotransposition in RTT patients can act as a driving force for disease phenotype or progression, or is a peripheral outcome of MeCP2 mutation.
Psychiatric diseases, such as autism, schizophrenia and bipolar disorder, have also been linked with retrotransposon activity [177,179,187,188,190–192]. The aetiology of these disorders involves polygenic and environmental risk factors. Recreational drugs that cause addiction, such as methamphetamines (METH) and cocaine, are major risk factors for the development of psychiatric disorders [214,215] and have been reported to affect L1 retrotransposition in cell lines and rodent brains [216,217]. One study reported elevated L1 mRNA and ORF2p levels after METH exposure in rat neurogenic areas [217]. Both METH and cocaine can induce L1 retrotransposition in cultured neuroblastoma cell lines, as determined by L1-EGFP [217] and L1-neomycin reporter assays [216]. In cell culture, METH and cocaine do not induce DNA breaks, as revealed by the absence of gamma H2A histone family X (H2AX) phosphorylation at serine 139, a marker for double-strand DNA breaks [216]. Instead, L1 mobilization in response to METH and cocaine treatment may occur via the cyclic AMP response element-binding protein pathway mediating ORF1p access to chromatin [216]. In a recent study, Doyle et al. [191] sought to further test the potential link between L1 insertions and cocaine in brain tissue of cocaine addicts and controls. The authors employed a TaqMan ddPCR assay and found no significant increase in the L1 mRNA transcript levels in the cocaine group [191]. WGS performed on pools of neuronal and non-neuronal nuclei, as well as blood samples from cases and controls, revealed no significant increase in the number of L1 insertions in cocaine samples compared with controls [191]. The authors reported gene ontology terms, potentially important for cocaine signalling pathways, to be enriched for genes harbouring L1 insertions found in the cocaine group and not observed in the control. However, the study had several caveats which could be addressed in future research. Samples were pooled across control or case individuals, relying on a ddPCR assay to determine the allele frequency of insertions in each individual. Furthermore, insertions were detected based only on the 3' end of L1 and 100 nt single-end Illumina reads, and therefore were not fully resolved and characterized. How and whether these new insertions ultimately contribute to addiction is still very much unclear. The relevant studies have to date not incorporated single-cell genomic analysis with full characterization of candidate somatic insertions.
Increased retrotransposon copy number and mRNA levels have been reported in schizophrenia and depression [187,192]. Bundo et al. [187] revealed a significant increase in L1 ORF2 copy number in neurons isolated from schizophrenia PFC when compared with matched control neurons. This result was replicated in iPSC-derived neurons from patients affected by 22q11 deletion, a mutation found in a number of schizophrenia patients and considered a high genetic risk factor [218]. A consistent increase in L1 copy number was also observed in the PFC tissue of two established schizophrenia animal models (i.e. maternal immune activation induced by polyinosinic : polycytidylic acid (PolyI : C) and epidermal growth factor (EGF)) [187]. WGS performed on neuronal nuclei and liver samples revealed no increase in L1 insertion number in patients when compared with controls. The authors reported that the L1 insertions in schizophrenia patients were more frequently present in genes important for synaptic function and schizophrenia-related genes [187]. However, the insertions were not validated and characterized. Moreover, many of the brain-specific insertions appeared from inactive, old L1 subfamilies, which could not have accounted for de novo retrotransposition. Another study reported significant L1 promotor hypomethylation in the PolyI : C model of maternal immune activation, which could explain elevated L1 transcription [219]. Very recently, Bedrosian et al. [79] proposed that low maternal care, another risk factor for schizophrenia and other psychiatric disorders, may elevate L1 retrotransposition in the hippocampus. This study, based largely on a qPCR L1 CNV assay, estimated higher L1 copy number and transcript levels in mice after low maternal care when compared with those reared in high maternal care conditions. The authors reported no change in neurogenesis between the two groups. However, the elevated L1 transcript levels correlated with hypomethylation of L1 at a YY1 TF binding site [79]. As a qPCR assay cannot prove L1 integration, it remains unclear whether the difference in L1 copy number and transcript abundance was reflected by an increase in retrotransposition. Consistent L1 upregulation across various schizophrenia experimental systems indicates a strong association between disease phenotype and L1 activity, although it remains unclear whether L1 is a driver or a passenger with respect to neuropathology and, again, single-cell genomic analyses supporting elevated L1 mobilization, as opposed to L1 CNV, have yet to be published.
Even if elevated somatic retrotransposition ultimately is not found to occur in a given neurological condition, a molecular signature of L1 regulatory disruption may indicate a wider landscape of epigenome abnormality, and prove useful as a diagnostic tool. For example, altered L1 and Alu epigenetic regulation has been reported in post-traumatic stress disorder (PTSD), an anxiety condition characterized by persistent re-experiences of a past traumatic event or events [192]. PTSD patients within a cohort of military personnel assessed post-deployment showed hypermethylation of L1 and Alu in serum when compared with healthy individuals [192]. Non-PTSD post-deployment individuals also exhibited a hypermethylation pattern when compared with the same individuals pre-deployment. These two observations are intriguing and might suggest an adaptive stress response mediated by retrotransposon repression or pre-existing methylation in PTSD cases compared with controls [220]. These findings could be buttressed by experiments performed using animal models where L1 methylation and CNV could be analysed in brain tissue. Overall, a theme has emerged of L1 activation in a wide range of neurological disorders. More research is required in this area to understand whether there is a functional consequence of L1 mobilization in schizophrenia and other psychiatric disorders.
9.2. Neurodegeneration
Neurodegenerative conditions, such as Parkinson's disease, Huntington's disease and Alzheimer's disease, represent a group of nervous system disorders characterized by the selective death of neuronal subsets in particular brain regions [221]. Experimental data showing somatic retrotransposition, or even the broad involvement of retrotransposons, in neurodegeneration are quite scarce. To speculate, dysfunctional epigenetic silencing, deficient DNA repair and oxidative stress could all lead to retrotransposon activation associated with neuronal degeneration or to the progression of degenerative processes. As an instructive example, L1 copy number was found via a qPCR-based assay to be elevated in hippocampal neurons from ataxia telangiectasia patients [54]. These patients presented a loss of function mutation in the ATM gene encoding a serine/threonine kinase [222], which leads to neuronal degeneration [223]. ATM responds to double-stranded DNA breaks by phosphorylating downstream factors and activating DNA damage checkpoints, thus leading to cell cycle arrest and to the repair of damaged DNA or p53-mediated apoptosis [223]. ATM mutation in cultured human NPCs and ATM knockout in L1-EGFP mice significantly increased L1-EGFP retrotransposition, measured by the number of GFP-positive cells, without affecting L1 promoter activity or endogenous ORF1p levels, and with no change in cell division rates or survival [54]. Notably, longer L1 insertions appeared to occur in ATM mutant cells [54], potentially owing to the role of ATM in cellular DNA repair, which may interfere with host defences against L1 retrotransposition under wild-type conditions. Further analysis, preferably in vivo, is required to corroborate these results, and identify the role of ATM in controlling L1 integration.
As many neurodegenerative conditions are associated with ageing, retrotransposition in neurodegenerative conditions may, in general, reflect conditions encountered in senescent cells. Accumulation of oxidative DNA damage and unrepaired DNA, as well as changes in methylation patterns commonly associated with ageing, might lead to reduced silencing of retrotransposition in the brain. As an example of this, TAR DNA-binding protein 43 (TDP-43) is a nucleic acid binding protein involved in transcriptional repression and RNA metabolism during stress response. Mutant TDP-43 is usually associated with neurodegenerative disorders, but it can also be present in healthy elderly people [224]. Accumulation of TDP-43 in tau-negative and ubiquitin-positive cytoplasmic inclusions is a neuropathological hallmark in neurodegenerative conditions, such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTLD) [225]. Transcriptomic analyses performed on brain tissue obtained from TDP-43 mutant mice and mice expressing human mutant TDP-43 showed an overall increase in retrotransposon expression [226]. Moreover, surveys of protein–RNA interactions and gene expression performed on FTLD brain samples versus matched controls found a significantly reduced association of mutant TDP-43 at its target retrotransposons in patients [226]. Another recent study, wherein human TDP-43 was expressed in Drosophila brains, recapitulated these results from patients and mouse models, demonstrating that TDP-43 dysfunction results in de-repression of retrotransposons [227]. Interestingly, the expression of human TDP-43 specifically in fly glial cells led to an early and significant increase in gypsy ERV transcript levels as previously reported to be elevated in ageing flies [228]. Human TDP-43 in the fly glia resulted in a high number of apoptotic nuclei, very significant motor impairment and shortened lifespan. Knockdown of gypsy ameliorated this phenotype, suggesting that gypsy may be involved in cell death, potentially by mediating DNA damage [227].
Many neurodegenerative disorders are strongly linked with mitochondrial dysfunction. In intriguing recent work, Larsen et al. [229] put forward the hypothesis that retrotransposons were involved in mitochondrial gene dysfunction observed in neurodegeneration. Analysis of the retrotransposon sequence content in over a thousand mitochondrial genes, in conjunction with randomly selected protein-coding genes, showed an enrichment for Alu within and adjacent to mitochondrial genes [229]. Previous studies have identified L1 and Alu retrotransposon insertions in a number of translocase of outer mitochondrial membrane genes [4,230,231]. Moreover, Alu insertions in two genes involved in mitochondria stabilization and function led to disorders with a neurodegenerative component [193,194]. More studies, in cell culture and animal models, are required to test if Alu activity can preferentially insert into and induce mitochondrial gene disruption.
10. Concluding remarks
It is now well established that retrotransposon-driven mosaicism can occur in the mammalian brain. Dynamism in TF activity, and that of other regulatory mechanisms described to act during embryonic and adult neurogenesis, provides a temporal and spatial niche amenable to retrotransposition. Additionally, retrotransposition may not be limited to NPCs; it may also take place in postmitotic neurons. Further proof is required to be certain that this can occur in vivo, and the regulatory mechanisms involved in allowing L1 retrotransposition in mature neurons require further investigation. Environmental stress, as well as genetic dysfunction in L1 silencing factors, can result in high retrotransposition levels, and possibly lead to neurological disease. This also requires extensive investigation, as we lack even basic understanding of the pathways on which stress factors or dysfunction in silencing factors can act to impact retrotransposition in neural cells. Our understanding of retrotransposition in the brain is still very much in its infancy.
Further advances in single-cell genomics (and perhaps taking Eric Kandel's reductionist approach of ‘one cell at a time’ to focus on very well-defined neuronal subtypes [232]) to understand the precise timing of retrotransposition, and its regulation, are required at this point, and will also advance our knowledge of which neuronal types and contexts best support endogenous L1 retrotransposition. Genome-editing tools, such as the CRISPR–Cas9 system [233], may be employed in the near future to resolve L1 insertions identified in patient samples and introduce these faithfully into neuronal cultures or animal models, to evaluate their impact on gene expression and, potentially, neurobiological phenotype. This approach may prove to be a viable alternative to the current transgenic approaches used to study engineered L1 mobilization in the brain, and would bring us closer to understanding whether endogenous retrotransposition has an impact upon neurobiology and neurological disease.
Acknowledgements
We thank Dr Sandra Richardson, Dr Adam Ewing and Dr Liviu-Gabriel Bodea for critical reading of the manuscript and comments. We thank Dr Sandra Richardson and Dr Francisco Jose Sanchez-Luque for constructive comments on the figures.
Data accessibility
This article has no additional data.
Authors' contributions
G.O.B., E.G.Z.M. and G.J.F. read the literature and drafted the text. E.G.Z.M. and G.O.B. drafted table 1. G.O.B. drafted the figures. All authors approved the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
We acknowledge the support of an NHMRC-ARC Research Fellowship (GNT1108258) to G.O.B. and a CSL Centenary Fellowship to G.J.F.
References
- 1.DeFelipe J. 1993. Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb. Cortex 3, 273–289. ( 10.1093/cercor/3.4.273) [DOI] [PubMed] [Google Scholar]
- 2.Nelson SB, Sugino K, Hempel CM. 2006. The problem of neuronal cell types: a physiological genomics approach. Trends Neurosci. 29, 339–345. ( 10.1016/j.tins.2006.05.004) [DOI] [PubMed] [Google Scholar]
- 3.Tognini P, Napoli D, Pizzorusso T. 2015. Dynamic DNA methylation in the brain: a new epigenetic mark for experience-dependent plasticity. Front. Cell. Neurosci. 9, 331 ( 10.3389/fncel.2015.00331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baillie JK, et al. 2011. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537. ( 10.1038/nature10531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evrony GD, et al. 2012. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496. ( 10.1016/j.cell.2012.09.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evrony GD, et al. 2015. Cell lineage analysis in human brain using endogenous retroelements. Neuron 85, 49–60. ( 10.1016/j.neuron.2014.12.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erwin JA, et al. 2016. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 19, 1583–1591. ( 10.1038/nn.4388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConnell MJ, et al. 2013. Mosaic copy number variation in human neurons. Science 342, 632–637. ( 10.1126/science.1243472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodato MA, et al. 2015. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98. ( 10.1126/science.aab1785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upton KR, et al. 2015. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161, 228–239. ( 10.1016/j.cell.2015.03.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson SR, et al. 2017. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res. 27, 1395–1405. ( 10.1101/gr.219022.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macia A, et al. 2017. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 27, 335–348. ( 10.1101/gr.206805.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gleeson JG, Minnerath S, Kuzniecky RI, Dobyns WB, Young ID, Ross ME, Walsh CA. 2000. Somatic and germline mosaic mutations in the doublecortin gene are associated with variable phenotypes. Am. J. Hum. Genet. 67, 574–581. ( 10.1086/303043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazen JL, et al. 2016. The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron 89, 1223–1236. ( 10.1016/j.neuron.2016.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7, e1002384 ( 10.1371/journal.pgen.1002384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills RE, Bennett EA, Iskow RC, Devine SE. 2007. Which transposable elements are active in the human genome? Trends Genet. 23, 183–191. ( 10.1016/j.tig.2007.02.006) [DOI] [PubMed] [Google Scholar]
- 17.Singer T, McConnell MJ, Marchetto MCN, Coufal NG, Gage FH. 2010. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 33, 345–354. ( 10.1016/j.tins.2010.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erwin J, Marchetto MC, Gage FH. 2014. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 15, 497–506. ( 10.1038/nrn3730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 20.Weiss RA. 2016. Human endogenous retroviruses: friend or foe? APMIS 124, 4–10. ( 10.1111/apm.12476) [DOI] [PubMed] [Google Scholar]
- 21.Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48. ( 10.1038/ng1223) [DOI] [PubMed] [Google Scholar]
- 22.Dewannieux M, Heidmann T. 2005. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J. Mol. Biol. 349, 241–247. ( 10.1016/j.jmb.2005.03.068) [DOI] [PubMed] [Google Scholar]
- 23.Raiz J, et al. 2012. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683. ( 10.1093/nar/gkr863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20, 3386–3400. ( 10.1093/hmg/ddr245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esnault C, Maestre J, Heidmann T. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367. ( 10.1038/74184) [DOI] [PubMed] [Google Scholar]
- 26.Ullu E, Tschudi C. 1984. Alu sequences are processed 7SL RNA genes. Nature 312, 171–172. ( 10.1038/312171a0) [DOI] [PubMed] [Google Scholar]
- 27.Deininger PL, Jolly DJ, Rubin CM, Friedmann T, Schmid CW. 1981. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J. Mol. Biol. 151, 17–33. ( 10.1016/0022-2836(81)90219-9) [DOI] [PubMed] [Google Scholar]
- 28.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH. 2003. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 73, 1444–1451. ( 10.1086/380207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. 2005. SVA elements: a hominid-specific retroposon family. J. Mol. Biol. 354, 994–1007. ( 10.1016/j.jmb.2005.09.085) [DOI] [PubMed] [Google Scholar]
- 30.Hancks DC, Kazazian HH. 2010. SVA retrotransposons: evolution and genetic instability. Semin. Cancer Biol. 20, 234–245. ( 10.1016/j.semcancer.2010.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes SE, Singer MF, Swergold GD. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267, 19 765–19 768. [PubMed] [Google Scholar]
- 32.Hohjoh H, Singer MF. 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639. ( 10.1002/j.1460-2075.1996.tb00395.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Q, Moran JV, Kazazian HH, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916. ( 10.1016/S0092-8674(00)81997-2) [DOI] [PubMed] [Google Scholar]
- 34.Mathias SL, Scott AF, Kazazian HH, Boeke JD, Gabriel A. 1991. Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810. ( 10.1126/science.1722352) [DOI] [PubMed] [Google Scholar]
- 35.Denli AM, et al. 2015. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163, 583–893. ( 10.1016/j.cell.2015.09.025) [DOI] [PubMed] [Google Scholar]
- 36.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605. ( 10.1016/0092-8674(93)90078-5) [DOI] [PubMed] [Google Scholar]
- 37.Cost GJ. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910. ( 10.1093/emboj/cdf592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowronski J, Fanning TG, Singer MF. 1988. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 8, 1385–1397. ( 10.1128/MCB.8.4.1385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit AF, Tóth G, Riggs AD, Jurka J. 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246, 401–417. ( 10.1006/jmbi.1994.0095) [DOI] [PubMed] [Google Scholar]
- 40.Boissinot S, Chevret P, Furano AV. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17, 915–928. ( 10.1093/oxfordjournals.molbev.a026372) [DOI] [PubMed] [Google Scholar]
- 41.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170. ( 10.1016/j.cell.2010.05.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA 100, 5280–5285. ( 10.1073/pnas.0831042100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konkel MK, Walker JA, Hotard AB, Ranck MC, Fontenot CC, Storer J, Stewart C, Marth GT, Batzer MA. 2015. Sequence analysis and characterization of active human Alu subfamilies based on the 1000 Genomes pilot project. Genome Biol. Evol. 7, 2608–2622. ( 10.1093/gbe/evv167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. 2016. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl Acad. Sci. USA 113, E2326–E2334. ( 10.1073/pnas.1602336113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. 1985. Ty elements transpose through an RNA intermediate. Cell 40, 491–500. ( 10.1016/0092-8674(85)90197-7) [DOI] [PubMed] [Google Scholar]
- 46.Boeke JD, Voytas DF. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae. In Mobile DNA II (eds Craig NL, Craigie R, Gellert M, Lambowitz AM), pp. 631–662. Washington, DC: American Society of Microbiology; ( 10.1128/9781555817954.ch26) [DOI] [Google Scholar]
- 47.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439. ( 10.1128/MCB.21.4.1429-1439.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doucet AJ, et al. 2010. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6, 1–19. ( 10.1371/journal.pgen.1001150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodier JL, Zhang L, Vetter MR, Kazazian HH. 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483. ( 10.1128/MCB.00332-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moran JVJ, Holmes SES, Naas TPT, DeBerardinis RJ, Boeke JD, Kazazian HH. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927. ( 10.1016/S0092-8674(00)81998-4) [DOI] [PubMed] [Google Scholar]
- 51.Jurka J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA 94, 1872–1877. ( 10.1073/pnas.94.5.1872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert N, Lutz S, Morrish TA, Moran JV. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25, 7780–7795. ( 10.1128/MCB.25.17.7780-7795.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers JS, et al. 2002. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 71, 312–326. ( 10.1086/341718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coufal NG, et al. 2011. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl Acad. Sci. USA 108, 20 382–20 387. ( 10.1073/pnas.1100273108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostertag EM, Kazazian H.H. J. 2001. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 11, 2059–2065. ( 10.1101/gr.205701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lutz SM, Vincent BJ, Kazazian HH, Batzer MA, Moran JV. 2003. Allelic heterogeneity in LINE-1 retrotransposition activity. Am. J. Hum. Genet. 73, 1431–1437. ( 10.1086/379744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seleme MdC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH. 2006. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc. Natl Acad. Sci. USA 103, 6611–6616. ( 10.1073/pnas.0601324103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH. 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16, 37–43. ( 10.1038/ng0597-37) [DOI] [PubMed] [Google Scholar]
- 59.Ewing AD, Kazazian HH. 2010. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 20, 1262–1270. ( 10.1101/gr.106419.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodier JL, Ostertag EM, Du K, Kazazian HH Jr. 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 11, 1677–1685. ( 10.1101/gr.198301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McClintock B. 1956. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 21, 197–216. ( 10.1101/SQB.1956.021.01.017) [DOI] [PubMed] [Google Scholar]
- 62.McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. USA 36, 344–355. ( 10.1073/pnas.36.6.344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166. ( 10.1038/332164a0) [DOI] [PubMed] [Google Scholar]
- 64.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH. 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312. ( 10.1101/gad.1803909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261. ( 10.1016/j.cell.2010.05.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott EC, Devine SE. 2017. The role of somatic L1 retrotransposition in human cancers. Viruses 9, 131 ( 10.3390/v9060131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Hurk JAJM, et al. 2007. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16, 1587–1592. ( 10.1093/hmg/ddm108) [DOI] [PubMed] [Google Scholar]
- 68.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. 1992. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645. [PubMed] [Google Scholar]
- 69.Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH. 2002. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum. Genet. 71, 327–336. ( 10.1086/341722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH. 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423. ( 10.1093/nar/28.6.1418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH. 2002. A mouse model of human L1 retrotransposition. Nat. Genet. 32, 655–660. ( 10.1038/ng1022) [DOI] [PubMed] [Google Scholar]
- 72.Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910. ( 10.1038/nature03663) [DOI] [PubMed] [Google Scholar]
- 73.Kuwabara T, et al. 2009. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105. ( 10.1038/nn.2360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. 2010. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446. ( 10.1038/nature09544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Savell KE, Day JJ. 2017. Applications of CRISPR/Cas9 in the mammalian central nervous system. Yale J. Biol. Med. 90, 567–581. [PMC free article] [PubMed] [Google Scholar]
- 76.Coufal NG, et al. 2009. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1158. ( 10.1038/nature08248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodier JL. 2014. Retrotransposition in tumors and brains. Mob. DNA 5, 11 ( 10.1186/1759-8753-5-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newkirk SJ, et al. 2017. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc. Natl Acad. Sci. USA 114, E5635–E5644. ( 10.1073/pnas.1701069114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bedrosian TA, Quayle C, Novaresi N, Gage FH. 2018. Early life experience drives structural variation of neural genomes in mice. Science 359, 1395–1399. ( 10.1126/science.aah3378) [DOI] [PubMed] [Google Scholar]
- 80.Kurnosov AA, et al. 2015. The evidence for increased L1 activity in the site of human adult brain neurogenesis. PLoS ONE 10, e0117854 ( 10.1371/journal.pone.0117854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanchez-Luque FJ, Richardson SR, Faulkner GJ. 2016. Retrotransposon capture sequencing (RC-Seq): a targeted, high-throughput approach to resolve somatic L1 retrotransposition in humans. Methods Mol. Biol. 1400, 47–77. ( 10.1007/978-1-4939-3372-3_4) [DOI] [PubMed] [Google Scholar]
- 82.Richardson SR, Morell S, Faulkner GJ. 2014. L1 Retrotransposons and somatic mosaicism in the brain. Annu. Rev. Genet. 48, 1–27. ( 10.1146/annurev-genet-120213-092412) [DOI] [PubMed] [Google Scholar]
- 83.Faulkner GJ, Garcia-Perez JL. 2017. L1 mosaicism in mammals: extent, effects, and evolution. Trends Genet. 33, 802–816. ( 10.1016/j.tig.2017.07.004) [DOI] [PubMed] [Google Scholar]
- 84.Azzarelli R, Hardwick LJA, Philpott A. 2015. Emergence of neuronal diversity from patterning of telencephalic progenitors. Wiley Interdiscip. Rev. Dev. Biol. 4, 197–214. ( 10.1002/wdev.174) [DOI] [PubMed] [Google Scholar]
- 85.Gao P, Sultan KT, Zhang XJ, Shi SH. 2013. Lineage-dependent circuit assembly in the neocortex. Development 140, 2645–2655. ( 10.1242/dev.087668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. 1998. Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317. ( 10.1038/3305) [DOI] [PubMed] [Google Scholar]
- 87.Jhaveri DJ, Tedoldi A, Hunt S, Sullivan R, Watts NR, Power JM, Bartlett PF, Sah P. 2018. Evidence for newly generated interneurons in the basolateral amygdala of adult mice. Mol. Psychiatry 23, 521–532. ( 10.1038/mp.2017.134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spalding KL, et al. 2013. Dynamics of hippocampal neurogenesis in adult humans. Cell 153, 1219–1227. ( 10.1016/j.cell.2013.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kubo S, Seleme MDC, Soifer HS, Perez JLG, Moran JV, Kazazian HH, Kasahara N. 2006. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA 103, 8036–8041. ( 10.1073/pnas.0601954103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi X, Seluanov A, Gorbunova V. 2007. Cell divisions are required for L1 retrotransposition. Mol. Cell. Biol. 27, 1264–1270. ( 10.1128/MCB.01888-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xie Y, Mates L, Ivics Z, Izsvák Z, Martin SL, An W. 2013. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob. DNA 4, 10 ( 10.1186/1759-8753-4-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mita P, et al. 2018. LINE-1 protein localization and functional dynamics during the cell cycle. Elife 7, e30058 ( 10.7554/eLife.30058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. 2011. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47. ( 10.1038/nrm3036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla M-E. 2017. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 49, 1502–1510. ( 10.1038/ng.3945) [DOI] [PubMed] [Google Scholar]
- 95.Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. 2014. Competing endogenous RNA: the key to posttranscriptional regulation. Sci. World J. 2014, 896206 ( 10.1155/2014/896206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barbash S, Simchovitz A, Buchman AS, Bennett DA, Shifman S, Soreq H. 2017. Neuronal-expressed microRNA-targeted pseudogenes compete with coding genes in the human brain. Transl. Psychiatry 7, e1199 ( 10.1038/tp.2017.163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chan W-L, Chang J-G. 2014. Pseudogene-derived endogenous siRNAs and their function. Methods Mol. Biol. 1167, 227–239. ( 10.1007/978-1-4939-0835-6_15) [DOI] [PubMed] [Google Scholar]
- 98.Muotri A, Zhao C, Marchetto M, Gage F. 2009. Environmental influence on L1 retrotransposon in the adult hippocampus. Hippocampus 19, 1002–1007. ( 10.1002/hipo.20564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han JS, Szak ST, Boeke JD. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429, 268–274. ( 10.1038/nature02536) [DOI] [PubMed] [Google Scholar]
- 100.Shukla R, et al. 2013. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111. ( 10.1016/j.cell.2013.02.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aneichyk T, et al. 2018. Dissecting the causal mechanism of X-Linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 172, 897–909.e21. ( 10.1016/j.cell.2018.02.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bragg DC, et al. 2017. Disease onset in X-linked dystonia-parkinsonism correlates with expansion of a hexameric repeat within an SVA retrotransposon in TAF1. Proc. Natl Acad. Sci. USA 114, E11020–E11028. ( 10.1073/pnas.1712526114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ito N, et al. 2016. Decreased N-TAF1 expression in X-linked dystonia-parkinsonism patient-specific neural stem cells. Dis. Model. Mech. 9, 451–462. ( 10.1242/dmm.022590) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. 2004. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 14, 2253–2260. ( 10.1101/gr.2745804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, Corbin A, Nigumann P, Cristofari G. 2016. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife 5, e13926. ( 10.7554/eLife.13926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hancks DC, Kazazian HH. 2016. Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 ( 10.1186/s13100-016-0065-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86. ( 10.1038/nrg.2016.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levin HL, Moran JV. 2011. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627. ( 10.1038/nrg3030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gilbert N, Lutz-Prigge S, Moran JV. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325. ( 10.1055/s-2008-1033461) [DOI] [PubMed] [Google Scholar]
- 110.Miné M, et al. 2007. A large genomic deletion in the PDHX gene caused by the retrotranspositional insertion of a full-length LINE-1 element. Hum. Mutat. 28, 137–142. ( 10.1002/humu.20449) [DOI] [PubMed] [Google Scholar]
- 111.Moran JV, DeBerardinis RJ, Kazazian HH. 1999. Exon shuffling by L1 retrotransposition. Science 283, 1530–1534. ( 10.1126/science.283.5407.1530) [DOI] [PubMed] [Google Scholar]
- 112.Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. 2007. Mobile DNA elements in primate and human evolution. Am. J. Phys. Anthropol. 134, 2–19. ( 10.1002/ajpa.20722) [DOI] [PubMed] [Google Scholar]
- 113.Perepelitsa-Belancio V, Deininger P. 2003. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 35, 363–366. ( 10.1038/ng1269) [DOI] [PubMed] [Google Scholar]
- 114.Mätlik K, Redik K, Speek M. 2006. L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 2006, 71753 ( 10.1155/JBB/2006/71753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Faulkner GJ, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41, 563–571. ( 10.1038/ng.368) [DOI] [PubMed] [Google Scholar]
- 116.Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21, 1973–1985. ( 10.1128/MCB.21.6.1973-1985.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Narita N, Nishio H, Kitoh Y, Ishikawa Y, Ishikawa Y, Minami R, Nakamura H, Matsuo M. 1993. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J. Clin. Invest. 91, 1862–1867. ( 10.1172/JCI116402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mülhardt C, Fischer M, Gass P, Simon-Chazottes D, Guenet J-L, Kuhse J, Betz H, Becker C-M. 1994. The spastic mouse: aberrant splicing of glycine receptor β subunit mRNA caused by intronic insertion of Ll element. Neuron 13, 1003–1015. ( 10.1016/0896-6273(94)90265-8) [DOI] [PubMed] [Google Scholar]
- 119.Kingsmore SF, Giros B, Suh D, Bieniarz M, Caron MG, Seldin MF. 1994. Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat. Genet. 7, 136–141. ( 10.1038/ng0694-136) [DOI] [PubMed] [Google Scholar]
- 120.Takahara T, et al. 1996. Dysfunction of the Orleans reeler gene arising from exon skipping due to transposition of a full-length copy of an active L1 sequence into the skipped exon. Hum. Mol. Genet. 5, 989–993. ( 10.1093/hmg/5.7.989) [DOI] [PubMed] [Google Scholar]
- 121.Kohrman DC, Harris JB, Meisler MH. 1996. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J. Biol. Chem. 271, 17 576–17 581. ( 10.1074/jbc.271.29.17576) [DOI] [PubMed] [Google Scholar]
- 122.de Bergeyck V, Nakajima K, de Rouvroit CL, Naerhuyzen B, Goffinet AM, Miyata T, Ogawa M, Mikoshiba K. 1997. A truncated Reelin protein is produced but not secreted in the ‘Orleans’ reeler mutation (Relnrl-Orl). Mol. Brain Res. 50, 85–90. ( 10.1016/S0169-328X(97)00166-6) [DOI] [PubMed] [Google Scholar]
- 123.D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. 1995. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374, 719–723. ( 10.1038/374719a0) [DOI] [PubMed] [Google Scholar]
- 124.Chang BS, et al. 2007. The role of RELN in lissencephaly and neuropsychiatric disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 58–63. ( 10.1002/ajmg.b.30392) [DOI] [PubMed] [Google Scholar]
- 125.Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. 2011. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol. Cell. Biol. 31, 300–316. ( 10.1128/MCB.00561-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goodier JL. 2016. Restricting retrotransposons: a review. Mob. DNA 7, 16 ( 10.1186/s13100-016-0070-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mortelmans K, Wang-Johanning F, Johanning GL. 2016. The role of human endogenous retroviruses in brain development and function. APMIS 124, 105–115. ( 10.1111/apm.12495) [DOI] [PubMed] [Google Scholar]
- 128.Jang HS, Shin WJ, Lee JE, Do JT. 2017. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes 8, 148 ( 10.3390/genes8060148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kozlenkov A, et al. 2016. Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic Acids Res. 44, 2593–2612. ( 10.1093/nar/gkv1304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hata K, Sakaki Y. 1997. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 189, 227–234. ( 10.1016/S0378-1119(96)00856-6) [DOI] [PubMed] [Google Scholar]
- 131.Murata Y, Bundo M, Ueda J, Kubota-Sakashita M, Kasai K, Kato T, Iwamoto K. 2017. DNA methylation and hydroxymethylation analyses of the active LINE-1 subfamilies in mice. Sci. Rep. 7, 13624 ( 10.1038/s41598-017-14165-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. 2003. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278, 4035–4040. ( 10.1074/jbc.M210256200) [DOI] [PubMed] [Google Scholar]
- 133.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. ( 10.1038/30764) [DOI] [PubMed] [Google Scholar]
- 134.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69, 905–914. ( 10.1016/0092-8674(92)90610-O) [DOI] [PubMed] [Google Scholar]
- 135.Yu F, Zingler N, Schumann G, Strätling WH. 2001. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 29, 4493–4501. ( 10.1093/nar/29.21.4493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lennartsson A, et al. 2015. Remodeling of retrotransposon elements during epigenetic induction of adult visual cortical plasticity by HDAC inhibitors. Epigenetics Chromatin 8, 55 ( 10.1186/s13072-015-0043-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. 2014. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5, 5011 ( 10.1038/ncomms6011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liao C-Y, Kennedy BK. 2016. SIRT6, oxidative stress and aging. Cell Res. 26, 143–144. ( 10.1038/cr.2016.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takasaki N, Kurokawa D, Nakayama R, Nakayama J-I, Aizawa S. 2007. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 26, 1649–1659. ( 10.1038/sj.emboj.7601619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kwon H-J, Chung H-M. 2003. Yin Yang 1, a vertebrate polycomb group gene, regulates antero-posterior neural patterning. Biochem. Biophys. Res. Commun. 306, 1008–1013. ( 10.1016/S0006-291X(03)01071-4) [DOI] [PubMed] [Google Scholar]
- 141.Rylski M, Amborska R, Zybura K, Konopacki FA, Wilczynski GM, Kaczmarek L. 2008. Yin Yang 1 expression in the adult rodent brain. Neurochem. Res. 33, 2556–2564. ( 10.1007/s11064-008-9757-y) [DOI] [PubMed] [Google Scholar]
- 142.Becker KG, Swergold GD, Ozato K, Thayer RE. 1993. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet. 2, 1697–1702. ( 10.1093/hmg/2.10.1697) [DOI] [PubMed] [Google Scholar]
- 143.Athanikar JN, Badge RM, Moran JV. 2004. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32, 3846–3855. ( 10.1093/nar/gkh698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yang N, Zhang L, Zhang Y, Kazazian HH. 2003. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 31, 4929–4940. ( 10.1093/nar/gkg663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Inoue K, Shiga T, Ito Y. 2008. Runx transcription factors in neuronal development. Neural Dev. 3, 20 ( 10.1186/1749-8104-3-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lallemend F, et al. 2012. Positional differences of axon growth rates between sensory neurons encoded by Runx3. EMBO J. 31, 3718–3729. ( 10.1038/emboj.2012.228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Graham V, Khudyakov J, Ellis P, Pevny L. 2003. SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765. ( 10.1016/S0896-6273(03)00497-5) [DOI] [PubMed] [Google Scholar]
- 148.Ring KL, et al. 2012. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109. ( 10.1016/j.stem.2012.05.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Heinrich C, Bergami M, Gascón S, Lepier A, Viganò F, Dimou L, Sutor B, Berninger B, Götz M. 2014. Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Rep. 3, 1000–1014. ( 10.1016/j.stemcr.2014.10.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tedeschi A, Di Giovanni S. 2009. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 10, 576–583. ( 10.1038/embor.2009.89) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wylie A, et al. 2015. p53 genes function to restrain mobile elements. Genes Dev. 30, 64–77. ( 10.1101/gad.266098.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J. 2009. p53 responsive elements in human retrotransposons. Oncogene 28, 3857–3865. ( 10.1038/onc.2009.246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Garcia-Perez JL, et al. 2010. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466, 769–773. ( 10.1038/nature09209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ewing AD, Kazazian HH. 2011. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 21, 985–990. ( 10.1101/gr.114777.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. 2014. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 3, e02008 ( 10.7554/eLife.02008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Buckley RM, Kortschak RD, Raison JM, Adelson DL. 2017. Similar evolutionary trajectories for retrotransposon accumulation in mammals. Genome Biol. Evol. 9, 2336–2353. ( 10.1093/gbe/evx179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.El-Sawy M, Kale SP, Dugan C, Nguyen TQ, Belancio V, Bruch H, Roy-Engel AM, Deininger PL. 2005. Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J. Mol. Biol. 354, 246–257. ( 10.1016/j.jmb.2005.09.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kale S, Carmichael M, Harris K, Roy-Engel A. 2006. The L1 retrotranspositional stimulation by particulate and soluble cadmium exposure is independent of the generation of DNA breaks. Int. J. Environ. Res. Public Health 3, 121–128. ( 10.3390/ijerph2006030015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kale S, Moore L, Deininger P, Roy-Engel A. 2005. Heavy metals stimulate human LINE-1 retrotransposition. Int. J. Environ. Res. Public Health 2, 14–23. ( 10.3390/ijerph2005010014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Giorgi G, Marcantonio P, Del Re B. 2011. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res. 346, 383–391. ( 10.1007/s00441-011-1289-0) [DOI] [PubMed] [Google Scholar]
- 161.Stribinskis V, Ramos KS. 2006. Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 66, 2616–2620. ( 10.1158/0008-5472.CAN-05-3478) [DOI] [PubMed] [Google Scholar]
- 162.Morales JF, Snow ET, Murnane JP. 2003. Environmental factors affecting transcription of the human L1 retrotransposon. II. Stressors. Mutagenesis 18, 151–158. ( 10.1093/mutage/18.2.151) [DOI] [PubMed] [Google Scholar]
- 163.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31, 159–165. ( 10.1038/ng898) [DOI] [PubMed] [Google Scholar]
- 164.Farkash EA, Kao GD, Horman SR, Prak ETL. 2006. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 34, 1196–1204. ( 10.1093/nar/gkj522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Teng SC, Kim B, Gabriel A. 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383, 641–644. ( 10.1038/383641a0) [DOI] [PubMed] [Google Scholar]
- 166.Perron H, et al. 2012. Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult. Scler. 18, 1721–1736. ( 10.1177/1352458512441381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.van Horssen J, van der Pol S, Nijland P, Amor S, Perron H. 2016. Human endogenous retrovirus W in brain lesions: rationale for targeted therapy in multiple sclerosis. Mult. Scler. Relat. Disord. 8, 11–18. ( 10.1016/j.msard.2016.04.006) [DOI] [PubMed] [Google Scholar]
- 168.Antony JM, Deslauriers AM, Bhat RK, Ellestad KK, Power C. 2011. Human endogenous retroviruses and multiple sclerosis: innocent bystanders or disease determinants? Biochim. Biophys. Acta 1812, 162–176. ( 10.1016/j.bbadis.2010.07.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shuvarikov A, et al. 2013. Recurrent HERV-H-mediated 3q13.2–q13.31 deletions cause a syndrome of hypotonia and motor, language, and cognitive delays. Hum. Mutat. 34, 1415–1423. ( 10.1002/humu.22384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Douville R, Liu J, Rothstein J, Nath A. 2011. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann. Neurol. 69, 141–151. ( 10.1002/ana.22149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li W, et al. 2015. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 7, 307ra153 ( 10.1126/scitranslmed.aac8201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Laska MJ, Brudek T, Nissen KK, Christensen T, Møller-Larsen A, Petersen T, Nexø BA. 2012. Expression of HERV-Fc1, a human endogenous retrovirus, is increased in patients with active multiple sclerosis. J. Virol. 86, 3713–3722. ( 10.1128/JVI.06723-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Frank O, Giehl M, Zheng C, Hehlmann R, Leib-Mösch C, Seifarth W. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 79, 10 890–10 901. ( 10.1128/JVI.79.17.10890-10901.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Karlsson H, Bachmann S, Schröder J, McArthur J, Torrey EF, Yolken RH. 2001. Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl Acad. Sci. USA 98, 4634–4639. ( 10.1073/pnas.061021998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Perron H, et al. 2012. Molecular characteristics of human endogenous retrovirus type-W in schizophrenia and bipolar disorder. Transl. Psychiatry 2, e201 ( 10.1038/tp.2012.125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Guffanti G, et al. 2016. LINE1 insertions as a genomic risk factor for schizophrenia: preliminary evidence from an affected family. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 534–545. ( 10.1002/ajmg.b.32437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Weis S, Llenos IC, Sabunciyan S, Dulay JR, Isler L, Yolken R, Perron H. 2007. Reduced expression of human endogenous retrovirus (HERV)-W GAG protein in the cingulate gyrus and hippocampus in schizophrenia, bipolar disorder, and depression. J. Neural Transm. 114, 645–655. ( 10.1007/s00702-006-0599-y) [DOI] [PubMed] [Google Scholar]
- 178.Christensen T. 2016. Human endogenous retroviruses in neurologic disease. APMIS 124, 116–126. ( 10.1111/apm.12486) [DOI] [PubMed] [Google Scholar]
- 179.Balestrieri E, et al. 2012. HERVs expression in autism spectrum disorders. PLoS ONE 7, e48831 ( 10.1371/journal.pone.0048831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Balestrieri E, et al. 2014. Human endogenous retroviruses and ADHD. World J. Biol. Psychiatry 15, 499–504. ( 10.3109/15622975.2013.862345) [DOI] [PubMed] [Google Scholar]
- 181.Kondo-Iida E, et al. 1999. Novel mutations and genotype–phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD). Hum. Mol. Genet. 8, 2303–2309. ( 10.1093/hmg/8.12.2303) [DOI] [PubMed] [Google Scholar]
- 182.Wimmer K, Callens T, Wernstedt A, Messiaen L. 2011. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 7, e1002371 ( 10.1371/journal.pgen.1002371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bernard V, Minnerop M, Bürk K, Kreuz F, Gillessen-Kaesbach G, Zühlke C. 2009. Exon deletions and intragenic insertions are not rare in ataxia with oculomotor apraxia 2. BMC Med. Genet. 10, 87 ( 10.1186/1471-2350-10-87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Carreira PE, et al. 2016. Evidence for L1-associated DNA rearrangements and negligible L1 retrotransposition in glioblastoma multiforme. Mob. DNA 7, 21 ( 10.1186/s13100-016-0076-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Achanta P, et al. 2016. Somatic retrotransposition is infrequent in glioblastomas. Mob. DNA 7, 22 ( 10.1186/s13100-016-0077-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martínez-Garay I, Ballesta M, Oltra S, Orellana C, Palomeque A, Moltó M, Prieto F, Martínez F. 2003. Intronic L1 insertion and F268S, novel mutations in RPS6KA3 (RSK2) causing Coffin–Lowry syndrome. Clin. Genet. 64, 491–496. ( 10.1046/j.1399-0004.2003.00166.x) [DOI] [PubMed] [Google Scholar]
- 187.Bundo M, et al. 2014. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron 81, 306–313. ( 10.1016/j.neuron.2013.10.053) [DOI] [PubMed] [Google Scholar]
- 188.Liu S, Du T, Liu Z, Shen Y, Xiu J, Xu Q. 2016. Inverse changes in L1 retrotransposons between blood and brain in major depressive disorder. Sci. Rep. 6, 37530 ( 10.1038/srep37530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Doyle GA, Crist RC, Karatas ET, Hammond MJ, Ewing AD, Ferraro TN, Hahn C-G, Berrettini WH. 2017. Analysis of LINE-1 elements in DNA from postmortem brains of individuals with schizophrenia. Neuropsychopharmacology 42, 2602–2611. ( 10.1038/npp.2017.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shpyleva S, Melnyk S, Pavliv O, Pogribny I, Jill James S. 2017. Overexpression of LINE-1 retrotransposons in autism brain. Mol. Neurobiol. 55, 1740–1749. ( 10.1007/s12035-017-0421-x) [DOI] [PubMed] [Google Scholar]
- 191.Doyle GA, Doucet-O'Hare TT, Hammond MJ, Crist RC, Ewing AD, Ferraro TN, Mash DC, Kazazian HH, Berrettini WH. 2017. Reading LINEs within the cocaine addicted brain. Brain Behav. 7, 1–12. ( 10.1002/brb3.678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rusiecki JA, Chen L, Srikantan V, Zhang L, Yan L, Polin ML, Baccarelli A. 2012. DNA methylation in repetitive elements and post-traumatic stress disorder: a case–control study of US military service members. Epigenomics 4, 29–40. ( 10.2217/epi.11.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gallus GN, et al. 2010. Alu-element insertion in an OPA1 intron sequence associated with autosomal dominant optic atrophy. Mol. Vis. 16, 178–183. [PMC free article] [PubMed] [Google Scholar]
- 194.Kutsche K, Ressler B, Katzera H-G, Orth U, Gillessen-Kaesbach G, Morlot S, Schwinger E, Gal A. 2002. Characterization of breakpoint sequences of five rearrangements in L1CAM and ABCD1 (ALD) genes. Hum. Mutat. 19, 526–535. ( 10.1002/humu.10072) [DOI] [PubMed] [Google Scholar]
- 195.Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS. 1991. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353, 864–866. ( 10.1038/353864a0) [DOI] [PubMed] [Google Scholar]
- 196.Okubo M, Horinishi A, Saito M, Ebara T, Endo Y, Kaku K, Murase T, Eto M. 2007. A novel complex deletion-insertion mutation mediated by Alu repetitive elements leads to lipoprotein lipase deficiency. Mol. Genet. Metab. 92, 229–233. ( 10.1016/j.ymgme.2007.06.018) [DOI] [PubMed] [Google Scholar]
- 197.Conceição Pereira M, et al. 2012. Alu elements mediate large SPG11 gene rearrangements: further spatacsin mutations. Genet. Med. 14, 143–151. ( 10.1038/gim.2011.7) [DOI] [PubMed] [Google Scholar]
- 198.Boone PM, Liu P, Zhang F, Carvalho CMB, Towne CF, Batish SD, Lupski JR. 2011. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 13, 582–592. ( 10.1097/GIM.0b013e3182106775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ricci V, Regis S, Di Duca M, Filocamo M. 2003. An Alu-mediated rearrangement as cause of exon skipping in Hunter disease. Hum. Genet. 112, 419–425. ( 10.1007/s00439-002-0900-6) [DOI] [PubMed] [Google Scholar]
- 200.Gu Y, Kodama H, Watanabe S, Kikuchi N, Ishitsuka I, Ozawa H, Fujisawa C, Shiga K. 2007. The first reported case of Menkes disease caused by an Alu insertion mutation. Brain Dev. 29, 105–108. ( 10.1016/j.braindev.2006.05.012) [DOI] [PubMed] [Google Scholar]
- 201.Bouchet C, et al. 2007. Detection of an Alu insertion in the POMT1 gene from three French Walker Warburg syndrome families. Mol. Genet. Metab. 90, 93–96. ( 10.1016/j.ymgme.2006.09.005) [DOI] [PubMed] [Google Scholar]
- 202.Brooks EM, Branda RF, Nicklas JA, O'Neill JP. 2001. Molecular description of three macro-deletions and an Alu–Alu recombination-mediated duplication in the HPRT gene in four patients with Lesch–Nyhan disease. Mutat. Res. 476, 43–54. ( 10.1016/S0027-5107(01)00065-3) [DOI] [PubMed] [Google Scholar]
- 203.Huie ML, Shanske AL, Kasper JS, Marion RW, Hirschhorn R. 1999. A large Alu-mediated deletion, identified by PCR, as the molecular basis for glycogen storage disease type II (GSDII). Hum. Genet. 104, 94–98. ( 10.1007/s004390050916) [DOI] [PubMed] [Google Scholar]
- 204.Vogt J, et al. 2014. SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints. Genome Biol. 15, R80 ( 10.1186/gb-2014-15-6-r80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Makino S, et al. 2007. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am. J. Hum. Genet. 80, 393–406. ( 10.1086/512129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Taniguchi-Ikeda M, et al. 2011. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature 478, 127–131. ( 10.1038/nature10456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Selig S, Bruno S, Scharf JM, Wang CH, Vitale E, Gilliam TC, Kunkel LM. 1995. Expressed cadherin pseudogenes are localized to the critical region of the spinal muscular atrophy gene. Proc. Natl Acad. Sci. USA 92, 3702–3706. ( 10.1073/pnas.92.9.3702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bagot RC, Labonté B, Peña CJ, Nestler EJ. 2014. Epigenetic signaling in psychiatric disorders: stress and depression. Dialogues Clin. Neurosci. 16, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Douville RN, Nath A. 2014. Human endogenous retroviruses and the nervous system. Handb. Clin. Neurol. 123, 465–485. ( 10.1016/B978-0-444-53488-0.00022-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Meyer TJ, Rosenkrantz JL, Carbone L, Chavez SL. 2017. Endogenous retroviruses: with us and against us. Front. Chem. 5, 23 ( 10.3389/fchem.2017.00023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Quinn JP, Bubb VJ. 2014. SVA retrotransposons as modulators of gene expression. Mob. Genet. Elements 4, e32102 ( 10.4161/mge.32102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gianfrancesco O, Bubb VJ, Quinn JP. 2017. SVA retrotransposons as potential modulators of neuropeptide gene expression. Neuropeptides 64, 3–7. ( 10.1016/j.npep.2016.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188. ( 10.1038/13810) [DOI] [PubMed] [Google Scholar]
- 214.Akindipe T, Wilson D, Stein DJ. 2014. Psychiatric disorders in individuals with methamphetamine dependence: prevalence and risk factors. Metab. Brain Dis. 29, 351–357. ( 10.1007/s11011-014-9496-5) [DOI] [PubMed] [Google Scholar]
- 215.Zhornitsky S, Tikàsz A, Rizkallah É, Chiasson J-P, Potvin S. 2015. Psychopathology in substance use disorder patients with and without substance-induced psychosis. J. Addict. 2015, 843762 ( 10.1155/2015/843762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Okudaira N, Ishizaka Y, Nishio H. 2014. Retrotransposition of long interspersed element 1 induced by methamphetamine or cocaine. J. Biol. Chem. 289, 25 476–25 485. ( 10.1074/jbc.M114.559419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Moszczynska A, Flack A, Qiu P, Muotri AR, Killinger BA. 2015. Neurotoxic methamphetamine doses increase LINE-1 expression in the neurogenic zones of the adult rat brain. Sci. Rep. 5, 14356 ( 10.1038/srep14356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bassett AS, Chow EWC, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R. 2003. The schizophrenia phenotype in 22q11 deletion syndrome. Am. J. Psychiatry 160, 1580–1586. ( 10.1176/appi.ajp.160.9.1580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Basil P, Li Q, Dempster EL, Mill J, Sham P-C, Wong CCY, McAlonan GM. 2014. Prenatal maternal immune activation causes epigenetic differences in adolescent mouse brain. Transl. Psychiatry 4, e434 ( 10.1038/tp.2014.80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li T-H, Schmid CW. 2001. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene 276, 135–141. ( 10.1016/S0378-1119(01)00637-0) [DOI] [PubMed] [Google Scholar]
- 221.Jellinger KA. 2001. Cell death mechanisms in neurodegeneration. J. Cell. Mol. Med. 5, 1–17. ( 10.1111/j.1582-4934.2001.tb00134.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Savitsky K, et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268, 1749–1753. ( 10.1126/science.7792600) [DOI] [PubMed] [Google Scholar]
- 223.Shiloh Y. 2001. ATM (ataxia telangiectasia mutated): expanding roles in the DNA damage response and cellular homeostasis. Biochem. Soc. Trans. 29, 661–666. ( 10.1042/bst0290661) [DOI] [PubMed] [Google Scholar]
- 224.Wilson AC, Dugger BN, Dickson DW, Wang D-S. 2011. TDP-43 in aging and Alzheimer's disease—a review. Int. J. Clin. Exp. Pathol. 4, 147–155. [PMC free article] [PubMed] [Google Scholar]
- 225.Cohen TJ, Lee VMY, Trojanowski JQ. 2011. TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol. Med. 17, 659–667. ( 10.1016/j.molmed.2011.06.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li W, Jin Y, Prazak L, Hammell M, Dubnau J. 2012. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE 7, e44099 ( 10.1371/journal.pone.0044099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Krug L, et al. 2017. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 13, e1006635 ( 10.1371/journal.pgen.1006635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Li W, Prazak L, Chatterjee N, Grüninger S, Krug L, Theodorou D, Dubnau J. 2013. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 16, 1–4. ( 10.1038/nn.3368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Larsen PA, Lutz MW, Hunnicutt KE, Mihovilovic M, Saunders AM, Yoder AD, Roses AD. 2017. The Alu neurodegeneration hypothesis: a primate-specific mechanism for neuronal transcription noise, mitochondrial dysfunction, and manifestation of neurodegenerative disease. Alzheimers. Dement. 13, 828–838. ( 10.1016/j.jalz.2017.01.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.de Andrade A, Wang M, Bonaldo MF, Xie H, Soares MB. 2011. Genetic and epigenetic variations contributed by Alu retrotransposition. BMC Genomics 12, 617 ( 10.1186/1471-2164-12-617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lin L, Jiang P, Park JW, Wang J, Lu Z-X, Lam MPY, Ping P, Xing Y. 2016. The contribution of Alu exons to the human proteome. Genome Biol. 17, 15 ( 10.1186/s13059-016-0876-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.2006. NerveCenter: (1) Eric Kandel on the path to the Nobel Prize: one cell at a time (2) short- and long-term memory in aplysia. Ann. Neurol. 59, A11–A13. ( 10.1002/ana.20909) [DOI] [PubMed] [Google Scholar]
- 233.Metzakopian E, et al. 2017. Enhancing the genome editing toolbox: genome wide CRISPR arrayed libraries. Sci. Rep. 7, 2244 ( 10.1038/s41598-017-01766-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.