The causal agent of white-nose syndrome in bats is Pseudogymnoascus destructans, a filamentous fungus recently introduced from its native range in Europe. Infections caused by P. destructans have progressed across the eastern parts of Canada and the United States over the last 10 years. It is not clear how the disease is spread, as the pathogen is unable to grow above 23°C and ambient temperature can act as a barrier when hosts disperse. Here, we explore the patterns of phenotypic diversity and the germination of the fungal asexual spores, arthroconidia, from strains across a sizeable area of the epizootic range. Our analyses revealed evidence of adaptation along geographic gradients during its expansion. The results have implications for understanding the diversification of P. destructans and the limits of WNS spread in North America. Given the rapidly expanding distribution of WNS, a detailed understanding of the genetic bases for phenotypic variations in growth, reproduction, and dispersal of P. destructans is urgently needed to help control this disease.
KEYWORDS: clonal expansion, fungal pathogen, adaptation, colony size, pigmentation, pigment diffusion, spore germination, genome sequencing
ABSTRACT
White-nose syndrome (WNS) is an ongoing epizootic affecting multiple species of North American bats, caused by epidermal infections of the psychrophilic filamentous fungus Pseudogymnoascus destructans. Since its introduction from Europe, WNS has spread rapidly across eastern North America and resulted in high mortality rates in bats. At present, the mechanisms behind its spread and the extent of its adaptation to different geographic and ecological niches remain unknown. The objective of this study was to examine the geographic patterns of phenotypic variation and the potential evidence for adaptation among strains representing broad geographic locations in eastern North America. The morphological features of these strains were evaluated on artificial medium, and the viability of asexual arthroconidia of representative strains was investigated after storage at high (23°C), moderate (14°C), and low (4°C) temperatures at different lengths of time. Our analyses identified evidence for a geographic pattern of colony morphology changes among the clonal descendants of the fungus, with trait values correlated with increased distance from the epicenter of WNS. Our genomic comparisons of three representative isolates revealed novel genetic polymorphisms and suggested potential candidate mutations that might be related to some of the phenotypic changes. These results show that even though this pathogen arrived in North America only recently and reproduces asexually, there has been substantial evolution and phenotypic diversification during its rapid clonal expansion.
IMPORTANCE The causal agent of white-nose syndrome in bats is Pseudogymnoascus destructans, a filamentous fungus recently introduced from its native range in Europe. Infections caused by P. destructans have progressed across the eastern parts of Canada and the United States over the last 10 years. It is not clear how the disease is spread, as the pathogen is unable to grow above 23°C and ambient temperature can act as a barrier when hosts disperse. Here, we explore the patterns of phenotypic diversity and the germination of the fungal asexual spores, arthroconidia, from strains across a sizeable area of the epizootic range. Our analyses revealed evidence of adaptation along geographic gradients during its expansion. The results have implications for understanding the diversification of P. destructans and the limits of WNS spread in North America. Given the rapidly expanding distribution of WNS, a detailed understanding of the genetic bases for phenotypic variations in growth, reproduction, and dispersal of P. destructans is urgently needed to help control this disease.
INTRODUCTION
White-nose syndrome (WNS) is an epidermal infection of North American (NA) bat species, caused by the psychrophilic fungal pathogen, Pseudogymnoascus destructans (formerly Geomyces destructans) (1). WNS infection is characterized by the presence of white mycelial growth on the muzzle or wing tissues of bats and eventually leads to the formation of ulcers and the erosion of epithelial tissues (2). Torpid bats with WNS experience electrolyte imbalance and evaporative water loss (3) and tend to rouse more frequently from hibernation than noninfected bats (4). These additional arousal events are energetically very taxing (5) and cause further depletion of fat reserves, starvation, and death (6, 7). Indeed, since its discovery in 2006 (8), P. destructans has caused an estimated 5.7 to 6.7 million cases of mortality (9) in 31 U.S. states and 5 Canadian provinces and resulted in population collapses within some regions of the northeastern United States (10). In contrast, the ancestral P. destructans populations from Europe do not cause obvious diseases in European bats (11, 12). The rapid spread and high mortality rate of NA bats associated with WNS is consistent with the novel pathogen hypothesis (13). However, despite significant efforts in monitoring and controlling the spread of the WNS epizootic, the timing, route, and mode of WNS expansion remain largely unknown.
Several factors have been proposed to influence WNS transmission, including the sociality of the host species, population density, and the local climate (11, 13, 14). Research has shown that WNS can be transmitted by direct bat-to-bat contact right before hibernation (12). Within NA, the current distribution of P. destructans and WNS covers a large latitudinal and longitudinal range and great variation in climate and other conditions. One of the factors is temperature. NA bat species hibernate in sites with temperatures that generally range between 3 and 15°C (15, 16). Within the hibernacula, the conditions conducive to P. destructans growth, 12.5 to 15.8°C and greater than 70% relative humidity (12), may mirror those outside the hibernacula, but perhaps with greater stability than those outside. However, this is only true of caves with limited air circulation (16). Generally, the variance of cave temperatures decreases with increasing latitude (17). Within individual hibernacula, a temperature gradient may also exist (16) (see Fig. S1A in the supplemental material). Overall, with decreasing seasonal temperatures at increasing latitudes (Fig. S1B), northern caves and hibernacula are more likely to maintain cooler temperatures for longer periods of time than southern caves and hibernacula. Since P. destructans growth is known to be extremely limited at temperatures greater than 20°C (18), such differences in temperature could impact the direction of adaptation and influence the transmission and spread of WNS differently between the southern and northern regions. The rapid expansion is especially intriguing, because the NA P. destructans population is known to be descended from a single clone through asexual reproduction (19). However, whether the different ecological conditions among regions in NA have impacted the adaptation and phenotypic characteristics of P. destructans remains unknown. Analyzing the geographic patterns of phenotypic changes in this pathogen and how phenotypic changes may be influenced by environmental factors will help understand the evolution of this pathogen and protect the bat populations (13).
In many filamentous ascomycetes such as P. destructans, arthroconidia are the propagules that prolong the survival of the population during environmental stress and facilitate the dispersal of the fungi to other ecological niches. Previous research has shown that P. destructans produces abundant asexual spores, arthroconidia (1), which likely play a significant role in the spread of this pathogen among geographic regions. While the methods by which WNS transmission could be facilitated have been suggested (20), the mechanisms behind the rapid geographic expansion of WNS in NA have not been adequately explained. For example, if 90 to 100% of infections cause mortality and the increased temperatures that these bats experience after the hibernation period ends could eliminate P. destructans (18), then how does this epizootic continue to spread? Also, given the large differences in ecological niche conditions across the latitudinal gradients in eastern NA, how might a clonal genotype respond to a diversity of ecological niches? For P. destructans to spread, arthroconidia must be able to withstand temperatures outside the optimal range for vegetative growth.
Here, we hypothesize that during the clonal expansion of P. destructans in eastern NA, there has been significant phenotypic diversification along geographic gradients and that some of the phenotypic variations are associated with or are indicative of local adaptations. To test these hypotheses, we explored the geographic patterns of phenotypic changes among representative isolates from the WNS epizootic in eastern NA. Our analyses identified evidence for geographic patterns of phenotypic variation and suggest that adaptation to low temperatures has likely occurred during its northern spread to eastern Canada. In addition, mutations potentially associated with such an adaptation were investigated using whole-genome sequencing of representative isolates.
RESULTS
We surveyed the colony morphology of 62 P. destructans isolates, including 47 Canadian isolates from four provinces and 15 isolates from six U.S. states (Fig. 1). The total area represented by these isolates covers 2 × 106 km2 (approximately one-fifth of the area of the United States). This collection of clonal strains has been preserved at −80°C since isolation from their natural environment, and the isolations spanned 5 years, from 2008 to 2013. Based on multilocus sequence typing and PCR fingerprinting (21), all these isolates were assumed to have descended clonally from a single genotype in NA, as represented by the early isolate, US15.
FIG 1.
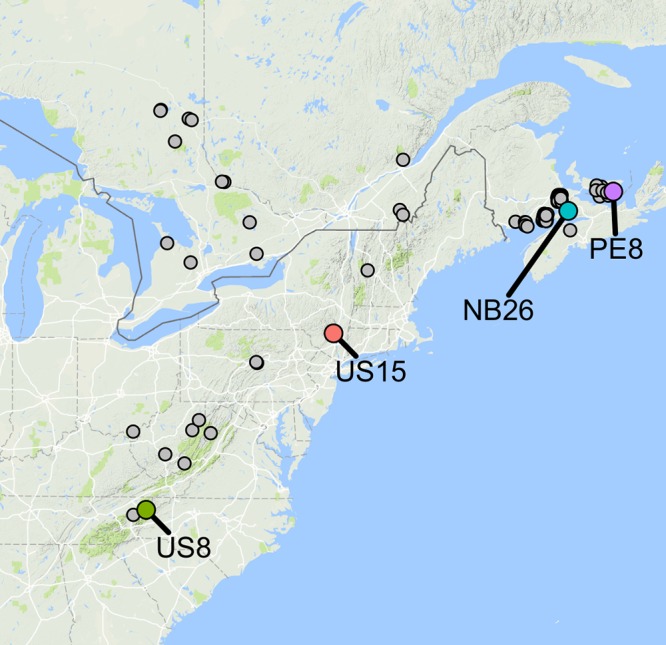
Geographic locations of the isolates analyzed in this study. While all isolates were examined for their colony phenotype, only the four highlighted ones (US8, US15, NB26, and PE8) were examined for their arthroconidial survival and genetic differences.
Geographical pattern of phenotypic differences.
The strains showed variation in all three colony morphology traits: colony size, colony surface pigmentation, and the extent of pigment diffusion through agar. To analyze the geographic patterns of phenotypic variations, we first considered the variation of each trait as a function of the relative distance from the location of the earliest P. destructans sample, William's Hotel mine in New York State (Fig. 2; see also Fig. S2 in the supplemental material). We then compared the variance/covariance of the collection of phenotypic traits using a multivariate linear mixed model (see Fig. S3). Overall, the colony phenotype divergences (colony size, pigment production, and pigment diffusion) were correlated with increased geographic distance from the epicenter. However, we also observed heterogeneity among isolates from the same geographical regions, especially within eastern Canadian provinces. Below, we describe the results for each of the traits.
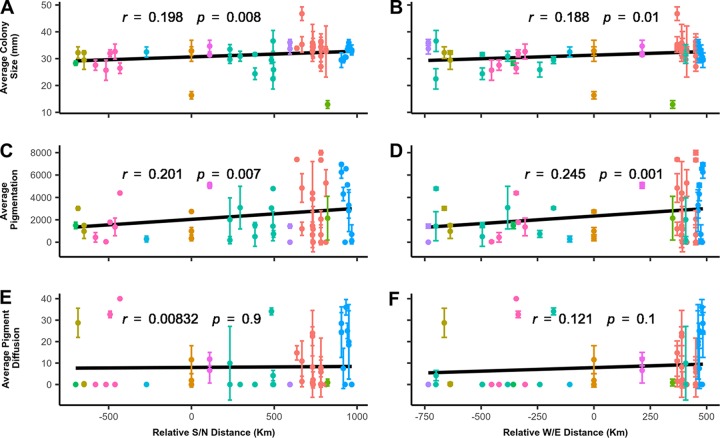
FIG 2.
Measurements of P. destructans colony phenotypic traits at 14°C after 28 days of growth. (A) Fungal colony area on agar plates over west/east (S/N) relative distances. (B) Fungal colony area on agar plates over west/east (W/E) relative distances. (C) Fungal colony surface pigmentation along the S/N relative distances. (D) Fungal colony surface pigmentation over W/E relative distances. (E) Fungal colony pigment diffusion over S/N relative distances. (F) Fungal colony pigment diffusion over W/E relative distances.
P. destructans colony size.
Mycelial growth can provide insights into the efficiency of substrate utilization and reproduction under specific environmental conditions. Overall, we found a positive correlation between the variability in measurements of colony size on petri plates and the geographic distance to the epicenter (Fig. S2A). Furthermore, the positive correlation was statistically significant along both the south/north axis (F = 2.8, r = 0.2, P = 0.008) (Fig. 2A) and the west/east axis (F = 3, r = 0.19, P = 0.01) (Fig. 2B). Isolates from eastern Canada, particularly cultures from New Brunswick, generally had larger colony sizes at 14°C than those from the United States (Fig. 2A). Though the change in colony size showed a significant correlation with the geographic distance from the epicenter of the outbreak, the amounts of variation among all strains at each distance bin (of every ∼50 km) were overall very similar (Fig. S2A). However, as the isolates spread further in the western direction, there was an increased variation among the strains in their mycelial growth (F = 0.4776, r = −0.6, P = 0.01) (Fig. S2B).
Surface pigmentation.
Pigmentation is considered a virulence factor in many pathogenic fungi and contributes to resistance toward multiple environmental stressors (22). In this study, the majority of the 62 isolates displayed dense pigmentation on the tested medium and under the incubation condition. However, significant variations in pigmentation were observed among strains. A small proportion of the isolates in this survey lacked surface pigmentation. Over a geographical gradient, the amounts of colony surface pigmentation showed moderate but statistically significant correlations with increasing northern/southern (F = 2.5, r = 0.2, P = 0.007) (Fig. 2C) and eastern/western distances (F = 3.5, r = 0.25, P = 0.001) (Fig. 2D) from the epicenter. Furthermore, we found a positive correlation between the variance in surface pigmentation and the distance from the epicenter as the pathogen spread northward (F = 6.7, r = 0.6, P = 0.02) (Fig. S2C) and eastward (F = 0.05835, r = 0.78, P = 0.04) (Fig. S2D). However, there was no increase in the variance of surface pigmentation in the pathogen's spread either westwards (F = 7.8, r = 0.1, P = 0.7) (Fig. S2D) or southwards (F = 0.06, r = 0, P = 0.8) (Fig. S2C).
Pigment diffusion through agar.
Whereas the secretion of pigments within hyphae of P. destructans likely confers some form of resistance to exogenous stresses, the deposition of pigments into surrounding medium may contribute to their competitiveness within the substrate. Although variation was observed among strains in the deposition of pigments into surrounding solid medium, in contrast to that observed for colony surface pigmentation, there was no correlation between the amounts of pigment diffusion through the agar medium and the relative geographic distances from the epicenter of the outbreak, either in total geographic distance or in longitudinal/latitudinal distance (F = 0.25, r < 0.1, P > 0.05) (Fig. 2E and F). Interestingly, though statistically not significant, as WNS spread, there were increasing variances among strains in pigment diffusion in all directions, with the eastward strains showing the strongest correlation (F = 2.5, r = 0.478, P = 0.3) (Fig. S2F). This result is consistent with continued divergence among strains.
Relationships among the three colony phenotypes.
A summary of the results from the multivariate linear mixed model is presented in a correlogram, shown in Fig. S3. Our analyses showed that colony surface pigmentation was positively correlated with pigment diffusion in agar (r = 0.5, P < 0.001). Similarly, colony size and pigment diffusion were positively correlated. However, their correlation was weak (r = 0.09, P = 0.04). Interestingly, although the correlation coefficient was larger than that between colony size and pigment diffusion, that between colony size and surface pigmentation was statistically insignificant (r = 0.15, P = 0.23).
Arthroconidial survival.
From the 62 isolates, we selected four representatives, on the basis of their geographical origins and divergent colony phenotypes, to further examine the effect of storage at various temperatures on arthroconidial viability. For each isolate, we created arthroconidial suspensions in sterile water, air-dried them, and exposed them to 4°C, 14°C, and 23°C for various lengths of time before they were rehydrated and plated on Sabouraud dextrose agar (SDA) plates for germination. A reference arthroconidial germination rate was obtained for each isolate using rehydrated asexual spores immediately after they were dehydrated, without any storage. The reference germination rates varied from 21.1% to 38.2% among the four isolates. In general, the storage of arthroconidia at all three temperatures led to steadily decreased germination rates (Table 1). Among the three temperatures, arthroconidia stored at 4°C had the highest germination rate for the greatest length of time. In general, the two isolates (NB26 and PE8) from further north fared better than the two from the south (US15 and US8) after long-term exposure to 4°C and 14°C temperatures. Lastly, exposure to 23°C resulted in no germination after 28 days of storage. Interestingly, arthroconidia from the early isolate in the WNS epizootic, US15, had the highest germination rate after storage at 23°C for 21 days. The detailed germination rates for all four isolates at the five specific time points after storage at each of the three temperatures are presented in Table 1. Also shown in Table 1 are the F statistic test values of the comparisons between pairs of strains in their arthroconidial germination rates at each of the 18 treatments (3 temperatures × 6 time points of storage).
TABLE 1.
Viability of arthroconidia of four strains of P. destructans stored at three different temperatures for various lengths of timea
| Temp (°C) | Day |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 |
1 |
7 |
14 |
21 |
28 |
|||||||||||||
| CFU | F | P value | CFU | F | P value | CFU | F | P value | CFU | F | P value | CFU | F | P value | CFU | F | P value | |
| 4 | ||||||||||||||||||
| NB26 | 42.8 ± 37.5 A | 1.1 | 0.36 | 59.3 ± 25.1 A | 3.7 | 0.02 | 35.9 ± 18.2 AB | 2.54 | 0.074 | 14.4 ± 2.6 AB | 3.92 | 0.017 | 31.8 ± 17.2 A | 5.53 | 0.004 | 9.4 ± 2.5 B | 30.39 | <0.001 |
| PE8 | 33.2 ± 8.5 A | 37.6 ± 14.3 A | 49.8 ± 26.1 A | 15.3 ± 2.7 A | 46.7 ± 7.4 A | 13.8 ± 1.9 A | ||||||||||||
| US15 | 36.9 ± 3.7 A | 52.3 ± 13.2 A | 27.9 ± 9.2 B | 10.3 ± 2.2 B | 15.1 ± 10.9 B | 2.7 ± 1.5 C | ||||||||||||
| US8 | 20.4 ± 6.2 A | 34.6 ± 10.4 A | 34 ± 8.1 AB | 11.9 ± 5.6 AB | 34.6 ± 12 A | 9 ± 3.9 B | ||||||||||||
| 14 | ||||||||||||||||||
| NB26 | 13.7 ± 3.2 B | 6.7 | 0.001 | 9.7 ± 3.5 A | 2.84 | 0.054 | 10.3 ± 3.1 A | 27.6 | <0.001 | 4.8 ± 4.1 BC | 11.9 | <0.001 | 6.9 ± 1.8 A | 8.1 | <0.001 | 2.4 ± 1.2 B | 14.8 | <0.001 |
| PE8 | 7 ± 2 C | 18.2 ± 10.6 A | 4.5 ± 2.6 B | 8 ± 1.9 A | 2.1 ± 0.4 B | 12.2 ± 7.5 A | ||||||||||||
| US15 | 6.5 ± 1.7 C | 9.5 ± 4.2 A | 2 ± 0.4 C | 2.3 ± 0.8 C | 1.7 ± 0.2 B | 0.3 ± 0.1 B | ||||||||||||
| US8 | 22.3 ± 5 A | 12.3 ± 3.1 A | 2 ± 0.4 C | 7.5 ± 1.8 AB | 4.2 ± 4.4 AB | 9.3 ± 7.2 A | ||||||||||||
| 23 | ||||||||||||||||||
| NB26 | 11.2 ± 7.7 C | 23 | <0.001 | 13.8 ± 7.9 B | 6.85 | 0.001 | 1.1 ± 0.2 B | 20 | <0.001 | 0.3 ± 0.2 B | 49.7 | <0.001 | 0.3 ± 0.2 B | 5.9 | 0.003 | 0 ± 0 A | 10.7 | <0.001 |
| PE8 | 81.4 ± 13.1 A | 26.9 ± 6.4 B | 3.8 ± 1.3 B | 0.5 ± 0.3 B | 0.2 ± 0 B | 0 ± 0 A | ||||||||||||
| US15 | 25.2 ± 5.7 B | 21.7 ± 6.1 AB | 10.4 ± 6.8 A | 2.7 ± 0.6 A | 1 ± 0.8 A | 0 ± 0 A | ||||||||||||
| US8 | 31.6 ± 13.4 B | 13.8 ± 8.7 B | 1 ± 0.5 B | 0.5 ± 0.1 B | 0.5 ± 0.4 AB | 0 ± 0 A | ||||||||||||
Arthroconidia were first extracted from colonies then stored on an inert substrate (filter paper) before being rehydrated and plated on fresh medium after the predetermined storage times. A linear model for each temperature group was run on CFU, adjusted for zero inflation. Statistical differences between groups were determined from an analysis of variance on the fit of each linear model and post hoc Tukey's honestly significant difference tests, indicated by uppercase letters.
Genotyping.
Of the four isolates analyzed for both their colony morphology traits and arthroconidial germination, the potential mutations were investigated using whole-genome sequencing for three isolates. For the fourth isolate, a high-quality whole-genome sequence was not obtained. Instead, we used Sanger sequencing to assay the mutations identified in the other three isolates. In total, we identified 23 mutations that differ between strain US15 and the other two sequenced strains, NB26 and PE8, including 16 single nucleotide polymorphisms (SNPs) and 7 indels. Fifteen of the twenty-three mutations were shared between isolates NB26 and PE8, while 8 mutations (1 indel and 7 nucleotide substitutions) were unique to PE8, including those found in genes encoding a putative COP9 signalosome, a retrotransposon, and a reverse transcriptase. The nucleotide substitutions are shown in Table 2. A single missense variant was found in all three derived isolates (NB26, PE8, and US8) in a gene putatively identified as a velvet factor, pertaining to a family of fungus-specific transcription factors linked to the regulation of morphological development and the production of secondary metabolites in other filamentous fungi (23). Of the sixteen SNPs, nine were G·C→A·T transitions and six were A·T→C·G transversions. Among the seven indels, six mutations affected protein translation, and one caused a nonsense mutation resulting in the truncation of its protein product.
TABLE 2.
Collection of SNPs from whole-genome sequencing conducted on three isolates (NB26, US15, and PE8)a
| Variant | Mutation type | Putative annotation | AAb |
Scaffold | Position | ||
|---|---|---|---|---|---|---|---|
| Change | Reference | Alternate | |||||
| NB26/PE8/US8 | Missense | Velvet factor | Asp309Glu | T | G | 4871 | 927 |
| NB26/PE8 | Stop gained | Nucleic acid binding; zinc ion binding; DNA integration | Arg749 | C | T | 720 | 2245 |
| Missense | Hydroxyacid-oxoacid transhydrogenase | Arg261Leu | G | T | 6034 | 782 | |
| NAc | Asp4453Asn | G | A | 355 | 13357 | ||
| Synonymous | Growth factor receptor cytosine rich domain | Ala142Ala | A | G | 322 | 426 | |
| Transposase (AF333034.1) | Gly957Gly | T | C | 1928 | 2871 | ||
| Related to high-affinity phosphodiesterase (PDE2); 3′,5′-cyclic-nucleotide phosphodiesterase | Thr188Thr | C | T | 3235 | 564 | ||
| Hypothetical protein (GMDG_02575) | Leu1526Leu | T | A | 1536 | 4578 | ||
| RNase H | Phe1700Phe | T | C | 1498 | 5100 | ||
| PE8 | Missense | DNA binding | Thr1813Lys | C | A | 104 | 5438 |
| Retrotransposon nucleocapsid: nucleic acid binding; DNA integration | Ala114Ser | G | T | 5792 | 340 | ||
| Nucleic acid binding | Lys131Thr | A | C | 7402 | 392 | ||
| Reverse transcriptase family | Gln57His | A | C | 5306 | 171 | ||
| Monooxygenase activity | Val1752Leu | G | T | 979 | 5254 | ||
| Probable COP9 signalosome subunit 2 | Arg1548Lys | G | A | 2133 | 4643 | ||
| Synonymous | NA | Lys198Lys | G | A | 4883 | 594 | |
These SNPs were all confirmed using custom primers and Sanger sequencing among the isolates.
AA, amino acid.
NA, not available.
A subsequent search of gene ontology (GO) terms provided functional annotation information for each of the genes impacted by the identified mutations. Our analysis revealed that several mutations were in genes involved in catalytic enzyme activity, cellular respiration, cell wall components, and other constituents of cell metabolism, among others (Table 3). The majority of regions containing variants were significantly enriched with GO terms.
TABLE 3.
Results from enrichment analysis using Fisher's exact testa
| SNP scaffold | GO |
P value | FDR | |
|---|---|---|---|---|
| IDb | Name | |||
| 203, 6125, 720 | 0015074 | DNA integration | 1.62E−07 | 2.02E−06 |
| 1316 | 0042450 | Arginine biosynthetic process via ornithine | 3.56E−03 | 5.85E−03 |
| 322 | 0070469 | Respiratory chain | 3.56E−03 | 5.85E−03 |
| 322, 592 | 0016021 | Integral component of membrane | 7.11E−03 | 1.13E−02 |
| 104, 1928 | 0003677 | DNA binding | 3.66E−05 | 2.29E−04 |
| 0 | 0016758 | Transferase activity, transferring hexosyl groups | 3.56E−03 | 5.85E−03 |
| 720 | 0008270 | Zinc ion binding | 3.56E−03 | 5.85E−03 |
| 322 | 0009916 | Alternative oxidase activity | 3.56E−03 | 5.85E−03 |
| 41 | 0008137 | NADH dehydrogenase (ubiquinone) activity | 3.56E−03 | 5.85E−03 |
| 1316 | 0004056 | Argininosuccinate lyase activity | 3.56E−03 | 5.85E−03 |
| 322 | 0016787 | Hydrolase activity | 1.06E−02 | 1.69E−02 |
Significantly enriched genes (P < 0.05) from regions containing SNPs and the associated GO term annotation information. Genomic regions were significantly enriched in terms relevant to various cellular functions, including metabolism and respiration pathways.
ID, identifier.
Aside from comparing the mutations among representative strains in our own collections, we also surveyed the sequenced genomes of other P. destructans strains at the specific variant sites by querying the short read archives on NCBI submitted from other studies (BioProjects PRJNA276926 and PRJNA400587) (4, 7, 24). Of the 60 strains with genome sequences in the archive, 16 had missing reads in at least one of those variant regions. None of the 16 genomes contained read information for 3 of the 23 variant sites located in three different scaffolds (322, 1498, and 1928), while the remaining 20 variant sites were found in at least 1 of the 16 genomes. These 16 genomes were excluded from Table S1. The remaining 44 were compared with our SNP variant set as described in Table 2. At these 23 variable sites, 12 of the 44 strains were identical to the US15 genome (Table S1). We found that only two samples (22884-4W and Pd_692102) contained one variant allele on scaffold 3235 that was shared with NB26 and PE8 (Table 2). The sample 22884-4W was isolated from an infected bat in Vermont (Table S1). Thus, most of the mutations identified here are novel, not reported in other sequenced genomes.
DISCUSSION
The WNS epizootic in NA is thought to have been initiated by the propagation and spread of a single clonal genotype of P. destructans (19, 25, 26). This genotype was among the many genotypes within its native ecosystem in Europe, and the low virulence of these strains toward European bats was likely the result of their long-term coevolution (26, 27). However, the situation differs in NA where P. destructans is exploiting naive hosts. Since the first detection of WNS in NA over 10 years ago, both phenotypic and genotypic changes have been reported among the NA strains of this species (21, 28). However, the broad geographic pattern of phenotypic changes and the potential genetic basis for the phenotypic diversity have not been investigated. Here we tested the hypothesis that the NA environments have impacted the evolution of certain traits of the WNS pathogen since its introduction. We postulated that environmental pressures might have driven the evolution of phenotypic variation for some of the clonal descendants of the original genotype. Our analyses demonstrated that (i) phenotypic variability in the NA population of P. destructans had a broad geographic pattern, (ii) differences in the responses of P. destructans arthroconidia to temperature stress have emerged among isolates, and (iii) several newly accumulated mutations may have contributed to the phenotypic variation among a subset of isolates. Below, we discuss the relevance of our results to those from previous studies and the implications of the results to the management of WNS and the conservation of bats in NA.
Phenotypic patterns in North America.
The ability to generate and maintain phenotypic diversity within a population can enhance the survival of the population under environmental stress and improve the persistence of a population within novel environments (29). In NA, the environmental conditions that P. destructans inhabits are highly heterogeneous, so far spanning 31 U.S. states and five Canadian provinces. Such heterogeneity could pose a significant challenge for the spread, reproduction, and survival of the pathogen, especially because the pathogen is psychrophilic and started as a single genotype in NA. A previous study reported the qualitative measures of phenotypic variation among strains of P. destructans in NA (21). Our results here showed the quantitative relationships between phenotypic variations and the geographic spread of this fungus in northeastern United States and eastern Canada.
Overall, our results demonstrated a positive correlation between the fungal colony area on artificial medium and the relative geographic distance/coordinates from the epicenter of WNS in North America. Isolates from more distant northern or eastern regions showed a greater variability and overall higher radial growth rates. In contrast, isolates from the southern expanse of the WNS range tend to have lower growth rates under the same lab condition (Fig. 2). Multiple environmental factors, such as changes in temperature, pH, or available nutrients, can trigger adaptive evolution in populations (30). Such a response has been demonstrated within P. destructans; when the incubation temperature is increased from 12 to 15°C, the cultures exhibited higher growth rates (31). In addition, the same study demonstrated that the growth rate of the establishing strain in New York State was higher than that of P. destructans from Europe (31). Taken together, these results suggest that the increasing growth rate in certain geographical directions likely represents a significant adaptive change of this fungus in NA. Furthermore, such adaptations may have included acclimatization to the array of environmental conditions, such as the air temperatures, in most NA regions (Fig. S1B in the supplemental material). An increased growth rate for such strains in vitro may also increase their virulence and invasiveness on bats.
The production of pigments in pathogenic fungi is a common virulence factor, contributing resistance to both host and environmental stresses (32–34). Within the northeastern range of P. destructans, the observed trend in colony surface pigment production mirrored that of the colony growth rate. In contrast, the diffusion of pigments through agar was not significantly correlated with increased distance from the William's Hotel mine (Fig. 2E and F). Interestingly, both measurements of colony pigmentation (colony surface excretions and the production and diffusion of pigments through substrates) have greater trait variances with increasing northern and eastern distances from early WNS isolates (Fig. S2). In addition, these two traits share a strong correlation and significant covariation (r = 0.5, P < 0.01) (Fig. S3). Although the pigmentation at the top of the colony and the pigment diffusion through agar likely differ in function, these results suggest that their production and secretion in P. destructans may have been impacted by similar environmental conditions as WNS spread into northeastern NA. In this context, selective forces acting on P. destructans populations might have come from multiple stressors in the hibernaculum environments, including from other competing environmental microbes and interactions with bats. In addition, while the conditions for the long-term persistence of P. destructans within the hibernacula are different from those outside the hibernacula during the summer months, environmental factors from outside the hibernacula could also act as selective forces, contributing to differences in pigment production and secretion. A recent study showed that interactions with a partitivirus influence the phenotypes in P. destructans (35). Indeed, multiple microbial species with antagonistic activities against P. destructans within the cave environment have been isolated (36). Similarly, NA Myotis species have shown various immune responses to P. destructans infections (37), which may provide additional selective pressure on pigment production and secretion. The benefits of such adaptations to pigment production might influence the survival and reproduction rates of P. destructans both within and outside NA hibernacula and potentially increase the probability of successful spread among niches.
The mechanism of WNS spread is among the most poorly understood aspects of the epizootic. The phenotypic variation observed here among strains and traits might be relevant to the patterns of spread and population adaptation over geographical ranges. The patterns of spread and adaptation of P. destructans in NA may be explained by one of two competing hypotheses: (i) the spread of WNS has continued in a stepwise fashion from the epicenter via clonal expansion and (ii) the spread of WNS is not stepwise and spread patterns are complex, involving multiple back and forth dispersals among hibernacula (38). If the spread is a stepwise process, we should see an increased accumulation of spontaneous mutations along geographic gradients and different mutations that accumulated among different isolates as the population expanded in different directions. The phenotypic expectation of this stepwise process is that phenotypic trait values should diverge more as the distance of the population from the epicenter increases. While there is some evidence for the increased variance, there was no consistent pattern (Fig. S2). Our results suggest that the spread may not follow a strict stepwise or linear fashion over a temporal or geographical gradient. Rather, the spread likely involves multiple back and forth transmissions between hibernacula, resulting in mixed phenotypic diversity within certain areas of the WNS epizootic. Recent genetic analyses of strains from NA suggested evidence for widespread mixings and long-distance dispersals (28, 38, 39), consistent with the second hypothesis and with what we observed here for phenotypic traits.
Suboptimal temperatures and environmental stress.
WNS has rapidly spread into NA habitats, reaching the eastern Canadian provinces and southern United States after approximately 7 years, most recently appearing on the western coast of the United States (4). The dispersal over long distances requires tolerance across various climates and environments. The long-term persistence of this species in the presence of minimal nutrients and water has already been demonstrated (40). Due to its psychrophilic nature, the exposure to suboptimal high temperatures likely represents a significant constraint on P. destructans spore viability (37). By controlling the exposure to suboptimal temperatures, a threshold for P. destructans spore germination can be determined. Overall, we found that a 4°C environment resulted in a higher germination rate of arthroconidia for a greater length of time than the 14°C and 23°C environments. The differential responses to these temperatures among strains from different environments are consistent with their adaptive significance. Specifically, we propose that such divergent responses may be related to coping with different environmental stresses and are associated with an increase in long-term persistence or spread potential of P. destructans in NA, as was suggested by Drees et al. (38) and Palmer et al. (24). Here, we discuss the consequences of the differences in arthroconidium temperature tolerance in the context of the dispersal of WNS in NA.
WNS was first reported in caves in New Brunswick, Nova Scotia, and Prince Edward Island (PE) as early as 2012 to 2013. The caves in northern areas of the WNS distribution (New Brunswick, Nova Scotia, etc.) experience mean air temperatures of 5.49°C (± 9.72°C) through the year (Fig. S2A). Our low-temperature treatment emulates a minimum level of temperature stress present in northern habitats for the majority of the winter months (November to March, −3.6 ± 4.7°C) (Fig. S2A). After long-term exposure to 4°C, both derived isolates germinated significantly better than the isolate US15 from around the epicenter in NA (Table 1). This result suggests that isolates from these areas may have adapted to extend the life span of arthroconidia by entering a state of dormancy that can be activated in times of stress (41). The activation of dormancy pathways was likely stimulated by the absence of proper nutrients, desiccation, and suboptimal temperatures. Cell dormancy at temperatures <4°C is relevant in the context of the seasonal variation that occurs in many regions in NA. Low temperatures around 4°C could occur in early autumn (Fig. S2A) and last until early the next summer. Being able to enter dormancy and maintain viability might offer a significant advantage to P. destructans populations in the long term.
In comparison to the northern reaches of this epizootic, the regions closer to the WNS epicenter (New York State) or the southern range of the distribution of our isolates (North Carolina) have higher average year-round air temperatures of 9.65°C (± 8.13°C) and 17.12°C (± 6.68°C), respectively (Fig. S2A). We induced heat stress in P. destructans by subjecting our spore populations to temperatures that exceeded the upper limit for growth (11, 31, 41). Compared to P. destructans, other species of the genus Pseudogymnoascus are found to be capable of growing at temperatures higher than 20°C (42). Different from the germination rate data for arthroconidia stored at 4°C, arthroconidia of strain US15 maintained viability for a longer period of time at the 23°C environment than all three derived isolates. These results suggest that a trade-off may have arisen since WNS has spread into novel environments, with arthroconidia gaining cold temperature tolerance while losing resistance to heat stress.
We observed that storage time can have a greater impact than expected on the germination potential of arthroconidia. For example, the spore viability of isolates PE8 and US8 dropped significantly after 24 h of storage at 23°C. Interestingly, a slightly prolonged exposure may result in unexpected increases in P. destructans germination. As demonstrated in other psychrophilic fungi, cold exposure can increase spore germination rates (43). Consistent with those observations, the storage at 4°C for 14 days resulted in significant increases in spore germination over the 7-day period. It should be noted that the activation of arthroconidia after prolonged dormancy likely depends on several factors, including water availability, nutrients, temperature, and other biotic and abiotic factors (44). Taken together, this suggests that the viability of arthroconidia can be extended after storage at such cold temperatures.
Our result indicates that arthroconidia can survive up to 3 weeks at a temperature of 23°C. This result suggests that the surviving bats with WNS that exit winter hibernacula during the spring/summer months can carry viable arthroconidia with them (45, 46). However, other factors may influence the effectiveness of this dispersal, including solar radiation. In natural environments, solar radiation includes UV that can negatively influence arthroconidial survival (24). Furthermore, an estimated 106 cells of P. destructans are required to cause WNS (4), and a successful dispersal event between hibernacula requires not only appropriate temperatures but also other conditions that are conducive for P. destructans germination and growth. A realistic model for the spread of WNS should consider these factors.
Genetic mutations and adaptation.
Adaptations provide some benefits to an organism's reproductive success under the sum of all environmental conditions (47). In their natural habitats, most fungi often experience rapidly changing conditions. Thus, having a robust system to cope with environmental changes might be highly beneficial. This study explored the potential relationships between the recently accumulated genetic variations within the NA clonal population of P. destructans and possible adaptations to local conditions during the spread of WNS. Our analyses identified several putative variants related to the morphological/physiological variations found among the isolates. We specifically considered mutations related to the regulation of growth, development, metabolism, and membrane integrity and those with enriched GO terms representing potential targets for studying adaptive diversification in P. destructans. Interestingly, mutations within genes relevant to these areas have not been reported in other recently sequenced strains of P. destructans (28, 44, 48, 49).
Of special note is the mutation in the homolog of a velvet protein in P. destructans. The velvet proteins are a family of transcriptional regulators widely distributed in fungi. They regulate the expressions of many genes and impact fungal growth, secondary metabolism, and cell differentiation such as conidia germination/maturation, sporogenesis, and toxigenesis (28, 50–52). Compared to US15, the early isolate close to the epicenter, all three derived isolates that we sequenced shared the same missense mutation (Asp→Glu) at amino acid position 309 within the velvet protein. At present, the influence of this mutation on the function of velvet protein in P. destructans is not known. Previous studies have found that interactions between members of the velvet family can impact conidia germination/maturation by stimulating dormancy pathways, trehalose biogenesis, and/or cell wall production (53–55).
An improved metabolism can convey obvious benefits to a population's survival. In the context of adaptation, the expansion of gene families pertaining to metabolic activity has been suggested as a major factor driving the transition to pathogenesis in other fungal pathogens (56). In the chytrid fungus (Batrachochytrium dendrobatidis), clonal spread and host shift were accompanied by the duplication and expansion of protease genes (57, 58). In this study, several mutations and regions with enriched GO terms pose particular relevance to metabolism/catabolic activity (Tables 1 and 2). However, only one, a homolog of the hydroxyacid-oxoacid transhydrogenase, was impacted by a missense mutation (Arg→Leu). While its function in P. destructans is presently not known, the gene encoding this protein shows a sequence similarity range of 46% to 54% with mammalian genes (59) in the category of glycoside hydrolase family 61 and part of the lytic polysaccharide monooxygenases (LPMOs). LPMOs cleave polysaccharides via a novel oxidative mechanism (60), and certain LPMOs are found only in fungi, acting preferentially on cellulose (61, 62). In the laboratory, P. destructans can grow in medium containing complex carbohydrates as carbon and energy sources. Thus, polysaccharides in the hibernacula could also be a significant carbon and energy source for P. destructans, and the altered amino acid in the putative hydroxyacid-oxoacid transhydrogenase may represent an adaptation in the specific strains (63).
Being able to maintain cell membrane integrity is critical for the dispersal and survival of all organisms. Increased membrane permeability or rigidity can result in cell death, especially in stressful conditions (64). As previously mentioned, mutations in the velvet protein in P. destructans might influence the regulation of trehalose and cell wall production, as well as spore dormancy (53, 65), all of which could contribute to P. destructans survival under suboptimal temperatures. In addition to this gene, one mutation found in the COP9 signalosome subunit 2 (CNS-2) was unique to strain PE8 and might be of importance for cold temperature tolerance (Table 2). Altered functioning of CNS-2 can cause a cascade of effects impacting components of cellular circadian rhythm development and the formation of fungal hydrophobin in asexual conidia cells (66–68). It is possible that the mutation in COP9 observed here may impact membrane integrity in P. destructans, contributing to the differences observed between strain PE8 and others.
Conclusions and perspectives.
Invasive fungal pathogens bring significant concerns for the conservation and health of our ecosystems (69, 70). Of the bat species infected with WNS, many play multiple ecological roles. For example, bats are predators of many insect pests. As bat populations decline, the ecosystem services that they provide to the agricultural industry could also disappear (71). Here, we explored the phenotypic diversity, temperature-dependent spore germination, and novel genetic variations on the fungal pathogen of bats from different areas of the WNS epidemic. Our results suggest that temperature has likely played an important role in the geographic diversification of P. destructans. However, temperature is only one of many factors in the proliferation of P. destructans in the hibernaculum environment; other factors such as relative humidity (72), nutrient availability (73), and interactions with other microbes within the soil community likely also hold significant influence. Our results suggest that the genetic changes accumulated during the spread of WNS and the impact they have on survival and reproduction should be other factors in our understanding of the transmission and spread of WNS. Additional studies involving experimental infections of bats or in vitro models of WNS infection are needed to determine how these genetic and morphological changes might have contributed to the expansion of WNS. Given the rapidly expanding distribution of WNS, a detailed understanding of these and other issues related to the growth, reproduction, and dispersal of P. destructans is urgently needed to help control this disease.
MATERIALS AND METHODS
P. destructans isolates.
Our experiments involved a total of 62 isolates from various areas of the WNS epizootic (Fig. 1; see also supplemental material). All isolates were obtained by the Department of Natural Resources (Canada) or the Fish and Wildlife Service (United States) and were subcultured to select for single-cell colonies. These purified cultures were preserved in Sabouraud-glycerol tubes at −80°C. We revived each isolate by spotting cell suspensions onto Sabouraud dextrose agar (SDA) and incubating at 14°C for 28 days. Cell suspensions were created in triplicates for each isolate; each suspension was then inoculated on the center of each separate plate of SDA for colony morphology observations.
Phenotypic characterizations.
After incubating at 14°C for 28 days, each strain was assessed for several phenotypic traits, including colony size, pigmentation on the surface of the colony, and pigment diffusion underneath the colony through the agar medium. To assess colony size of each P. destructans isolate, we recorded digital images of all 62 isolates, and these images were subsequently analyzed using the spot densitometry function in FluorChem 8900 (Alpha Innotech, San Leandro, CA). Colony size was recorded as an estimate of the total amount of visible culture on the agar plate, from which the total area was calculated on the basis of the shape of each colony. Similarly, pigmentation on both the colony surface and its diffusion in agar was approximated using the spot densitometry function using the tools available with FluorChem 8900, according to the protocols described by Vogan et al. (74).
Arthroconidial survival.
Here we examined the arthroconidium spore survival of four (US15, PE8, NB26, and US8) of the 62 isolates at 4°C, 14°C, and 23°C environments. These four isolates came from different geographic areas and represented different colony phenotypes. Briefly, we first grew these isolates on SDA medium at 14°C for 2 months, allowing enough time for cultures to sporulate to produce arthroconidia. These cells were then collected from the surfaces of the colonies and adjusted to a density of 5 × 104 arthroconidia per ml. Three separate samples of 20 μl each for each isolate were transferred onto an 8-mm-diameter disk of sterile Whatman filter paper no. 5 (2.5-μm pore size). These paper disks were allowed to dry at 14°C under sterile conditions and were subsequently exposed to suboptimal temperatures of either 4°C or 23°C. The paper disks containing the arthroconidia were held at these two temperatures for various lengths of time: 0 days, 1 day, 7 days, 14 days, 21 days, and 28 days. After each respective time period, the filter paper disks were rehydrated with 200 μl of sterile water, with the arthroconidia released into the water and calibrated to a spore density of 2 × 104 arthroconidia per ml. Samples of 100 μl were taken from the suspension created from each paper disk in triplicates; each sample was then spread onto fresh SDA medium. These plates containing the final cultures were incubated at 14°C, the optimal temperature for spore germination and growth, for a minimum of 7 days before they were scored for arthroconidial germination. Each plate was imaged, and the number of CFU was counted using the multipoint function on ImageJ (75). In total, the experimental design consisted of 432 data points (4 isolates × 2 temperature levels × 6 exposure times × 3 tubes [per treatment] × 3 plates [per tube]).
Genotyping.
To identify the potential genetic changes associated with phenotypic differences (including arthroconidial viability), three of the four isolates (NB26, US15, and PE8) were chosen for initial whole-genome sequencing. Cells were harvested from SDA plates of 1-month-old cultures growing in a 14°C incubator. DNA was extracted using a standard cetyl trimethylammonium bromide (CTAB) and phenol-chloroform protocol for filamentous fungal cultures (76). Sequencing was conducted with a paired-end library on an Illumina MiSeq platform. A de novo assembly of the isolate US15 (NHWC 20631-21) was constructed using an open source pipeline, specifically designed for use with MiSeq data and microbial genomes (77). Raw reads from all isolates were aligned to our de novo build (see supplemental material), which had a total size of 30.9 Mb, consisting of 8,145 scaffolds with an N50 of 7,747 bp, at approximately 50× coverage.
Variant calling was conducted on concatenated BAM files using Freebayes V1.1.0 (78). Variants that lacked coverage and quality scores were excluded, and were filtered on the basis of quality and depth (QUAL & DP; >20) using bcf-tools V1.3 (79). With this final set, we performed a sequence homology search of a fungal protein database, mapping of GO terms, and functional annotation using the tools available on the Blast2GO platform (80). We then conducted an enrichment analysis using Fisher's exact test to identify statistically significant enrichment within genes that contain variants. We investigated 30 loci that contained putative mutations on the basis of genome sequence comparisons. Through locus-specific PCR and sequencing, we identified 7 of the 30 loci as false positives (a rate of 30%). while 23 mutations were confirmed among the three isolates. In addition, the same sites were also investigated in the isolate US8, as its genome was not sequenced in the original Illumina MiSeq run.
In addition to our three whole-genome sequences, the genomic data from 60 P. destructans isolates were obtained from NCBI and were examined for the variants that we found in our MiSeq data described above. Genomic reads obtained from NCBI were aligned to whole scaffolds containing variant sequences from the US15 reference genome, using the same alignment and variant calling protocol described above. A variant site was considered robust if the quality and depth scores exceeded 20.
Statistical analyses.
All statistical tests on phenotypic data were carried out in R (v3.3.2) (81). To evaluate the patterns of phenotypic variation across geographical regions in the WNS epizootic, we used linear models for each trait. Relative geographical distances (in km) to the closest isolate from the epicenter of WNS outbreak (in the William's Hotel mine, NY, near Albany) were converted from geographical coordinates using the haversine method (available in the R package geosphere) (82). When measuring trait variability, results from each trait were binned separately for every 50 km of relative distance in each cardinal direction. In our analyses and figure presentations, relative to the early isolate US15, the geographic distance to the north and east is labeled a positive change along the x axis, whereas the distance to the south and west is labeled a negative relative change. The variation among strains within each geographic distance class was calculated using these bins, and a separate linear model was conducted for each trait and geographic direction.
To compare phenotypic patterns among multiple traits, we used a mixed-effects linear model approach to perform a multivariate analysis of variance. Results from all three phenotypic measures (colony area, pigment, and pigment diffusion) were first scaled prior to constructing each model using the lme4 R package (83) (see supplemental material).
The proportions of arthroconidia that germinated after storing at various temperatures for different lengths of time were analyzed using linear models for each temperature group. We log-transformed the raw data points to normalize our data set, applied an offset value to adjust for data points that were zero, and excluded outliers that exceeded three standard deviations from the mean for each isolate/temperature/day grouping. We then computed an analysis of variance table for each linear model fit and extracted significant groups for each factor via multiple-comparison Tukey tests using default settings (R package agricolae) (84).
An enrichment analysis determines the probability that the GO terms within a given subset of genes, compared to a background set of genes, were assigned by chance. Using the built-in functions in Blast2GO, we compared the set of genes containing variants (Table 1) to all other genes that were assigned GO terms. The results from Fisher's exact test were corrected for false discovery rate (FDR), with a significance cutoff of <0.05.
Accession number(s).
Raw reads used for genomic analyses were submitted to the NCBI SRA database and can be found under the BioSample identifiers SAMN09233324, SAMN09233325, and SAMN09233326.
Supplementary Material
ACKNOWLEDGMENTS
The following researchers contributed strains from Canada: Karen Vanderwolf, Donald McAlpine, Scott McBurney, David Overy, and Durda Slavic. We thank Vishnu Chaturvedi for sharing his isolates from the United States.
The research presented here is supported by the Natural Sciences and Engineering Research Council of Canada (J.X.), McMaster Institute of Infectious Disease Research Fellowship (J.A.), and by Ontario Graduate Scholarship (A.F.).
The authors declare no competing interests.
J.X. designed the study, A.F. conducted the arthroconidial germination experiment, genome sequencing, bioinformatics, and statistical analysis, V.G. performed part of the arthroconidial germination experiment, J.A. conducted colony phenotype assays, and A.F. and J.X. drafted the manuscript. All authors contributed to the writing of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00863-18.
REFERENCES
- 1.Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS. 2009. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108:147–154. doi: 10.5248/108.147. [DOI] [Google Scholar]
- 2.Meteyer CU, Buckles EL, Blehert DS, Hicks AC, Earl Green D, Shearn-Bochsler V, Thomas NJ, Gargas A, Behr MJ. 2009. Histopathologic criteria to confirm white-nose syndrome in bats. J Vet Diagn Invest 21:411–414. doi: 10.1177/104063870902100401. [DOI] [PubMed] [Google Scholar]
- 3.Willis CKR, Menzies AK, Boyles JG, Wojciechowski MS. 2011. Evaporative water loss is a plausible explanation for mortality of bats from white-nose syndrome. Integr Comp Biol 51:364–373. doi: 10.1093/icb/icr076. [DOI] [PubMed] [Google Scholar]
- 4.Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, Ballmann AE, Coleman JTH, Redell DN, Reeder DM, Blehert DS. 2011. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480:376–378. doi: 10.1038/nature10590. [DOI] [PubMed] [Google Scholar]
- 5.Thomas DW, Dorais M, Bergeron J-M. 1990. Winter energy budgets and cost of arousals for hibernating little brown bats, Myotis lucifugus. J Mammal 71:475–479. doi: 10.2307/1381967. [DOI] [Google Scholar]
- 6.Reeder DM, Frank CL, Turner GG, Meteyer CU, Kurta A, Britzke ER, Vodzak ME, Darling SR, Stihler CW, Hicks AC, Jacob R, Grieneisen LE, Brownlee SA, Muller LK, Blehert DS. 2012. Frequent arousal from hibernation linked to severity of infection and mortality in bats with white-nose syndrome. PLoS One 7:e38920. doi: 10.1371/journal.pone.0038920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cryan PM, Meteyer CU, Boyles JG, Blehert DS. 2010. Wing pathology of white-nose syndrome in bats suggests life-threatening disruption of physiology. BMC Biol 8:135. doi: 10.1186/1741-7007-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, Coleman JTH, Darling SR, Gargas A, Niver R, Okoniewski JC, Rudd RJ, Stone WB. 2009. Bat White-nose syndrome: an emerging fungal pathogen? Science 323:227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 9.U.S. Fish And Wildlife Service. 2012. North American bat death toll exceeds 5.5 million from white-nose syndrome. News release. U.S. Fish and Wildlife Service, Washington, DC. [Google Scholar]
- 10.Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, Butchkoski CM, Kunz TH. 2010. An emerging disease causes regional population collapse of a common North American bat species. Science 329:679–682. doi: 10.1126/science.1188594. [DOI] [PubMed] [Google Scholar]
- 11.Langwig KE, Frick WF, Bried JT, Hicks AC, Kunz TH, Kilpatrick AM. 2012. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecol Lett 15:1050–1057. doi: 10.1111/j.1461-0248.2012.01829.x. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds HT, Ingersoll T, Barton HA. 2015. Modeling the environmental growth of Pseudogymnoascus destructans and its impact on the white-nose syndrome epidemic. J Wildl Dis 51:318–331. doi: 10.7589/2014-06-157. [DOI] [PubMed] [Google Scholar]
- 13.Flory AR, Kumar S, Stohlgren TJ, Cryan PM. 2012. Environmental conditions associated with bat white-nose syndrome mortality in the north-eastern United States. J Appl Ecol 49:680–689. doi: 10.1111/j.1365-2664.2012.02129.x. [DOI] [Google Scholar]
- 14.Davis WH. 1970. Hibernation: ecology and physiological ecology, p 265–300. In Wimsatt W. (ed), Biology of bats. Academic Press, Cambridge, MA. [Google Scholar]
- 15.McNab BK. 1974. The behavior of temperate cave bats in a subtropical environment. Ecology 55:943–958. doi: 10.2307/1940347. [DOI] [Google Scholar]
- 16.Swezey C, Garrity C. 2011. Geographical and geological data from caves and mines infected with white-nose syndrome (WNS) before September 2009 in the eastern United States. J Caves Karst Stud 73:125–157. [Google Scholar]
- 17.Moore GW, Sullivan GN. 1978. Speleology: the study of caves. Zephyrus Press, Teaneck, NJ. [Google Scholar]
- 18.Langwig KE, Frick WF, Reynolds R, Parise KL, Drees KP, Hoyt JR, Cheng TL, Kunz TH, Foster JT, Kilpatrick AM. 2015. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc Biol Sci 282:20142335. doi: 10.1098/rspb.2014.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajkumar SS, Li X, Rudd RJ, Okoniewski JC, Xu J, Chaturvedi S, Chaturvedi V. 2011. Clonal genotype of Geomyces destructans among bats with white nose syndrome, New York, USA. Emerg Infect Dis 17:1273–1276. doi: 10.3201/eid1707.102056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lučan RK, Bandouchova H, Bartonička T, Pikula J, Zahradníková A Jr, Zukal J, Martínková N. 2016. Ectoparasites may serve as vectors for the white-nose syndrome fungus. Parasit Vectors 9:16. doi: 10.1186/s13071-016-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khankhet J, Vanderwolf KJ, McAlpine DF, McBurney S, Overy DP, Slavic D, Xu J. 2014. Clonal expansion of the Pseudogymnoascus destructans genotype in North America is accompanied by significant variation in phenotypic expression. PLoS One 9:e104684. doi: 10.1371/journal.pone.0104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson ES. 2000. Pathogenic roles for fungal melanins. Clin Microbiol Rev 13:708–717. doi: 10.1128/CMR.13.4.708-717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvo AM, Lohmar JM, Ibarra B, Satterlee T. 2016. Chapter 18. Velvet regulation of fungal development, p 475–497. In Wendland J. (ed), Growth, differentiation and sexuality. Springer, Berlin, Germany. [Google Scholar]
- 24.Palmer JM, Drees KP, Foster JT, Lindner DL. 2018. Extreme sensitivity to ultraviolet light in the fungal pathogen causing white-nose syndrome of bats. Nat Commun 9:35. doi: 10.1038/s41467-017-02441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren P, Haman KH, Last LA, Rajkumar SS, Keel MK, Chaturvedi V. 2012. Clonal spread of Geomyces destructans among bats, midwestern and southern United States. Emerg Infect Dis 18:883–885. doi: 10.3201/eid1805.111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leopardi S, Blake D, Puechmaille SJ. 2015. White-nose syndrome fungus introduced from Europe to North America. Curr Biol 25:R217–R219. doi: 10.1016/j.cub.2015.01.047. [DOI] [PubMed] [Google Scholar]
- 27.Wibbelt G, Kurth A, Hellmann D, Weishaar M, Barlow A, Veith M, Prüger J, Görföl T, Grosche L, Bontadina F, Zöphel U, Seidl HP, Seidl HP, Blehert DS. 2010. White-nose syndrome fungus (Geomyces destructans) in bats, Europe. Emerg Infect Dis 16:1237–1243. doi: 10.3201/eid1608.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trivedi J, Lachapelle J, Vanderwolf KJ, Misra V, Willis CKR, Ratcliffe JM, Ness RW, Anderson JB, Kohn LM. 2017. Fungus causing white-nose syndrome in bats accumulates genetic variability in North America with no sign of recombination. mSphere 2:e00271-17. doi: 10.1128/mSphereDirect.00271-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pigliucci M. 2001. Phenotypic plasticity: beyond nature and nurture. JHU Press, Baltimore, MD. [Google Scholar]
- 30.Ernst JF. 2000. Transcription factors in Candida albicans - environmental control of morphogenesis. Microbiology 146:1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- 31.Verant ML, Boyles JG, Waldrep W Jr, Wibbelt G, Blehert DS. 2012. Temperature-dependent growth of Geomyces destructans, the fungus that causes bat white-nose syndrome. PLoS One 7:e46280. doi: 10.1371/journal.pone.0046280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosas AL, Casadevall A. 2001. Melanization decreases the susceptibility of Cryptococcus neoformans to enzymatic degradation. Mycopathologia 151:53–56. doi: 10.1023/A:1010977107089. [DOI] [PubMed] [Google Scholar]
- 33.Casadevall A, Steenbergen JN, Nosanchuk JD. 2003. “Ready made” virulence and “dual use” virulence factors in pathogenic environmental fungi—the Cryptococcus neoformans paradigm. Curr Opin Microbiol 6:332–337. doi: 10.1016/S1369-5274(03)00082-1. [DOI] [PubMed] [Google Scholar]
- 34.Jahn B, Koch A, Schmidt A, Wanner G, Gehringer H, Bhakdi S, Brakhage AA. 1997. Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect Immun 65:5110–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thapa V, Turner GG, Hafenstein S, Overton BE, Vanderwolf KJ, Roossinck MJ. 2016. Using a novel partitivirus in Pseudogymnoascus destructans to understand the epidemiology of white-nose syndrome. PLoS Pathog 12:e1006076. doi: 10.1371/journal.ppat.1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamm PS, Caimi NA, Northup DE, Valdez EW, Buecher DC, Dunlap CA, Labeda DP, Lueschow S, Porras-Alfaro A. 2017. Western bats as a reservoir of novel Streptomyces species with antifungal activity. Appl Environ Microbiol 83:e03057-16. doi: 10.1128/AEM.03057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JS, Reeder DM, Lilley TM, Czirják GÁ Voigt CC, McMichael JW, Meierhofer MB, Seery CW, Lumadue SS, Altmann AJ, Toro MO, Field KA. 2015. Antibodies to Pseudogymnoascus destructans are not sufficient for protection against white-nose syndrome. Ecol Evol 5:2203–2214. doi: 10.1002/ece3.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drees KP, Lorch JM, Puechmaille SJ, Parise KL, Wibbelt G, Hoyt JR, Sun K, Jargalsaikhan A, Dalannast M, Palmer JM, Lindner DL, Marm Kilpatrick A, Pearson T, Keim PS, Blehert DS, Foster JT. 2017. Phylogenetics of a fungal invasion: origins and widespread dispersal of white-nose syndrome. mBio 8:e01941-17. doi: 10.1128/mBio.01941-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drees KP, Parise KL, Rivas SM, Felton LL, Bastien S, Puechmaille J, Keim P, Foster JT. 2017. Characterization of microsatellites in Pseudogymnoascus destructans for white-nose syndrome genetic analysis. J Wildl Dis 53:869–874. doi: 10.7589/2016-09-217. [DOI] [PubMed] [Google Scholar]
- 40.Hoyt JR, Langwig KE, Okoniewski J, Frick WF, Stone WB, Kilpatrick AM. 2015. Long-term persistence of Pseudogymnoascus destructans, the causative agent of white-nose syndrome, in the absence of bats. Ecohealth 12:330–333. doi: 10.1007/s10393-014-0981-4. [DOI] [PubMed] [Google Scholar]
- 41.Mysyakina IS, Kochkina GA, Ivanushkina NE, Bokareva DA, Feofilova EP. 2016. Germination of spores of mycelial fungi in relation to exogenous dormancy. Microbiology 85:290–294. doi: 10.1134/S0026261716030085. [DOI] [Google Scholar]
- 42.Samson RA. 1972. Notes on Pseudogymnoascus, Gymnoascus, and related genera. Acta Bot Neerl 21:517–527. doi: 10.1111/j.1438-8677.1972.tb00804.x. [DOI] [Google Scholar]
- 43.Juge C, Samson J, Bastien C, Vierheilig H, Coughlan A, Piché Y. 2002. Breaking dormancy in spores of the arbuscular mycorrhizal fungus Glomus intraradices: a critical cold-storage period. Mycorrhiza 12:37–42. doi: 10.1007/s00572-001-0151-8. [DOI] [PubMed] [Google Scholar]
- 44.Wyatt TT, Wösten HAB, Dijksterhuis J. 2013. Fungal spores for dispersion in space and time. Adv Appl Microbiol 85:43–91. doi: 10.1016/B978-0-12-407672-3.00002-2. [DOI] [PubMed] [Google Scholar]
- 45.Ballmann AE, Torkelson MR, Bohuski EA, Russell RE, Blehert DS. 2017. Dispersal hazards of Pseudogymnoascus destructans by bats and human activity at hibernacula in summer. J Wildl Dis 53:725–735. doi: 10.7589/2016-09-206. [DOI] [PubMed] [Google Scholar]
- 46.Bernard RF, Willcox EV, Parise KL, Foster JT, McCracken GF. 2017. White-nose syndrome fungus, Pseudogymnoascus destructans, on bats captured emerging from caves during winter in the southeastern United States. BMC Zool 2:12. doi: 10.1186/s40850-017-0021-2. [DOI] [Google Scholar]
- 47.Fisher RA. 2004. The nature of adaptation, p 85–91. In Ridley M. (ed), Evolution, 2nd ed Oxford University Press, New York, NY. [Google Scholar]
- 48.Chibucos MC, Crabtree J, Nagaraj S, Chaturvedi S, Chaturvedi V. 2013. Draft genome sequences of human pathogenic fungus Geomyces pannorum sensu lato and bat white nose syndrome pathogen Geomyces (Pseudogymnoascus) destructans. Genome Announc 1:e01045-13. doi: 10.1128/genomeA.01045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drees KP, Palmer JM, Sebra R, Lorch JM, Chen C, Wu C-C, Bok W, Keller NP, Blehert DS, Cuomo CA, Lindner DL, Foster JT. 2016. Use of multiple sequencing technologies to produce a high-quality genome of the fungus Pseudogymnoascus destructans, the causative agent of bat white-nose syndrome. Genome Announc 4:e00445-16. doi: 10.1128/genomeA.00445-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayram O, Braus GH. 2012. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 51.Ni M, Yu J-H. 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2:e970. doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed YL, Gerke J, Park H-S, Bayram Ö Neumann P, Ni M, Dickmanns A, Kim SC, Yu J-H, Braus GH, Ficner R. 2013. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-κB. PLoS Biol 11:e1001750. doi: 10.1371/journal.pbio.1001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park H-S, Ni M, Jeong KC, Kim YH, Yu J-H. 2012. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS One 7:e45935. doi: 10.1371/journal.pone.0045935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP, Yu J-H, Braus GH. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 55.Sarikaya Bayram O, Bayram O, Valerius O, Park HS, Irniger S, Gerke J, Ni M, Han K-H, Yu J-H, Braus GH. 2010. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet 6:e1001226. doi: 10.1371/journal.pgen.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gladieux P, Ropars J, Badouin H, Branca A, Aguileta G, de Vienne DM, Rodríguez de la Vega RC, Branco S, Giraud T. 2014. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol 23:753–773. doi: 10.1111/mec.12631. [DOI] [PubMed] [Google Scholar]
- 57.Joneson S, Stajich JE, Shiu S-H, Rosenblum EB. 2011. Genomic transition to pathogenicity in chytrid fungi. PLoS Pathog 7:e1002338. doi: 10.1371/journal.ppat.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenblum EB, Poorten TJ, Joneson S, Settles M. 2012. Substrate-specific gene expression in Batrachochytrium dendrobatidis, the chytrid pathogen of amphibians. PLoS One 7:e49924. doi: 10.1371/journal.pone.0049924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kardon T, Noël G, Vertommen D, Schaftingen EV. 2006. Identification of the gene encoding hydroxyacid-oxoacid transhydrogenase, an enzyme that metabolizes 4-hydroxybutyrate. FEBS Lett 580:2347–2350. doi: 10.1016/j.febslet.2006.02.082. [DOI] [PubMed] [Google Scholar]
- 60.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beeson WT, Van VV, Span EA, Phillips CM, Marletta MA. 2015. Cellulose degradation by polysaccharide monooxygenases. Annu Rev Biochem 84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 62.Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S, Fanuel M, Ropartz D, Rogniaux H, Gimbert I, Record E, Berrin J-G. 2015. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels 8:90. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reynolds HT, Barton HA. 2013. White-nose syndrome: human activity in the emergence of an extirpating mycosis. Microbiol Spectr 1:OH-0008-2012. doi: 10.1128/microbiolspec.OH-0008-2012. [DOI] [PubMed] [Google Scholar]
- 64.Gervais P, Martinez de Marañon I. 1995. Effect of the kinetics of temperature variation on Saccharomyces cereuisiae viability and permeability. Biochim Biophys Acta 1235:52–56. doi: 10.1016/0005-2736(94)00299-5. [DOI] [PubMed] [Google Scholar]
- 65.Fillinger S, Chaveroche MK, van Dijck P, de Vries R, Ruijter G, Thevelein J, d'Enfert C. 2001. Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungus Aspergillus nidulans. Microbiology 147:1851–1862. doi: 10.1099/00221287-147-7-1851. [DOI] [PubMed] [Google Scholar]
- 66.He Q, Cheng P, He Q, Liu Y. 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev 19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lakin-Thomas PL, Coté GG, Brody S. 1990. Circadian rhythms in Neurospora crassa: biochemistry and genetics. Crit Rev Microbiol 17:365–416. doi: 10.3109/10408419009114762. [DOI] [PubMed] [Google Scholar]
- 68.Bell-Pedersen D, Dunlap JC, Loros JJ. 1992. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev 6:2382–2394. doi: 10.1101/gad.6.12a.2382. [DOI] [PubMed] [Google Scholar]
- 69.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fisher M, Gow N, Gurr S. 2016. Tackling emerging fungal threats to animal health, food security and ecosystem resilience. Philos Trans R Soc Lond B Biol Sci 371:20160332. doi: 10.1098/rstb.2016.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kunz TH, de Torrez EB, Bauer D, Lobova T, Fleming TH. 2011. Ecosystem services provided by bats. Ann N Y Acad Sci 1223:1–38. doi: 10.1111/j.1749-6632.2011.06004.x. [DOI] [PubMed] [Google Scholar]
- 72.Marroquin CM, Lavine JO, Windstam ST. 2017. Effect of humidity on development of Pseudogymnoascus destructans, the causal agent of bat white-nose syndrome. Northeast Nat (Steuben) 24:54–64. doi: 10.1656/045.024.0105. [DOI] [Google Scholar]
- 73.Raudabaugh DB, Miller AN. 2013. Nutritional capability of and substrate suitability for Pseudogymnoascus destructans, the causal agent of bat white-nose syndrome. PLoS One 8:e78300. doi: 10.1371/journal.pone.0078300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogan AA, Khankhet J, Samarasinghe H, Xu J. 2016. Identification of QTLs associated with virulence related traits and drug resistance in Cryptococcus neoformans. G3 (Bethesda) 6:2745–2759. doi: 10.1534/g3.116.029595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katz ME, Cheetham BF. 2009. Isolation of nucleic acids from filamentous fungi, p 191 In Liu D. (ed), Handbook of nucleic acid purification. CRC Press, Boca Raton, FL. [Google Scholar]
- 77.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 78.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 http://arxiv.org/abs/1207.3907.
- 79.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conesa A., Götz S. 2008. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org/. [Google Scholar]
- 82.Hijmans RJ, Williams E, Vennes C. 2012. Geosphere: spherical trigonometry. R package version. https://CRAN.R-project.org/package=geosphere.
- 83.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67:v067.i01. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 84.De Mendiburu F. 2014. Agricolae: statistical procedures for agricultural research. R package version. https://CRAN.R-project.org/package=agricolae.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.