Finfish aquaculture is the fastest growing global food production sector. Infectious disease, particularly emergent pathogens, pose a significant threat to established and nascent aquaculture industries worldwide. Herein, we characterize a novel pathogen isolated from mortality events in cultured spotted rose snapper in Central America. The bacteria recovered from these outbreaks were genetically and phenotypically dissimilar from other known Francisella spp. from fish, representing a previously unrecognized member of the genus Francisella, for which the name Francisella marina sp. nov. is proposed.
KEYWORDS: aquaculture, Francisella, snapper, fish pathogens
ABSTRACT
Historically, piscine francisellosis in various warm-, temperate-, and cold-water fish hosts has been attributed to Francisella noatunensis. From 2015 to 2016, an undescribed Francisella sp. was recovered during mortality events in cultured spotted rose snapper (Lutjanus guttatus) off the Pacific coast of Central America. Despite high mortality and emaciation, limited gross findings were observed in affected fish. Histological examination revealed multifocal granulomatous lesions, with the presence of numerous small, pleomorphic coccobacilli, predominantly in the peritoneum, spleen, kidneys, liver, pancreas, heart, and intestine. Sequencing of an ∼1,400-bp fragment of the 16S rRNA gene demonstrated these isolates to be most similar (99.9% identity) to Francisella sp. isolate TX077308 cultured from seawater in the Gulf of Mexico, while sharing <99% similarity to other Fransicella spp. Biochemical analysis, multilocus sequence comparisons of select housekeeping genes, repetitive extragenic palindromic PCR fingerprinting, matrix-assisted laser desorption ionization–time of flight mass spectrometry, and fatty acid methyl ester analysis revealed marked differences between these isolates and other described members of the genus. Koch's postulates were fulfilled by experimental intracoelomic injection and immersion trials using Nile (Oreochromis niloticus) and blue (Oreochromis aureus) tilapia. Based on observed phenotypic and genotypic differences from recognized Francisella spp., the name Francisella marina sp. nov. (NRRL B-65518) is proposed to accommodate these novel strains.
IMPORTANCE Finfish aquaculture is the fastest growing global food production sector. Infectious disease, particularly emergent pathogens, pose a significant threat to established and nascent aquaculture industries worldwide. Herein, we characterize a novel pathogen isolated from mortality events in cultured spotted rose snapper in Central America. The bacteria recovered from these outbreaks were genetically and phenotypically dissimilar from other known Francisella spp. from fish, representing a previously unrecognized member of the genus Francisella, for which the name Francisella marina sp. nov. is proposed.
INTRODUCTION
Aquaculture is one of the world's fastest developing food production sectors, largely in response to worldwide declines in wild fisheries and global fish catches (1). However, infectious disease poses a significant threat to aquaculture production and economic viability (2, 3). The rapid expansion of worldwide aquaculture, globalization, and the increased international transport of fish and fish products have facilitated the emergence and rapid dissemination of several potentially devastating disease agents, including members of the genus Francisella. Furthermore, intensive aquaculture practices provide an environment favorable to epizootics of disease resulting in significant economic losses (2, 3).
The family Francisellaceae, order Thiotrichales of the subclass Gammaproteobacteria, contains the single genus Francisella. Two subspecies, the cold-water pathogen Francisella noatunensis subsp. noatunensis (syn. Francisella philomiragia subsp. noatunensis and Francisella piscicida) and the temperate/warm-water pathogen Francisella noatunensis subsp. orientalis (syn. Francisella asiatica) have been associated with significant losses in a range of cultured and wild fish species from the Americas, Europe, and Asia (4). Francisella spp. are Gram-negative, nonmotile, pleomorphic coccobacilli, ranging from 0.2 to 0.4 μm wide and 0.4 to 1.9 μm long. They are catalase-positive, oxidase-negative, facultative anaerobes with a growth requirement for cysteine. While F. noatunensis subsp. noatunensis and F. noatunensis subsp. orientalis have been well characterized and their impacts on fish health broadly described, uncharacterized Francisella spp. have been isolated from marine environments (5), suggesting that additional species will be recognized in the future.
Francisellosis in cold- and warm-water fish results in similar nonspecific clinical signs, including abnormal swimming and anorexia. Catastrophic outbreaks can occur, with mortality rates exceeding 90%. Disease incidence is associated with a variety of fish sizes and ages, water temperature, water quality, and handling stress (4). External changes include pale gills, pale or darkened skin, petechial to ecchymotic hemorrhages, scale loss, skin ulceration, frayed fins, and exophthalmia. Internally, ascites and marked organomegaly characterize the disease. Pale tan to cream-colored granulomas are dispersed primarily in the kidneys and spleen but may occur in the gills, liver, gastrointestinal tract, and mesenteric fat. The small bacteria are found within vacuoles in macrophage cytoplasm (4).
Herein we report findings from investigations of mortality events that occurred in cultured spotted rose snapper (Lutjanus guttatus) on the Pacific coast of Central America in 2015 and 2016. A previously unrecognized Francisella sp. was recovered and characterized by multiple molecular and phenotypic techniques. Histopathologic findings are described, and Koch's postulates were fulfilled.
RESULTS
Clinical signs and histopathology of affected snapper.
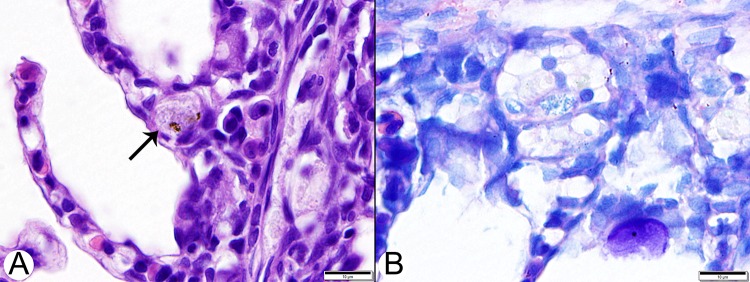
Clinically affected fish weighed 0.05 to 0.3 g and were 2 to 3 cm in length. The diseased fish were underconditioned and anorexic and swam lethargically just beneath the water's surface, often with darkened skin and distended coeloms. No specific external signs were evident, and no ectoparasites were present on gill clips or skin wet mounts. Mortality was estimated at 3 to 8% per day. Some survivors developed disproportionately large heads and small bodies. Histologically, the most severe changes involved the spleen, kidneys, liver, pancreas, heart, peritoneum, and intestine (Fig. 1), consisting of granulomatous inflammation, with large numbers of macrophages containing small pleomorphic coccobacilli. Bacteria were most evident with Giemsa and toluidine blue stains (Fig. 1D). Unaffected fish had normal growth and development in sea cages, particularly if larger than 1.5 g at the time of stocking.
FIG 1.
Histologic lesions in a spotted rose snapper (Lutjanus guttatus) naturally infected with a novel Francisella sp. (A) Low-magnification image of liver with a large focus of necrosis and granulomatous inflammatory infiltrates (arrows). H&E stain. Bar, 200 μm. (B) The spleen (lower left) is enlarged by inflammatory infiltrates dominated by macrophages. An arrow indicates an area of submucosal inflammation. The pale-staining cells are macrophages with cytoplasmic vacuoles containing bacteria. H&E stain. Bar, 50 μm. (C) Higher-magnification image of liver with a small focal area of necrosis and granulomatous inflammation. Arrowheads indicate vacuolated macrophages with intracellular bacteria. H&E stain. Bar, 50 μm. (D) Giemsa-stained section of liver with an irregular, pale-staining focus composed of macrophages with small intracytoplasmic bacteria typical of Francisella spp. Arrowheads indicate vacuolated macrophages with intracellular bacteria. Bar, 50 μm.
Isolation, growth, and biochemical characteristics.
A bacterium with a single-colony morphology was recovered from the coelomic cavities of diseased snapper. Growth of isolates E103-15 and E95-16 was visible after 24 h at 28 to 30°C, producing whitish-gray, smooth, convex colonies on blood agar. Growth at 20 to 28°C was slow, requiring 4 days of incubation, and no growth was observed at 37°C (Table 1). Under light microscopy, the bacterium was pleomorphic, Gram negative, nonmotile, and ∼0.5 to 1 μm in diameter.
TABLE 1.
Growth analysis of a novel Francisella sp. (E95-16) in Central Americaa
| Temp (°C) | Growthb |
|||
|---|---|---|---|---|
| 24 h |
96 h |
|||
| MTM | Blood agar | MTM | Blood agar | |
| 20 | − | − | + | ++ |
| 25 | + | ++ | ++ | +++ |
| 30 | ++ | +++ | +++ | ++++ |
| 37 | − | − | − | − |
Growth was on modified Thayer-Martin agar (MTM) and Columbia agar supplemented with 5% sheep blood (blood agar) at different temperatures (20, 25, 30 and 37°C) for 24 and 96 h.
+, growth only in first quadrant of inoculated plate; ++, growth in first and second quadrants of inoculated plate; +++, growth in first, second, and third quadrants of inoculated plate; ++++, growth in all quadrants; −, no growth observed.
All novel Francisella isolates were catalase positive and spot indole and cytochrome oxidase negative. Reactions in the triple sugar iron (TSI) medium were alkaline slant and alkaline butt, with no H2S or gas production. All isolates were negative for urease, citrate utilization, and β-galactosidase. The only positive tests produced by the novel Francisella sp. on API 20E identification strips were acetoin production from glucose (Voges-Proskauer test) and gelatinase. Comparably, F. noatunensis isolates were spot indole and cytochrome oxidase negative, failed to grow on any of the tube media, and produced no positive reactions in the identification strip.
Molecular identification.
Isolates E103-15 and E95-16 yielded PCR products of 1,150 bp when the Francisella genus-specific primers F11 and F5 were used (6), consistent with other members of the genus Francisella. However, isolates were negative for the iglC gene of F. noatunensis subsp. orientalis by quantitative PCR (7). Initial sequencing of a 1,357-bp fragment of the 16S rRNA gene demonstrated high homology (99.9%) to Francisella sp. isolate TX077308 (GenBank accession no. NC_015696) isolated from seawater in the Gulf of Mexico (5), while demonstrating <99% identity to other validated Francisella congeners (Table 2).
TABLE 2.
Francisella species genomes used in MLSA primer design and Bayesian analysis and 16S rRNA gene homology (1,357 bp) to case isolates E95-16 and E103-15
| Isolate | GenBank accession no. | 16S rRNA gene homology (%) |
|---|---|---|
| Francisella sp. TX077308 | NC_015696 | 99.9 |
| Francisella sp. FSC1006 | NZ_CP009574 | 99.6 |
| F. noatunensis subsp. orientalis F1 | NZ_CP018051 | 98.9 |
| F. noatunensis subsp. orientalis FNO24 | NZ_CP011922 | 98.9 |
| F. noatunensis subsp. orientalis LADL 07-185A | NC_023029 | 98.9 |
| Francisella sp. TX077310 | NZ_CP016796 | 98.9 |
| F. noatunensis subsp. noatunensis FSC772 | NZ_CP022207 | 98.8 |
| F. philomiragia subsp. philomiragia 0#319-067 | NZ_CP009436 | 98.7 |
| F. philomiragia subsp. philomiragia ATCC 25015 | NZ_CP010019 | 98.7 |
| F. philomiragia subsp. philomiragia ATCC 25017 | NC_010336 | 98.7 |
| F. tularensis subsp. tularensis NE061598 | NC_017453 | 97.5 |
| F. tularensis subsp. tularensis SCHU S4 | NC_006570 | 97.5 |
| F. tularensis subsp. novicida AL97-2214 | NZ_CP009653 | 97.4 |
| F. tularensis subsp. novicida F6168 | NZ_CP009353 | 97.4 |
| F. tularensis subsp. holarctica F92 | NC_019537 | 97.3 |
| F. tularensis subsp. holarctica LVS | NC_CP009694 | 97.3 |
| F. tularensis subsp. holarctica OSU18 | NC_008369 | 97.3 |
| F. tularensis subsp. novicida PA-7858 | NZ_CP016635 | 97.3 |
| F. tularensis subsp. tularensis WY96-3418 | NC_009257 | 97.3 |
DNA fingerprinting and MLSA.
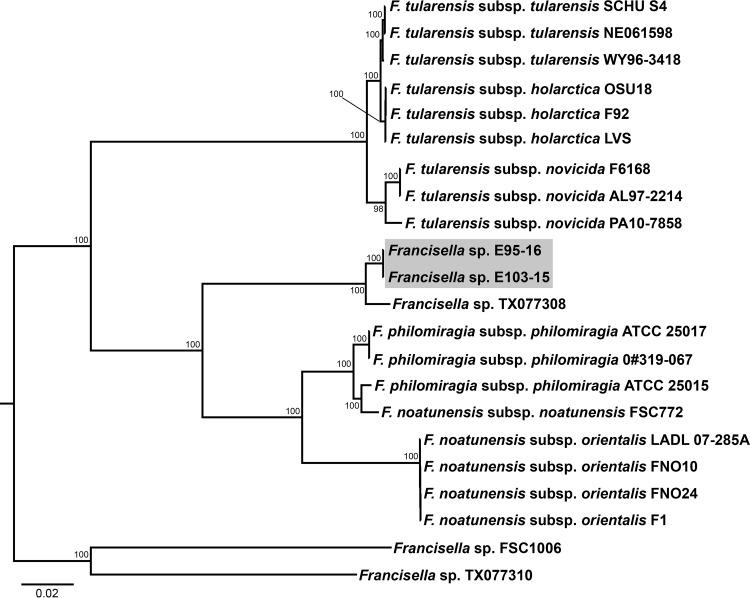
Repetitive extragenic palindromic PCR (Rep-PCR) analysis supported the initial 16S rRNA gene sequencing, which revealed isolates E103-15 and E95-16 to be divergent from archived F. noatunensis subsp. orientalis isolates from other fish species (Fig. 2 and 3). Multilocus sequence analysis (MLSA) showed that isolates recovered from rose snapper were clonal, demonstrating 100% homology at all gene targets, and were most similar to Francisella sp. TX077308, forming a discrete phyletic haplogroup, sister to the F. noatunensis subsp. orientalis/F. noatunensis subsp. noatunensis/F. philomiragia cluster, within a larger monophyletic clade separate from the F. tularensis subspecies cluster (Fig. 4). Sequence similarities (%) for select housekeeping genes of the snapper isolates (E103-15 and E95-16) to genomes of F. noatunensis subsp. orientalis (LADL 07-285) from tilapia in Costa Rica (GenBank NC_023029), F. noatunensis subsp. noatunensis (strain FSC772) from Atlantic salmon (Salmo salar) in Chile (GenBank NZ_CP022207), and Francisella sp. TX077308 from seawater in the Gulf of Mexico (GenBank accession no. NC_015696) are listed in Tables 2 and 3.
FIG 2.
Repetitive extragenic palindromic PCR (Rep-PCR) analysis of unknown Francisella sp. isolates from cultured spotted rose snapper and archived F. noatunensis subsp. orientalis isolates. Amplification was performed using the ERIC I-ERIC II primer set. Lanes L, HyperLadder, 50 bp; lanes 1 to 10, F. noatunensis subsp. orientalis isolates 1 to 10, respectively (Table 7); lane 11, E95-16; lane 12, E103-15; N, no-template control.
FIG 3.
Bayesian inference tree for Francisella spp. based on 1,357 bp of the 16S rRNA gene sequence. Numbers adjacent to branches represent posterior probability values (values of <0.50 are not shown). Francisella sp. isolates recovered from mortality events in snapper mariculture in Central America are highlighted in gray. GenBank accession numbers for reference genomes used in this study are listed in Tables 2 and 3.
FIG 4.
Bayesian inference tree for Francisella spp. based on concatenated 16S rRNA > dnaK > gyrB > mutS > pgm > prfB > rpoB > sodB sequences. Numbers adjacent to branches represent posterior probability values (values of <0.50 are not shown). Francisella sp. isolates recovered from mortality events in snapper mariculture in Central America are highlighted in gray. GenBank accession numbers for reference genomes used in this study are listed in Tables 2 and 3.
TABLE 3.
Sequence identity of select housekeeping genes for Francisella sp. isolates recovered from disease outbreaks in cultured spotted rose snapper in Central America and representative Francisella genomesa
| Outbreak isolateb | % sequence identity to the indicated housekeeping gene (size, bp) of the genome of: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fno | Fnn | TX077308 | Fno | Fnn | TX077308 | Fno | Fnn | TX077308 | Fno | Fnn | TX077308 | |
| 16S rRNA gene (1,357) | dnaK (875) | gyrB (1,091) | mutS (859) | |||||||||
| Fno 10 | 100.0 | 99.0 | 98.8 | 100.0 | 87.4 | 87.7 | 100.0 | 96.0 | 85.9 | 100.0 | 92.9 | 81.3 |
| E95-16 | 98.9 | 98.8 | 99.9 | 87.1 | 93.4 | 98.5 | 85.2 | 85.7 | 97.6 | 80.7 | 80.6 | 98.7 |
| E103-15 | 98.9 | 98.8 | 99.9 | 87.1 | 93.4 | 98.5 | 85.2 | 85.7 | 97.6 | 80.7 | 80.6 | 98.7 |
| pgm (702) | prfB (765) | rpoB (887) | sodB (334) | |||||||||
| Fno 10 | 100.0 | 95.0 | 94.3 | 100.0 | 94.1 | 85.6 | 100.0 | 93.9 | 89.4 | 100.0 | 89.5 | 84.1 |
| E95-16 | 94.4 | 96.0 | 98.1 | 87.5 | 88.8 | 94.9 | 89.4 | 88.4 | 100.0 | 83.5 | 84.4 | 99.1 |
| E103-15 | 94.4 | 96.0 | 98.1 | 87.5 | 88.8 | 94.9 | 89.4 | 88.4 | 100.0 | 83.5 | 84.4 | 99.1 |
Sequence identity was determined for Francisella sp. isolates recovered from disease outbreaks in cultured spotted rose snapper (Lutjanus guttatus) in Central America (E95-16 and E103-15) and representative genomes of Francisella noatunensis subsp. orientalis LADL 07-285 (Fno) from tilapia (Oreochromis sp.) in Costa Rica (GenBank accession no. NC_023029), Francisella noatunensis subsp. noatunensis FSC772 (Fnn) from Atlantic salmon (Salmo salar) in Chile (GenBank accession no. NZ_CP022207), and Francisella sp. TX077308 from seawater in the Gulf of Mexico (NC_015696).
Francisella noatunensis subsp. orientalis isolate 10 (Fno 10) was included as a sequencing-positive control.
Antimicrobial susceptibility testing.
MICs for 18 different antimicrobials against isolates E103-15 and E95-16 were identical. Consistent with other Francisella spp., the two isolates had low MICs to florfenicol, enrofloxacin, gentamicin, neomycin, oxytetracycline, and tetracycline and high MICs to amoxicillin, erythromycin, ceftiofur, spectinomycin, sulfadimethoxine, and trimethoprim-sulfamethoxazole. Complete MIC findings are presented in Table 4.
TABLE 4.
Antimicrobial MICs of novel Francisella sp. isolates determined by broth microdilutiona
| Antimicrobial | MIC (μg/ml) |
|
|---|---|---|
| E95-16 | E103-15 | |
| Enrofloxacin | <0.12 | <0.12 |
| Gentamicin | <0.5 | <0.5 |
| Cetiofur | >4 | >4 |
| Neomycin | <2 | <2 |
| Erythromycin | >4 | >4 |
| Oxytetracycline | <0.25 | <0.25 |
| Tetracycline | <0.25 | <0.25 |
| Amoxicillin | >16 | >16 |
| Spectinomycin | >64 | >64 |
| Sulfadimethoxine | >256 | >256 |
| Trimethoprim-sulfamethoxazole | >2/38 | >2/38 |
| Florfenicol | <1 | <1 |
| Sulfathiazole | >256 | >256 |
| Penicillin | >8 | >8 |
| Streptomycin | <8 | <8 |
| Novobiocin | 2 | 2 |
| Tylosin tartrate | >20 | >20 |
| Clindamycin | >4 | >4 |
Broth microdilution was performed according to the methods of Soto et al. (21).
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis.
Both snapper isolates and all archived F. noatunensis subsp. orientalis isolates (n = 5) yielded high-quality spectra, adequate for protein spectrum analysis and comparisons, differentiating the snapper isolates from spectra of other known Francisella spp. A dendrogram was constructed using main spectrum profiles (MSPs) from isolates of the two snappers, five F. noatunensis subsp. orientalis profiles, and reference main spectrum profiles, including six F. philomiragia profiles and six F. tularensis spectra (Fig. 5). The novel Francisella isolates were more similar to the F. noatunensis subsp. orientalis isolates than to F. philomiragia or F. tularensis yet formed their own discrete cluster.
FIG 5.
Dendrogram constructed from MALDI-TOF MS main spectrum profiles of 19 Francisella sp. strains, including five Francisella noatunensis subsp. orientalis (Fno) strains (Table 7) and two novel Francisella marina isolates recovered from cultured snapper (Lutjanus guttatus) (E103-15 and E95-16).
FAME analysis.
Fatty acid methyl ester (FAME) analysis of the two snapper isolates and archived F. noatunensis subsp. orientalis isolates (n = 5) demonstrated a similar core group of shared fatty acids in all isolates (9:0, 10:0, 11:0, 12:0, 13:0, 14:0, 16:0, 17:1 ω8c, 16:0 3OH, 18:1 ω9c, 18:0, 17:0 3OH, 18:0 3OH, 20:0, and summed feature 3 [Table 5]). However, Francisella sp. isolates E103-15 and E95-16 exhibited a lower percentage of the fatty acid 12:0 and a higher percentage of the fatty acid 16:0 3OH than the F. noatunensis subsp. orientalis isolates. Although the fatty acid compositions were low (<1%), isolates E103-15 and E95-16 did not produce fatty acids 17:0 and 20:1 ω7c and were the only isolates to produce the fatty acids 16:1 ω9c, summed feature 2, and summed feature 7 (Table 5). The dendrogram generated from the cluster analysis segregated the isolates into two clusters, one composed of the F. noatunensis subsp. orientalis isolates and one composed of isolates E103-15 and E95-16 (Fig. 6).
TABLE 5.
Fatty acid composition of five F. noatunensis subsp. orientalis isolates and two Francisella species isolatesa
| Fatty acid | Fatty acid composition (%)a |
||||||
|---|---|---|---|---|---|---|---|
|
Francisella sp. |
F. noatunensis subsp. orientalis |
||||||
| E103-15 | E95-16 | Isolate 1 | Isolate 2 | Isolate 3 | Isolate 4 | Isolate 5 | |
| 9:0 | 0.40 ± 0.35 | 0.65 ± 0.02 | 1.28 ± 0.38 | 1.19 ± 0.10 | 1.32 ± 0.28 | 1.39 ± 0.06 | 1.14 ± 0.15 |
| 10:0 | 54.2 ± 0.81 | 52.4 ± 2.20 | 51.4 ± 3.52 | 45.6 ± 8.21 | 45.3 ± 7.62 | 46.1 ± 2.39 | 51.8 ± 1.16 |
| 11:0 | 0.26 ± 0.23 | 0.37 ± 0.03 | 1.45 ± 0.47 | 1.38 ± 0.15 | 1.60 ± 0.36 | 1.66 ± 0.08 | 1.40 ± 0.16 |
| 12:0 | 0.77 ± 0.02 | 0.77 ± 0.02 | 4.30 ± 0.10 | 4.00 ± 0.53 | 4.09 ± 0.49 | 3.97 ± 0.10 | 4.15 ± 0.08 |
| 13:0 | 0.35 ± 0.32 | 0.34 ± 0.07 | 1.18 ± 0.26 | 1.53 ± 0.45 | 1.73 ± 0.24 | 1.86 ± 0.10 | 1.40 ± 0.14 |
| 14:0 | 16.7 ± 1.00 | 16.9 ± 0.48 | 17.1 ± 2.40 | 19.4 ± 0.83 | 19.0 ± 2.14 | 18.2 ± 0.35 | 17.3 ± 1.37 |
| 16:1 ω9c | 0.55 ± 0.02 | 0.64 ± 0.06 | ND | ND | ND | ND | ND |
| 16:0 | 3.20 ± 0.19 | 3.33 ± 0.29 | 2.75 ± 0.52 | 3.58 ± 1.24 | 3.54 ± 1.28 | 3.55 ± 0.58 | 2.94 ± 0.04 |
| 17:1 ω8c | 0.19 ± 0.16 | 0.33 ± 0.03 | 0.42 ± 0.32 | 0.71 ± 0.24 | 0.76 ± 0.10 | 0.73 ± 0.04 | 0.52 ± 0.03 |
| 17:0 | ND | ND | 0.44 ± 0.07 | 0.36 ± 0.62 | 0.67 ± 0.14 | 0.28 ± 0.48 | 0.51 ± 0.12 |
| 16:0 3OH | 6.49 ± 0.63 | 6.49 ± 0.10 | 2.65 ± 0.31 | 3.04 ± 0.51 | 3.40 ± 0.29 | 3.40 ± 0.29 | 3.14 ± 0.08 |
| 18:1 ω9c | 5.57 ± 0.64 | 5.80 ± 0.66 | 6.74 ± 0.89 | 7.94 ± 2.02 | 7.83 ± 2.04 | 7.75 ± 1.16 | 6.66 ± 0.07 |
| 18:1 ω5c | ND | ND | 0.12 ± 0.22 | 0.20 ± 0.35 | 0.21 ± 0.36 | 0.20 ± 0.35 | ND |
| 18:0 | 0.66 ± 0.23 | 0.56 ± 0.05 | 2.00 ± 1.05 | 1.54 ± 0.14 | 1.28 ± 0.54 | 1.27 ± 0.17 | 1.24 ± 0.06 |
| 17:0 3OH | 0.71 ± 0.12 | 0.83 ± 0.07 | 0.76 ± 0.25 | 0.84 ± 0.35 | 0.92 ± 0.17 | 0.96 ± 0.07 | 0.75 ± 0.19 |
| 19:0 | ND | ND | ND | 0.08 ± 0.13 | 0.07 ± 0.12 | ND | ND |
| 18:0 3OH | 7.80 ± 0.54 | 8.27 ± 0.29 | 4.31 ± 0.32 | 5.62 ± 1.97 | 5.87 ± 1.83 | 6.27 ± 0.73 | 4.80 ± 0.14 |
| 20:1 ω7c | ND | ND | 0.55 ± 0.08 | 0.84 ± 0.53 | 0.09 ± 0.16 | 0.22 ± 0.20 | 0.35 ± 0.30 |
| 20:0 | 0.39 ± 0.06 | 0.42 ± 0.04 | 0.51 ± 0.09 | 0.68 ± 0.09 | 0.59 ± 0.07 | 0.55 ± 0.06 | 0.56 ± 0.16 |
| Summed feature 2b | 0.15 ± 0.13 | 0.21 ± 0.02 | ND | ND | ND | ND | ND |
| Summed feature 3c | 0.82 ± 0.14 | 0.78 ± 0.05 | 1.36 ± 0.09 | 1.08 ± 0.35 | 1.21 ± 0.05 | 1.09 ± 0.05 | 1.25 ± 0.04 |
| Summed feature 6d | ND | ND | ND | 0.06 ± 0.10 | 0.09 ± 0.15 | ND | ND |
| Summed feature 7e | 0.67 ± 0.21 | 0.76 ± 0.07 | ND | ND | ND | ND | ND |
Values are means ± standard deviations of results of triplicate FAME extractions per isolate. ND, not detected.
The fatty acids 16:1 iso I/14:0 3OH and 14:0 3OH/16:1 iso I could not be separated from each other and together were considered summed feature 2.
The fatty acids 16:1 ω7c/16:1 ω6c and 16:1 ω6c/16:1 ω7c could not be separated from each other and together were considered summed feature 3.
The fatty acids 19:1 ω11c/19:1 ω9c and 19:1 ω9c/19:1 ω11c could not be separated from each other and together were considered summed feature 6.
The fatty acids 19:1 ω7c/19:1 ω6c and 19:1 ω6c/ω7c/19cy could not be separated from each other and together were considered summed feature 7.
FIG 6.

Dendrogram of fatty acid profiles of five Francisella noatunensis subsp. orientalis (Fno) isolates and two Francisella marina isolates (E103-15 and E95-16) (Table 7).
Experimental challenge.
No clinical signs of diseases were observed in the challenged fish. Limited mortality was observed during experimental challenges. Intracoelomic (IC) injection of ∼107 CFU/fish resulted in 40% (4/10) mortality in naive Nile tilapia (Oreochromis niloticus) 21 days postinoculation. Challenged fish died on days 5 (n = 1), 7 (n = 2), and 20 (n = 1), although postmortem autolysis precluded histological analysis. No naive blue tilapia (O. aureus) died. Similarly, no mortality was found in either tilapia species challenged via immersion routes. At the completion of the experimental challenge, all control fish were alive, and no bacterial infection was detected in spleen homogenates. Digests of harvested spleens from surviving fish in each group of animals challenged by IC injection and immersion routes with isolate E103-15 were plated on modified Thayer-Martin agar for CFU determination per milligram of organ weight. One of three Nile tilapia from the immersion challenge yielded CFU from its spleen, with an estimated bacterial load of 1 CFU/mg spleen, while two of three Nile tilapia from the injection challenge group had positive splenic cultures, with estimated bacterial loads of 120 and 1 CFU/mg spleen, respectively. Comparably, three of three blue tilapia from the IC injection challenge group had positive splenic cultures, with estimated bacterial loads of 1, 1, and 4 CFU/mg spleen. The spleens of these animals were grossly nondescript. No viable bacteria were recovered from Nile or blue tilapia control fish or blue tilapia challenged by immersion.
Incidental findings in both control and treatment fish groups included scattered eosinophilic granular cells, mild nephrocalcinosis, mineralization in the pseudobranch, and mild bile duct hyperplasia. Lesions in surviving Nile and blue tilapia challenged by IC injection were dominated by variably sized, often irregular, loosely organized collections of pale-staining macrophages with vacuolated cytoplasm that often contained pale eosinophilic to golden or black cytoplasmic granules. Scattered throughout were small numbers of macrophages with a large cytoplasmic vacuole containing small to large numbers of small coccoid bacteria (Fig. 7A). Some foci contained small numbers of lymphocytes, primarily on their periphery. One blue tilapia had extensive coalescing lesions in the head kidney, with abundant central necrotic debris and apoptotic cells (Fig. 7B). Well-organized epithelioid granulomas with caseous centers were seen in only two Nile tilapia (Fig. 7C). Comparable lesions were present in the kidney and liver of one immersion-challenged Nile tilapia, but no bacteria were observed.
FIG 7.
Hematoxylin and eosin (H&E)-stained tissue sections from Nile and blue tilapia challenged with Francisella marina isolate E95-16 by intracoelomic injection. Lesions were similar in both species and most commonly affected the head kidney and spleen. (A) Head kidney with a small lesion composed of vacuolated macrophages. The arrow indicates a cluster of pleomorphic intracellular bacteria. Bar, 10 μm. (B) More advanced head kidney lesion with a central region containing apoptotic cells and necrotic debris. Bar, 50 μm. (C) Head kidney lesion dominated by typical vacuolated macrophages. A small granuloma with central caseation surrounded by epithelioid cells is visible in the upper right. Granuloma formation was rare and interpreted as a final stage in lesion progression. Bar, 20 μm. (D) Low-magnification image of head kidney with extensive, frequently coalescing lesions. Bar, 500 μm. (E) Splenic lesions began with expansion of periarteriolar sheaths by vacuolated macrophages (arrows). Bar, 50 μm. (F) Liver with a small perivascular cluster of macrophages containing intracellular bacteria. Bar, 10 μm.
In both tilapia species, the head kidney and spleen were most commonly and seriously affected. Head kidney lesions were randomly distributed (Fig. 7D), while those in the spleen expanded ellipsoids (Fig. 7E). Liver lesions were infrequent, small, and usually located adjacent to vascular or biliary structures (Fig. 7F). Other commonly affected sites included the ocular choroid rete and the gills. Gill lesions were widely distributed, but inconspicuous, usually involving only individual or small numbers of macrophages within the lumens of or immediately adjacent to the bases of lamellar capillaries (Fig. 8A). Infrequent lesion sites included the trunk kidney, where only hematopoietic tissues were affected, the meninges, gastric submucosa, and skeletal muscle. In all lesions, bacteria were best visualized using Giemsa stains (Fig. 8B). Lesion distribution findings are summarized in Table 6.
FIG 8.
Gill lesions in blue tilapia challenged with Francisella marina isolate E95-16 by intracoelomic injection. (A) Gill changes were inconspicuous, often involving only single macrophages laden with bacteria (arrow) within capillary lumens at the bases of lamellae. H&E stain. Bar, 10 μm. (B) In all tissue locations, the small bacteria were best visualized using Giemsa stains. Bar, 10 μm.
TABLE 6.
Lesion distribution in Nile and blue tilapia challenged with Francisella E95-16 from Central Americaa
| Fish and challenge | No. of tissue samples with lesions/total no.b |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BRN | OCR | GIL | HKD | TKD | LVR | SPL | STM | INT | HRT | SKM | GND | |
| Nile tilapia | ||||||||||||
| Control | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| Immer | 0/3 | 0/3 | 0/3 | 1/3c | 1/3c | 1/3c | 0/3 | 0/3 | 0/3 | 0/2d | 0/3 | 0/3 |
| IC inj | 1/3 | 2/3 | 3/3 | 3/3 | 2/3 | 2/3 | 3/3 | 0/3 | 0/3 | 0/3 | 1/3 | 0/3 |
| Blue tilapia | ||||||||||||
| Control | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| Immer | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| IC inj | 1/3 | 2/3 | 2/3 | 3/3 | 0/3 | 2/3 | 0/2d | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 |
Tilapia were challenged by the immersion (Immer) and intracoelomic injection (IC inj) routes.
Abbreviations: BRN, brain; OCR, ocular choroid rete; GIL, gill; HKD, head kidney; TKD, trunk kidney; LVR, liver; SPL, spleen; STM, stomach; INT, intestine; HRT, heart; SKM, skeletal muscle; GND, gonad.
Comparable lesions, but no bacteria were visualized.
One tissue was not present in histological sections.
DISCUSSION
Since its first description in 2005, piscine francisellosis has emerged as an important disease of cultured fish, affecting a wide range of freshwater and marine fish species around the world. Historically, francisellosis has been attributed to two subspecies of Francisella noatunensis, namely Francisella noatunensis subsp. noatunensis and Francisella noatunensis subsp. orientalis, the latter of which has been linked to epizootics in cultured tilapia in Central America (8). The current study reports the isolation and characterization of a novel marine Francisella sp. causing mortality in cultured spotted rose snapper in Central America.
Multiple techniques, including Rep-PCR DNA fingerprinting, MLSA, MALDI-TOF MS, and FAME, were employed to compare archived F. noatunensis subsp. orientalis isolates, typically associated with warm- and temperate-water fish species, and the Francisella sp. investigated here. Rep-PCR DNA fingerprinting has been previously used to discriminate closely related bacterial strains (9–11). The ERIC I-ERIC II primer set revealed marked differences between the snapper isolates and F. noatunensis subsp. orientalis. MLSA is also used to resolve interspecific relationships not readily determined by a single genetic marker, such as the multicopy 16S rRNA gene. In addition to gene targets commonly utilized in bacterial systematics (12–16), the analysis included the genes mutS, pgm, and prfB, used specifically to characterize Francisella spp. (17). Gene targets were chosen to represent diverse chromosomal loci and a wide distribution among different prokaryote taxa to facilitate PCR primers that would amplify all Francisella spp. The MLSA method was adequately robust and discriminated among Francisella isolates to the species and subspecies level and, similar to analysis based on the 16S rRNA gene, segregated isolates recovered from spotted rose snapper from other recognized Francisella spp. This genetic divergence was supported by MALDI-TOF MS and FAME analysis, which demonstrated notable differences in spectral and fatty acid profiles between the snapper isolates and other Francisella spp.
Histological lesions were consistent with francisellosis and, in particular, those described in tilapia challenged with F. noatunensis subsp. orientalis (18). However, pathogenicity of the Francisella marina isolated from the snapper epizootics was less than that caused by F. noatunensis subsp. orientalis in controlled challenges with tilapia, particularly via immersion (8). Future in vivo challenges using a marine fish model of infection, such as snapper, are warranted to investigate the pathogenesis and virulence of the novel isolates.
The MIC profiles of the Francisella sp. were consistent with those of F. noatunensis subsp. orientalis and F. noatunensis subsp. noatunensis, including susceptibility to oxytetracycline, which reduced mortality in the epizootics described previously (19, 20), and florfenicol (21). This is noteworthy, as oxytetracycline and florfenicol are approved therapeutants against piscine francisellosis in food fish in several fish-producing countries.
The source of the bacteria in these outbreaks is unknown, but recent descriptions of Francisella endociliophora and Francisella halioticida from marine invertebrates (17, 22, 23) suggest that marine environments may harbor additional Francisella spp., some of which may cause disease in aquatic organisms. While the techniques employed herein are reliable methods of bacterial typing, more robust genetic studies consisting of either phylogenomics and/or DNA-DNA hybridization analysis are warranted to elucidate the intrageneric and interspecific relationships between the case isolates and other members of the Francisella genus. However, based on these results, it is evident that the isolates recovered from cultured spotted rose snapper in Central America are phylogenetically divergent from any described species of Francisella and represent a genetically distinct yet previously unrecognized taxon of Francisella, most similar to Francisella sp. isolate TX077308 isolated from seawater in the Gulf of Mexico. Based on phenotypic distinctiveness and molecular genetic evidence, it is proposed these strains be classified as representatives of a novel species of the genus Francisella, known hereafter as Francisella marina sp. nov.
Description of Francisella marina sp. nov.
Francisella marina sp. nov. [ma.ri'na. L. fem. adj. marina, marine, referring to where the type strain was first isolated]. Cells are aerobic, Gram negative, pleomorphic, nonmotile, and ∼0.5 to 1 μm in diameter. Growth observed at 20 to 30°C, no growth at 37°C, optimal growth at 30°C. Growth was visible after 24 h at 28 to 30°C, producing whitish-gray, smooth, convex colonies on blood agar. Isolates are spot indole and cytochrome oxidase negative and catalase positive. Reactions in TSI medium are alkaline slant and alkaline butt, with no H2S or gas production. Isolates are negative for urease, citrate utilization, and β-galactosidase, produce acetoin from sodium pyruvate, and hydrolyze gelatin. Sequencing of an ∼1,400-bp fragment of the 16S rRNA gene demonstrated F. marina to be most similar (99.9% identity) to Francisella sp. isolate TX077308 cultured from seawater in the Gulf of Mexico, while sharing <99% similarity to other Fransicella spp. Additional phenotypic and genotypic properties obtained by multilocus sequence analysis of housekeeping genes, repetitive extragenic palindromic PCR fingerprinting, matrix-assisted laser-desorption ionization–time of flight mass spectrometry, and fatty acid methyl ester analysis revealed marked differences between these isolates and other described members of the genus. The type strain, Francisella sp. E95-16 (NRRL B-65518), was isolated from a marine fish, spotted rose snapper (Lutjanus guttatus), in Puntarenas, Costa Rica.
MATERIALS AND METHODS
Fish history.
In 2015 and 2016, a spotted rose snapper (Lutjanus guttatus) hatchery on the Pacific coast of Central America reported disease outbreaks with high mortality at the beginning of the rainy season. Most affected fish were undersized fry (2 to 3 cm in length), and disease occurred 30 to 40 days posthatching, during the transition from feeding live Artemia sp. to a pelleted commercial diet. Some batches of fish were empirically treated with 2-h immersion baths of 20 ppm tetracycline for several days. While these bath treatments reduced fish losses, mortality persisted.
Isolation and histological examination.
Swabs from internal organs and ascitic fluid from the coelomic cavity were used to inoculate Columbia agar supplemented with 5% sheep blood (blood agar). Plates were incubated at 25°C for 5 days. Single colonies, subcultured for purity from primary isolation plates, were suspended in modified Mueller-Hinton II cation-adjusted broth supplemented with 2% IsoVitaleX (BD BBL, Sparks, MD, USA) and 0.1% glucose (MMH) (24). The broth cultures were expanded overnight at 25°C with shaking at 175 rpm. Aliquots of expanded cultures (1 ml) were supplemented with 20% (vol/vol) glycerol and cryogenically stored (−80°C) until further characterization. Additional whole fish were fixed in 10% neutral buffered formalin, processed routinely, and stained with hematoxylin and eosin (H&E), Ziehl-Neelsen, Grocott, toluidine blue, and Giemsa stains for light microscopic examination.
Two isolates (E103-15 and E95-16) recovered from snapper cultured in different outbreaks in 2015 and 2016 were used in this analysis. The optimal in vitro growth temperatures of isolates E103-15 and E95-16 were determined by 7-day incubations at 20, 25, 30, and 37°C on blood and modified Thayer-Martin agars (MTM) (BD BBL, Sparks, MD, USA).
Additionally, archived Francisella noatunensis subsp. orientalis strains used for comparison in this study were isolated from a range of fish species and geographical locations, several of which were described previously (21). Other isolates were generously shared by John Hansen from the Interdisciplinary Program in Pathobiology, University of Washington, Seattle, WA, and Duncan J. Colquhoun at the Section for Bacteriology, Norwegian Veterinary Institute, Oslo, Norway (Table 7).
TABLE 7.
Francisella isolates used in this study
| Species and isolate | Yr of isolation | Location | Fish | Environment | Reference |
|---|---|---|---|---|---|
| F. noatunensis subsp. orientalis | |||||
| 1a,b | 2001 | Japan | Three-line grunt (Parapristipoma trilineatum) | Seawater | 33 |
| 2a,b | 2011 | Texas, USA | Tilapia (Oreochromis sp.) | Freshwater | 21 |
| 3a,b | 2012 | Costa Rica | Nile tilapia (Oreochromis niloticus) | Freshwater | 21 |
| 4a,b | 2012 | Hawaii, USA | Tilapia (Oreochromis sp.) | Freshwater | 34 |
| 5a,b | 2013 | Florida, USA | French grunt (Haemulon flavolineatum) | Seawater | 20 |
| 6b | 2012 | Europe | Nile tilapia (O. niloticus) | Freshwater | 35 |
| 7b | 2010 | Midwest, USA | Nile tilapia (O. niloticus) | Freshwater | 36 |
| 8b | 2007 | Costa Rica | Nile tilapia (O. niloticus) | Freshwater | 8 |
| 9b | 2000–2005 | Costa Rica | Tilapia (Oreochromis sp.) | Freshwater | 21 |
| 10b | Unknown | California, USA | Hybrid striped bass (Morone chrysops x M. saxatilis) | Freshwater | 37 |
| Francisella sp. | |||||
| E95-16a,b | 2016 | Central America | Snapper (Lutjanus guttatus) | Seawater | Present study |
| E103-15a,b | 2015 | Central America | Snapper (L. guttatus) | Seawater | Present study |
Isolates used for biochemical, MALDI-TOF MS, and FAME comparisons.
Isolates used for Rep-PCR.
Biochemical analysis.
Isolates (Table 7) were tested for biochemical reactivity using spot tests, tube media, and API 20E identification strips (bioMérieux, Durham, NC). Isolates were spot tested for cytochrome oxidase (Becton Dickinson and Company, Sparks, MD), indole production from tryptophan (indole reagent; Anaerobe Systems, Morgan Hill, CA), and catalase using a 3% H2O2 solution (Optima Fisher Chemical).
Tubed medium prepared by the University of California Biological Media Services, Davis, CA (BMS), was used to evaluate the urease test using Christensen's urea agar, sugar fermentation, and hydrogen sulfide production using triple sugar iron (TSI) agar, citrate utilization, and motility using sulfur-indole motility agar. Production of β-galactosidase was tested using the ONPG (o-nitrophenyl-β-d-galactopyranoside) (rapid test (Hardy Diagnostics, Santa Maria, CA). API 20E test strips were inoculated and developed according to the manufacturer's instructions. Incubation of all tubed media and test strips was performed at 28°C in ambient air with a 24-h incubation period.
Molecular analysis.
For the isolation of genomic DNA, a loop of bacteria was suspended in 400 μl of sterile water, washed, centrifuged at 3,000 × g for 5 min, and resuspended in 200 μl phosphate-buffered saline (PBS). Genomic DNA was isolated using the High Pure PCR template preparation kit (Roche, Mannheim, Germany) by following the manufacturer's protocol. Isolated DNA was stored at 4°C until further use. Francisella sp.-specific PCR (6), universal eubacterial 16S rRNA PCR (24), and F. noatunensis subsp. orientalis iglC-specific quantitative PCR (7) were performed by following established protocols (Table 8). Endpoint PCRs were deemed positive by the presence of appropriate sized bands, visualized by agarose electrophoresis in the presence of ethidium bromide (0.05 μg/μl) and UV illumination. Amplicons were purified with the QiaQuick PCR cleanup kit (Qiagen), and the resulting sequences were compared by BLASTN searches of the National Center for Biotechnology Information's nonredundant nucleotide database. For quantitative PCR, only samples presenting a threshold cycle (Ct) of <40 were considered positive.
TABLE 8.
PCR primers used in this study
| Assay and primera | Sequence (5′–3′) | Reference |
|---|---|---|
| Initial molecular diagnostics | ||
| Francisella sp. PCR | ||
| F11 | TACCAGTTGGAAACGACTGT | 6 |
| F5 | CCTTTTTGAGTTTCGCTCC | |
| F. noatunensis subsp. orientalis iglC qPCR | ||
| iglC forward | GGGCGTATCTAAGGATGGTATGAG | 7 |
| iglC reverse | AGCACAGCATACAGGCAAGCTA | |
| iglC probe (FAM-BHQ-1) | ATCTATTGATGGGCTCACAACTTCACAA | |
| MLSA | ||
| 16S | ||
| 27F | GAGTTTGATCCTGGCTCAG | 24 |
| 1525R | AGAAAGGAGGTGATCCAGCC | |
| dnaK | ||
| FranDnaK872F | TCTTGTACTAGAGGCATACG | This paper |
| FranDnaK1889R | ACTACTAACTCTTGTCTTGCTAT | |
| gyrB | ||
| FranGyrB130F | GAGGTTGTTGATAATGCTATCG | This paper |
| FranGyrB1321R | TTTTACGATCACGAGCCTG | |
| mutS | ||
| FranmutS634F | ATCGGATCAATACTTGCTTATTT | This paper |
| FranmutS1610R | TGATAGCTCTTTCTGCAAAGT | |
| pgm | ||
| Franpgm576F | CGTTTTGACTCTATGAGTGC | This paper |
| Franpgm1330R | ATTGGATCTGTATAACTAAAATCATCT | |
| prfB | ||
| FranprfB54F | SGGCAGACAAAAAGTAGADCT | This paper |
| FranprfB856R | AGARCGAATCTGRCTWCCCCA | |
| rpoB | ||
| FranrpoB812F | ACACCTTTGTTACCATGACG | This paper |
| FranrpoB2052R | CATGATGATGCTAACAGGGT | |
| sodB | ||
| FransodB202F | GCCCAACCAGAWCCAAATG | This paper |
| FransodB563R | TTTGAATTACCAAAACTACCTT | |
| Rep-PCR | ||
| ERIC I | ATGTAAGCTCCTGGGGATTCAC | 25 |
| ERIC II | AAGTAAGTGACTGGGGTGAGCG | 25 |
qPCR, quantitative PCR; MLSA, multilocus sequence analysis; Rep-PCR, repetitive extragenic palindromic PCR.
DNA fingerprinting.
Repetitive extragenic palindromic PCR (Rep-PCR) fingerprinting was performed using the ERIC I-ERIC II primer set on isolates E103-15 and E95-16 and archived F. noatunensis subsp. orientalis isolates (Tables 7 and 8) as described previously (10, 25). The 50-μl reaction mixtures consisted of 25 μl of IQ Supermix (Bio-Rad Laboratories, Inc.), 10 pmol of each primer, 50 ng of template DNA, and nuclease-free water to volume. Amplifications were performed on a C1000 Touch thermal cycler (Bio-Rad Laboratories, Inc.) with the following temperature profiles: 1 cycle at 95°C for 10 min; 5 cycles of 95°C for 1 min, 40°C for 1 min, and 72°C for 5 min; and 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 5 min. Aliquots of each amplification reaction mixture (10 μl each) and a molecular weight standard (HyperLadder, 50 bp; Bioline) were electrophoresed through a 1.5% (wt/vol) agarose gel with ethidium bromide (0.5 μg/ml) and visualized under UV light.
Multilocus sequence analysis.
Isolates E103-15 and E95-16 were subjected to multilocus sequence typing and analysis (MLSA), with an archived F. noatunensis subsp. orientalis isolate (F. noatunensis subsp. orientalis 10) included as a sequence quality control. A combination of slow- and fast-evolving genes were chosen as targets, including a 16S small-subunit rRNA gene (16S) and those encoding molecular chaperone DnaK (dnaK), DNA gyrase subunit B (gyrB), DNA mismatch repair protein (mutS), alpha-d-glucose phosphate-specific phosphoglucomutase (pgm), peptide chain release factor 2 (prfB), DNA-directed RNA polymerase subunit β (rpoB), and superoxide dismutase (sodB). Primers were designed using the Primer3 utility in Geneious v.9.1.8 (32) based on complete Francisella species genomes available in GenBank (Tables 2 and 3). With the exception of the sodB locus, primers were designed to amplify approximately 700- to 1,000-bp fragments.
All MLSA gene targets were amplified in 50-μl reaction mixtures containing 25 μl of Platinum PCR supermix high fidelity, 20 pmol of each primer (Table 8), ∼50 ng of template DNA, and nuclease-free water to volume. Amplifications were performed on a Bio-Rad C1000 Touch thermal cycler with the following conditions: 94°C for 3 min, followed by 45 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1.5 min, with a final extension step of 68°C for 5 min. Amplification products were visualized under UV light after electrophoretic migration through a 0.8% agarose gel in the presence of ethidium bromide (0.5 μg/ml). Amplicons were excised and purified using the Qiagen Qiaquick gel extraction kit and sequenced commercially (Eurofins Genomics, Louisville, KY) using the same primers that generated the amplicons. Ambiguous base calls were manually annotated in Geneious using corresponding chromatograms, and contiguous sequences were used in subsequent BLASTn searches and phylogenetic analysis.
The sequenced MLSA targets were aligned with corresponding sequences obtained from complete genomes of select Francisella spp. in GenBank (Tables 2 and 3). Sequences for each respective gene target were initially aligned using the default settings of the global alignment implementation built-in to Geneious. All positions in the alignment containing at least one gap were eliminated, and alignments were concatenated in the following order: 16S > dnaK > gyrB > mutS > pgm > prfB > rpoB > sodB. The resulting concatenated alignment contained 6,809 sites excluding gaps (6,871 bp, including gaps). The best-fit nucleotide substitution model for each gene was determined based on the Bayesian Information Criterion in MEGA 7: 16S rRNA (HKY + G + I; 1,348 positions), dnaK (GTR + G; 875 positions), gyrB (T92 + I; 1,083 positions), mutS (TN93 + G; 844 positions), pgm (T92 + G; 692 positions), prfB (T92 + G; 755 positions), rpoB (GTR + G + I; 887 positions), and sodB (T92 + G; 325 positions) (26, 27). Alignments of 16S rRNA gene sequences, as well as alignments of concatenated sequences, were subjected to partitioned Bayesian inference analysis in MrBayes 3.2.6 by Markov chain Monte Carlo searches of two simultaneous runs of four chains (28). Chains were sampled every 100th tree for 1,000,000 generations (29), at which point convergence was reached (standard deviation of split frequencies was <0.01). The first 25% of trees were discarded as “burn-in,” and posterior probability values were calculated from the remaining trees. Consensus trees were visualized using FigTree 1.4.2 (30) and refined in Adobe Illustrator (Adobe, San Jose, CA, USA).
Antimicrobial susceptibility.
Antimicrobial susceptibility was assessed using a broth microdilution method adapted from Francisella tularensis to F. noatunensis subsp. orientalis (21, 31). MICs of antimicrobial agents against isolates E103-15 and E95-16 were tested using the Sensititre AVIANF plate system (Sensititre; ThermoFisher, Cleveland, Ohio). MICs were determined as the lowest concentration that resulted in no visible growth.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry.
Five archived F. noatunensis subsp. orientalis isolates and isolates E103-15 and E95-16 (Table 7) were grown on MTM agar at 25°C under ambient atmospheric conditions for 96 h. Protein extraction was performed using the Bruker Daltonik Revision 1 formic acid extraction procedure (Bruker Daltonics, Inc., Billerica, MA). Briefly, 5 to 10 bacterial colonies were suspended in 300 μl deionized water and 900 μl of high-performance liquid chromatography (HPLC)-grade absolute ethanol (Sigma-Aldrich), vortexed, and then centrifuged at 12,000 × g for 2 min. The supernatant was removed, the pellet recentrifuged as described above, residual ethanol was removed, and the pellet was air dried for 2 min. For all isolates, 40 μl 70% LC-MS formic acid (Sigma-Aldrich) was added to the pellet and thoroughly mixed by pipetting, followed by the addition of 40 μl of pure acetonitrile (Fluka, ThermoFisher) and additional gentle mixing. The sample was then centrifuged as described above and frozen at −80°C until analysis.
A main spectrum profile (MSP) was created for each isolate using a Biotyper MALDI-TOF MS (Microflex LT/SH; Bruker Daltonics, Inc., Billerica, MA). One microliter of sample supernatant was applied to each of 8 spots on a 96-spot polished steel target (Bruker Daltonics) and air dried, followed by 1 μl of α-cyano-4-hydroxycinnamic acid (Bruker Daltonics) and air drying. The instrument was calibrated using a bacterial test standard (Bruker Daltonics) and 24 MS spectra were collected for each isolate, making three measurements per spot by using the manufacturer's default settings. Using Flex analysis software (version 3.4), spectra were smoothed and baselines subtracted. Up to three spectra per isolate were removed to eliminate flatline or outlier spectra. MSPs were created in Biotyper OC (version 3.1) according to the manufacturer's directions. Analysis of spectra, including direct comparisons of MSPs, and dendrogram creation were performed using default settings within Biotyper OC and included reference MSPs of F. philomiragia and F. tularensis from the Biotyper OC database.
FAME analysis.
The same F. noatunensis subsp. orientalis and case isolates used for MALDI-TOF MS were included in the fatty acid methyl ester (FAME) analysis and grown under the same conditions (Table 7). Each isolate was streaked onto 15 MTM plates and incubated for 3 days at 25°C. In triplicate for each isolate, an average of 40 mg of bacteria was removed by carefully scraping the surface of the agar plates. Bacteria were coated onto the bottoms of individual Pyrex glass tubes and centrifuged for 1 min at 5,250 rpm. Saponification of bacteria, methylation, FAME extraction, and gas chromatography were performed as previously described (11). Chromatography results were analyzed using the Sherlock microbial identification system (MIS) RCLIN6 6.2 library (version 6.2; MIDI, Inc.). The triplicate FAME results were averaged for each isolate and imported into BioNumerics (version 6.6; Applied Maths) for cluster analysis. A similarity matrix was calculated using Euclidian distance, and the matrix was converted into a dendrogram using the unweighted pair group method using average linkages (UPGMA) algorithm.
Experimental challenges.
All challenges were conducted under protocols approved by the Institutional Animal Care and Use Committee of the University of California Davis School of Veterinary Medicine. Experimental challenges were performed by intracoelomic (IC) injection and immersion with isolate E103-15. Bacteria were grown on MTM agar at 25°C for 24 h. Three single colonies were suspended in 50 ml of MMH broth and incubated in a shaking incubator (150 rpm) overnight at 25°C to a final concentration of ∼1.33 × 1010 (CFU/ml). Enumeration of bacteria was done by the drop plate method with 10-μl drops of each 10-fold dilution placed on agar. The approximate number of CFU per ml of inoculum was determined after 24 h of incubation at 25°C.
Naive Nile tilapia and blue tilapia fingerlings were obtained from an aquaculture farm with no history of Francisella infection. Before challenge, five fish from each species were collected arbitrarily and confirmed negative for francisellosis by bacterial culture and quantitative PCR analysis of excised spleens (7). Fish (10/tank) were stocked into eight separate 135-liter aquaria containing 60 liters of flowthrough well water maintained at 20 to 22°C with supplemental aeration. Each treatment group (species × exposure method) consisted of one exposed tank and one unexposed control. Prior to challenge, the fish were anesthetized with buffered MS-222 (100 mg/liter). Fish challenged by IC injection received 0.1 ml of bacterial suspension (∼5.33 × 107 CFU/fish). For the immersion challenge, doses equating to ∼5.5 × 105 CFU/ml of bacteria were added to tanks, which were held static for 1 h, after which water flow was resumed. Mortality was recorded daily for 21 days. Moribund fish or those showing signs of abnormal swimming, lethargy, exophthalmia, skin lesions, or coelomic distension were euthanized with an overdose of buffered MS-222 (500 mg/liter) and necropsied. After 21 days, the remaining fish were euthanized and the spleens of three fish from each treatment group were harvested to determine approximate bacterial burdens. Organs were weighed, homogenized in 0.2 ml sterile PBS, plated in triplicate on MTM agar plates, and incubated at 25°C for 48 h prior to CFU determinations.
Random surviving fish from each group (n = 3 per group) designated for histological evaluation were incised along their ventral midlines and fixed whole in 10% neutral buffered formalin. Bodies were decalcified in Kristensen's solution and transected in 2- to 3-mm sections along the length of the body. Tissues were processed routinely, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin (H&E). Select sections were also stained by the Ziehl-Neelsen and Giemsa methods.
Accession number(s).
MLSA sequences have been submitted to GenBank under accession numbers MH057668 to MH057691. The type strain, Francisella sp. strain E95-16, has been deposited in the U.S. Department of Agriculture Agricultural Research Service (ARS) Culture Collection under the number NRRL B-65518. The type strain is also being deposited at the American Type Culture Collection (ATCC), but a culture number is not available at this time.
ACKNOWLEDGMENTS
We thank Bernhard Schink (University of Konstanz, Germany) and Stanislava Králová (Czech Collection of Microorganisms, Czech Republic) for assistance in naming the organism.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Food Agriculture Organization, United Nations. 2011. The state of world fisheries and aquaculture 2010. Food Agriculture Organization, United Nations, Rome, Italy. [Google Scholar]
- 2.Cressey D. 2009. Aquaculture: future fish. Nature 458:398–400. doi: 10.1038/458398a. [DOI] [PubMed] [Google Scholar]
- 3.Krkosek M. 2010. Host density thresholds and disease control for fisheries and aquaculture. Aquac Environ Interact 1:21–32. doi: 10.3354/aei0004. [DOI] [Google Scholar]
- 4.Colquhoun DJ, Duodu S. 2011. Francisella infections in farmed and wild aquatic organisms. Vet Res 42:47. doi: 10.1186/1297-9716-42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen JM, Carlson J, Yockey B, Pillai S, Kuske C, Garbalena G, Pottumarthy S, Chalcraft L. 2009. Direct isolation of Francisella spp. from environmental samples. Lett Appl Microbiol 48:663–667. doi: 10.1111/j.1472-765X.2009.02589.x. [DOI] [PubMed] [Google Scholar]
- 6.Forsman M, Sandstrom G, Sjostedt A. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int J Syst Bacteriol 44:38–46. doi: 10.1099/00207713-44-1-38. [DOI] [PubMed] [Google Scholar]
- 7.Soto E, Bowles K, Fernandez D, Hawke JP. 2010. Development of a real-time PCR assay for identification and quantification of the fish pathogen Francisella noatunensis subsp. orientalis. Dis Aquat Organ 89:199–207. doi: 10.3354/dao02204. [DOI] [PubMed] [Google Scholar]
- 8.Soto E, Hawke JP, Fernandez D, Morales JA. 2009. Francisella sp., an emerging pathogen of tilapia, Oreochromis niloticus (L), in Costa Rica. J Fish Dis 32:713–722. doi: 10.1111/j.1365-2761.2009.01070.x. [DOI] [PubMed] [Google Scholar]
- 9.Soto E, Griffin M, Wiles J, Hawke JP. 2012. Genetic analysis and antimicrobial susceptibility of Francisella noatunensis subsp. orientalis (syn. F. asiatica) isolates from fish. Vet Microbiol 154:407–412. doi: 10.1016/j.vetmic.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 10.Griffin MJ, Quiniou SM, Cody T, Tabuchi M, Ware C, Cipriano RC, Mauel MJ, Soto E. 2013. Comparative analysis of Edwardsiella isolates from fish in the eastern United States identifies two distinct genetic taxa amongst organisms phenotypically classified as E. tarda. Vet Microbiol 165:358–372. doi: 10.1016/j.vetmic.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Reichley SR, Ware C, Steadman J, Gaunt PS, García JC, LaFrentz BR, Thachil A, Waldbieser GC, Stine CB, Buján N, Arias CR, Loch T, Welch TJ, Cipriano RC, Greenway TE, Khoo LH, Wise DJ, Lawrence ML, Griffin MJ. 2017. Comparative phenotypic and genotypic analysis of Edwarsdiella isolates from different hosts and geographic origins, with emphasis on isolates formerly classified as E. tarda, and evaluation of diagnostic methods. J Clin Microbiol 55:3466–3491. doi: 10.1128/JCM.00970-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang WM. 1996. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet 30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 13.Yamada Y, Wakabayashi H. 1999. Identification of fish-pathogenic strains belonging to the genus Edwardsiella by sequence analysis of sodB. Fish Pathol 34:145–150. doi: 10.3147/jsfp.34.145. [DOI] [Google Scholar]
- 14.Dahllof I, Baillie H, Kjelleberg S. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl Environ Microbiol 66:3376–3380. doi: 10.1128/AEM.66.8.3376-3380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stepkowski T, Czaplinska M, Miedzinska K, Moulin L. 2003. The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha Proteobacteria. Syst Appl Microbiol 26:483–494. doi: 10.1078/072320203770865765. [DOI] [PubMed] [Google Scholar]
- 16.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brevik OJ, Ottem KF, Kamaishi T, Watanabe K, Nylund A. 2011. Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantea) in Japan. J Appl Microbiol 111:1044–1056. doi: 10.1111/j.1365-2672.2011.05133.x. [DOI] [PubMed] [Google Scholar]
- 18.Soto E, Kidd S, Mendez S, Marancik D, Revan F, Hiltchie D, Camus A. 2013. Francisella noatunensis subsp. orientalis pathogenesis analyzed by experimental immersion challenge in Nile tilapia, Oreochromis niloticus (L). Vet Microbiol 164:77–84. doi: 10.1016/j.vetmic.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Mauel MJ, Miller DL, Frazier K, Liggett AD, Styer L, Montgomery-Brock D, Brock J. 2003. Characterization of a piscirickettsiosis-like disease in Hawaiian tilapia. Dis Aquat Organ 53:249–255. doi: 10.3354/dao053249. [DOI] [PubMed] [Google Scholar]
- 20.Soto E, Primus AE, Pouder DB, George RH, Gerlach TJ, Cassle SE, Johnson T, Boyd S, Handsel T, Yanong RP. 2014. Identification of Francisella noatunensis in novel host species French grunt (Haemulon flavolineatum) and Caesar grunt (Haemulon carbonarium). J Zoo Wildl Med 45:727–731. doi: 10.1638/2014-0059R.1. [DOI] [PubMed] [Google Scholar]
- 21.Soto E, Halliday-Simmonds I, Francis S, Fraites T, Martinez-Lopez B, Wiles J, Hawke JP, Endris RD. 2016. Improved broth microdilution method for antimicrobial susceptibility testing of Francisella noatunensis orientalis. J Aquat Anim Health 28:199–207. doi: 10.1080/08997659.2016.1185051. [DOI] [PubMed] [Google Scholar]
- 22.Sjodin A, Ohrman C, Backman S, Larkeryd A, Granberg M, Lundmark E, Karlsson E, Nilsson E, Vallesi A, Tellgren-Roth C, Stenberg P, Thelaus J. 2014. Complete genome sequence of Francisella endociliophora strain FSC1006, isolated from a laboratory culture of the marine ciliate Euplotes raikovi. Genome Announc 2:e01227-14. doi: 10.1128/genomeA.01227-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer GR, Lowe GJ, Gilmore SR, Bower SM. 2017. Disease and mortality among Yesso scallops Patinopecten yessoensis putatively caused by infection with Francisella halioticida. Dis Aquat Organ 125:79–84. doi: 10.3354/dao03130. [DOI] [PubMed] [Google Scholar]
- 24.Rainey FA, Ward-Rainey NL, Janssen PH, Hippe H, Stackebrandt E. 1996. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology 142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 25.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press, New York, NY. [Google Scholar]
- 28.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 29.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 30.Rambut A. 2014. FigTree: Tree Figure Drawing Tool v1.4.2. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Scotland. [Google Scholar]
- 31.Baker CN, Hollis DG, Thornsberry C. 1985. Antimicrobial susceptibility testing of Francisella tularensis with a modified Mueller-Hinton broth. J Clin Microbiol 22:212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamaishi T, Fukuda Y, Nishiyama H, Kawakami H, Matsuyama T, Yoshinaga T, Oseko N. 2005. Identification and pathogenicity of intracellular Francisella bacterium in three-lined grunt Parapristipoma trilineatum. Fish Pathol 40:67–71. doi: 10.3147/jsfp.40.67. [DOI] [Google Scholar]
- 34.Soto E, McGovern-Hopkins K, Klinger-Bowen R, Fox BK, Brock J, Antonio N, Waal Z, Rushton S, Mill A, Tamaru CS. 2013. Prevalence of Francisella noatunensis subsp. orientalis in cultured tilapia on the island of Oahu, Hawaii. J Aquat Anim Health 25:104–109. doi: 10.1080/08997659.2013.781554. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez-Paredes JG, Larsson P, Wehner S, Bekaert M, Ohrman C, Metselaar M, Thompson KD, Richards RH, Penman DJ, Adams A. 2017. Draft genome sequence of Francisella noatunensis subsp. orientalis STIR-GUS-F2f7, a highly virulent strain recovered from diseased red Nile tilapia farmed in Europe. Genome Announc 5:e01555-16. doi: 10.1128/genomeA.01555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soto E, Baumgartner W, Wiles J, Hawke JP. 2011. Francisella asiatica as the causative agent of piscine francisellosis in cultured tilapia (Oreochromis sp.) in the United States. J Vet Diagn Invest 23:821–825. doi: 10.1177/1040638711407058. [DOI] [PubMed] [Google Scholar]
- 37.Vojtech LN, Sanders GE, Conway C, Ostland V, Hansen JD. 2009. Host immune response and acute disease in a zebrafish model of Francisella pathogenesis. Infect Immun 77:914–925. doi: 10.1128/IAI.01201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]