The ecological success of bifidobacteria relies on the activity of extracellular proteins that are involved in the metabolism of nutrients and the interaction with the environment. To date, information on secreted proteins encoded by bifidobacteria is incomplete and just related to few species. In this study, we reconstructed the bifidobacterial pan-secretome, revealing extracellular proteins that modulate the interaction of bifidobacteria with their natural environment. Furthermore, a survey of the secretion systems between bifidobacterial genomes allowed the identification of a conserved Sec-dependent secretion machinery in all the analyzed genomes and the Tat protein translocation system in the chromosomes of 23 strains belonging to Bifidobacterium longum subsp. longum and Bifidobacterium aesculapii.
KEYWORDS: Bifidobacterium, bifidobacteria, genomics, metagenomics, secretome
ABSTRACT
The repertoire of secreted proteins decoded by a microorganism represents proteins released from or associated with the cell surface. In gut commensals, such as bifidobacteria, these proteins are perceived to be functionally relevant, as they regulate the interaction with the gut environment. In the current study, we screened the predicted proteome of over 300 bifidobacterial strains among the currently recognized bifidobacterial species to generate a comprehensive database encompassing bifidobacterial extracellular proteins. A glycobiome analysis of this predicted bifidobacterial secretome revealed that a correlation exists between particular bifidobacterial species and their capability to hydrolyze human milk oligosaccharides (HMOs) and intestinal glycoconjugates, such as mucin. Furthermore, an exploration of metatranscriptomic data sets of the infant gut microbiota allowed the evaluation of the expression of bifidobacterial genes encoding extracellular proteins, represented by ABC transporter substrate-binding proteins and glycoside hydrolases enzymes involved in the degradation of human milk oligosaccharides and mucin. Overall, this study provides insights into how bifidobacteria interact with their natural yet highly complex environment, the infant gut.
IMPORTANCE The ecological success of bifidobacteria relies on the activity of extracellular proteins that are involved in the metabolism of nutrients and the interaction with the environment. To date, information on secreted proteins encoded by bifidobacteria is incomplete and just related to few species. In this study, we reconstructed the bifidobacterial pan-secretome, revealing extracellular proteins that modulate the interaction of bifidobacteria with their natural environment. Furthermore, a survey of the secretion systems between bifidobacterial genomes allowed the identification of a conserved Sec-dependent secretion machinery in all the analyzed genomes and the Tat protein translocation system in the chromosomes of 23 strains belonging to Bifidobacterium longum subsp. longum and Bifidobacterium aesculapii.
INTRODUCTION
The translocation of proteins across the cytoplasmic membrane is one of the most conserved and essential mechanisms occurring in nature, involving both unicellular and multicellular forms of life (1, 2). While higher organisms developed complex transmembrane systems to translocate proteins across their multiple membranes, microorganisms possess more basic pathways (3). For this reason, bacteria are commonly used in commercial processes to extracellularly and heterologously produce proteins, for example, antimicrobial peptides (4). Escherichia coli is arguably the most commonly exploited protein expression system (5).
Members of the bacterial communities residing in the human gut produce extracellular proteins exerting crucial roles in establishment and maintenance, i.e., colonization, of these bacteria in the gut environment (6–8). Members of the genus Bifidobacterium are among the key bacterial components of the human gut microbiota, especially during early life. In fact, this genus constitutes one of the main representatives of the mammalian gut microbiota, and its members are also known to colonize the avian and insect intestinal tract (7, 9, 10). The Bifidobacterium genus currently consists of 56 species and 9 subspecies, members of which have been isolated from human, other mammalian, and certain insect intestinal contents (11, 12). Recently, it has been demonstrated that members of the genus Bifidobacterium interact with other bacterial species and the host using extracellular structures, such as sortase-dependent pili and type IVb tight adherence (Tad) pili (13–15). Furthermore, extracellular anti-inflammatory proteins, such as the serine protease inhibitor (serpin) protein, were identified in several bifidobacterial species (16, 17), as well as extracellular macromolecules, like exopolysaccharides (EPSs), which form a glycan slime layer that is loosely attached to the cell (18, 19). The ecological success of bifidobacteria is believed to partly rely on the activities of extracellular proteins that are involved in the metabolism of nutrients (breakdown and uptake of metabolic substrates). Numerous bifidobacterial species have been shown to metabolize different high-molecular-weight glycans derived from both the host, such as human milk oligosaccharides (HMO) and mucins, and the diet, e.g., starch and various plant-derived polysaccharides (20–22). Notably, the metabolism of these complex glycans is facilitated by extracellular enzymes (23). The ability to utilize complex carbohydrates provides a selective advantage for bifidobacteria and allows them to effectively compete for nutrients in their specific niche (21, 24). Altogether, these findings indicate that secreted proteins are fundamental for the establishment and maintenance of bifidobacteria in the human gut.
So far, information on the secreted proteins produced by bifidobacteria is rather fragmentary. Exported proteins have been identified in Bifidobacterium breve UCC2003, and several extracellular proteins have been characterized in strains belonging to Bifidobacterium longum subsp. longum, Bifidobacterium adolescentis, and Bifidobacterium animalis subsp. lactis (25–27). Furthermore, a phytase-based assay has relatively recently been employed to provide an insight into protein secretion and the associated signal peptides of bifidobacteria (28).
In the current study, we generated a complete database of the predicted pan-secretome of the genus Bifidobacterium based on in silico analyses of the genomes of over 300 different bifidobacterial strains distributed among the currently recognized bifidobacterial species. This secretome database was exploited for the purpose of screening metagenomic data sets, thus allowing us to assess the bifidobacterial contribution to the overall arsenal of extracellular proteins encoded by the human gut microbiome.
RESULTS AND DISCUSSION
Bifidobacterial pan-secretome.
Bifidobacteria, like other bacteria, differentiate cytosolic and extracellular proteins based on the presence of a signal peptide in their primary amino acid sequence. In detail, secreted proteins are synthesized as precursors with a cleavable amino-terminal signal sequence that guides the precursor to a molecular machinery responsible for translocation of the protein and concomitant removal of the signal peptide (1). In order to predict extracellular proteins encoded by bifidobacterial genomes, we employed multiple in silico programs that rely on the identification of signal peptide-specific sequence motifs. For this purpose, 311 bifidobacterial genome sequences were retrieved from the National Center for Biotechnology Information (NCBI) and analyzed. Notably, partial genome sequences with >200 contigs were excluded (see Table S1 in the supplemental material). The retrieved bifidobacterial genomes included at least one genome for each of the 62 sequenced (sub)species of the genus, thus allowing the investigation of the so-far-established genetic diversity of the genus Bifidobacterium. Over 600,000 genes were predicted and functionally annotated, starting from the genomic sequences of the 311 collected strains, and their amino acidic sequences were used to explore the predicted pan-proteome of the Bifidobacterium genus.
Using the subcellular localization prediction tool PSORTb version 3.0.2, we were able to identify 6,763 putative extracellular proteins and 4,925 proteins that were predicted to be associated with the cell wall (Table S1). Additionally, proteins whose putative cellular localization was not clearly predicted were manually evaluated, and 1,644 additional genes were added to the pool of (potentially) secreted proteins of the bifidobacterial genus, for a total of 13,332 proteins constituting the bifidobacterial secretome database (BSD). Notably, the selected cutoff value chosen for subcellular localization prediction favored specificity above sensitivity, thereby minimizing the number of false positives.
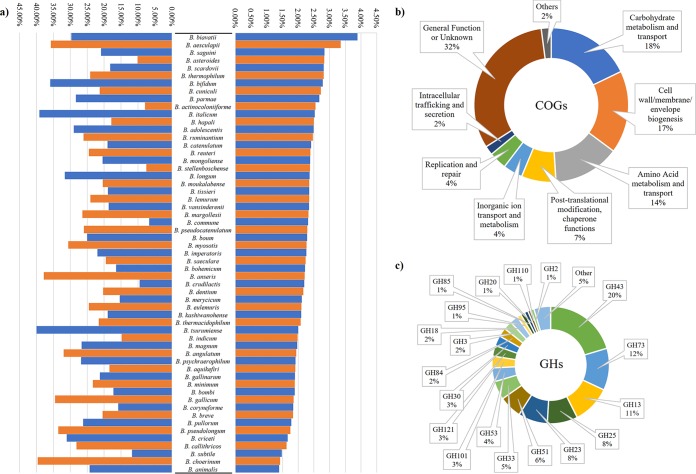
As expected from previous studies based on the prediction of extracellular proteins (29), the most common gene classes of BSD include solute-binding proteins of ABC transporter systems, amidases related to the peptidoglycan hydrolysis, a variety of glycosyl hydrolases, cell surface proteins that make up pilus subunits, and cell wall-degrading peptidases, such as d-alanyl–d-alanine carboxypeptidases. Interestingly, normalization of the number of predicted extracellular proteins for the number of encoding genes for each strain revealed that Bifidobacterium biavatii and Bifidobacterium aesculapii genomes encode the largest proportion of secreted proteins in the genus Bifidobacterium (Fig. 1 and Table 1). In contrast, chromosomes of Bifidobacterium animalis, Bifidobacterium choerinum, and Bifidobacterium subtile strains encompass the smallest percentage of secreted proteins (Fig. 1 and Table 1).
FIG 1.
Bifidobacterial extracellular protein overview. (a) Percentage of extracellular proteins normalized between strains of the same species and the total amount of proteome (right) and the percentage of extracellular GHs normalized between strains of the same species and the total amount of secreted proteins (left). (b) COG classifications of the whole secretome of the genus Bifidobacterium. (c) GH organization of the secreted enzymes of the genus.
TABLE 1.
Bifidobacterial secretome
| Phylogenetic groupa | Bifidobacterial species | Predicted extracellular proteinsb |
Predicted extracellular glycoside hydrolasesc |
||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| B. asteroides | B. actinocoloniiforme | 38 | 2.6 | 3 | 7.9 |
| B. asteroides | 48 | 2.8 | 4.8 | 10.1 | |
| B. coryneforme | 26 | 1.8 | 4 | 15.8 | |
| B. indicum | 27 | 2.0 | 4 | 14.8 | |
| B. psychraerophilum | B. aquikefiri | 38 | 1.9 | 7 | 18.4 |
| B. crudilactis | 42 | 2.2 | 4 | 9.5 | |
| B. psychraerophilum | 41 | 1.9 | 11 | 26.8 | |
| B. tissieri | 53 | 2.4 | 10 | 18.9 | |
| B. vansinderenii | 59 | 2.3 | 11 | 18.6 | |
| B. margollesii | 53 | 2.3 | 14 | 26.4 | |
| B. pseudolongum | B. italicum | 46 | 2.5 | 18 | 39.1 |
| B. anseris | 37 | 2.2 | 14 | 37.8 | |
| B. choerinum | 25 | 1.4 | 10 | 39.6 | |
| B. criceti | 29 | 1.7 | 9 | 31.0 | |
| B. pseudolongum | 29 | 1.8 | 9.6 | 33.5 | |
| B. cuniculi | 61 | 2.7 | 13 | 21.3 | |
| B. animalis | 22 | 1.4 | 5.4 | 24.3 | |
| B. gallicum | 29 | 1.9 | 10 | 34.5 | |
| B. magnum | 30 | 2.0 | 8 | 26.7 | |
| B. adolescentis | B. adolescentis | 46 | 2.5 | 13.3 | 29.0 |
| B. ruminantium | 46 | 2.5 | 12 | 26.1 | |
| B. catenulatum | 42 | 2.4 | 8 | 19.0 | |
| B. kashiwanohense | 42 | 2.1 | 8 | 18.9 | |
| B. pseudocatenulatum | 44 | 2.3 | 11.3 | 26.0 | |
| B. dentium | 47 | 2.2 | 9.5 | 20.3 | |
| B. moukalabense | 49 | 2.4 | 10 | 20.4 | |
| B. bifidum | B. biavatii | 101 | 3.9 | 30 | 29.7 |
| B. hapali | 56 | 2.5 | 10 | 17.9 | |
| B. scardovii | 72 | 2.8 | 13 | 18.2 | |
| B. bifidum | 51 | 2.8 | 18.3 | 35.9 | |
| B. longum | B. aesculapii | 70 | 3.4 | 25 | 35.7 |
| B. stellenboschense | 53 | 2.4 | 4 | 7.5 | |
| B. parmae | 60 | 2.7 | 17 | 28.3 | |
| B. callitrichos | 39 | 1.6 | 11 | 28.2 | |
| B. angulatum | 31 | 1.9 | 10 | 32.0 | |
| B. merycicum | 39 | 2.1 | 6 | 15.4 | |
| B. breve | 37 | 1.8 | 7.5 | 20.4 | |
| B. longum | 50 | 2.4 | 15.9 | 31.6 | |
| B. myosotis | 49 | 2.3 | 15 | 30.6 | |
| B. reuteri | 53 | 2.4 | 13 | 24.5 | |
| B. saguini | 67 | 2.8 | 14 | 20.9 | |
| B. imperatoris | 50 | 2.3 | 11 | 22.0 | |
| B. eulemuris | 49 | 2.1 | 12 | 24.5 | |
| B. lemurum | 54 | 2.4 | 13 | 24.1 | |
| B. pullorum | B. gallinarum | 33 | 1.9 | 7 | 21.2 |
| B. saeculare | 41 | 2.2 | 8 | 19.5 | |
| B. pullorum | 31 | 1.8 | 8 | 26.2 | |
| B. boum | B. boum | 40 | 2.3 | 10 | 25.0 |
| B. thermacidophilum | 35 | 2.1 | 7.5 | 21.6 | |
| B. thermophilum | 49 | 2.8 | 11.8 | 24.1 | |
| B. tsurumiense | 35 | 2.0 | 14 | 40.0 | |
| B. mongoliense | 44 | 2.4 | 9 | 20.5 | |
| B. subtile | 34 | 1.5 | 4 | 11.8 | |
| B. bombi | B. bohemicum | 37 | 2.2 | 6 | 16.4 |
| B. commune | 30 | 2.3 | 2 | 6.7 | |
| B. bombi | 29 | 1.9 | 5 | 17.2 | |
| B. minimum | 30 | 1.9 | 7 | 23.3 | |
Phylogenetic groups based on Lugli et al. (11).
Average number of extracellular proteins within the species, and the percentage of extracellular proteins related to the proteome of the species, i.e., based on the number of predicted proteins of that species.
Average number of extracellular GHs within the species, and the percentage related to the secreted proteins of the species.
Proteins constituting the BSD were further compared with a Clusters of Orthologous Groups (COG) database, i.e., the eggNOG database (30). Notably, 74% of the predicted bifidobacterial secreted proteins were distributed among groups M, G, and E, whose predicted functions are related to cell wall/membrane/envelope biogenesis, carbohydrate metabolism and transport, and amino acid metabolism and transport, respectively (Fig. 1). Notably, members of COG groups M, G, and E are predicted to exert a key role in modulating the interaction with the environment and are indispensable for the acquisition of nutrients (31, 32).
Bifidobacterial secretion systems.
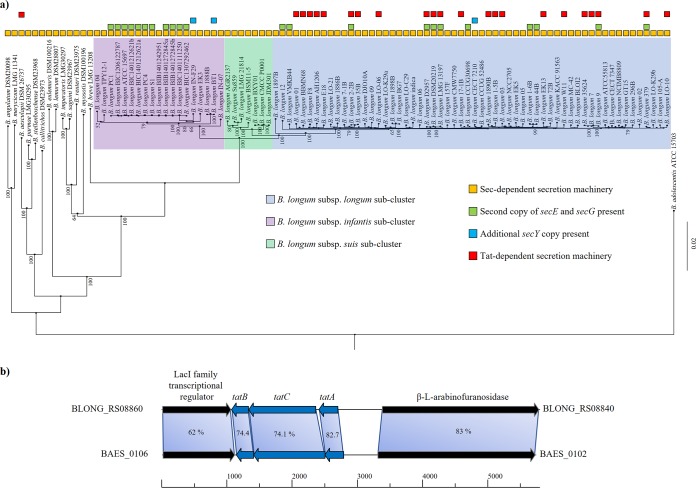
Bifidobacteria are Gram-positive bacteria possessing one lipid bilayer membrane surrounded by a thick cell wall (1). Thus, they employ the general secretion (Sec) pathway and, for some strains, the twin-arginine translocation (Tat) pathway to transport proteins across the cytoplasmic membrane (28). While the Sec pathway primarily translocates proteins in their unfolded state, experimental evidence showed that the Tat pathway secretes mainly (partially) folded proteins (33). In order to identify the secretion systems encoded by the 311 analyzed bifidobacterial genomes, we exploited the gene sequences of the characterized systems of B. longum E18 (28) as a query for the identification of homologs in the BSD database. Additionally, the collected genes were further used as query sequences to identify putative distant homologs with the same function. This analysis allowed the verification that all 311 bifidobacterial genomes encompass genes encoding the Sec translocase system, including the integral membrane complex encoded by secY, secE, and secG, as well as the ATPase-encoding gene secA. In addition, 28 bifidobacterial genomes, of which 22 are classified as B. longum, were shown to possess an additional copy of the secE and secG genes (Fig. 2). Furthermore, among sequenced members of the B. longum species, three strains were identified to possess an additional copy of secY encoding the membrane-associated secretion complex (Fig. 2).
FIG 2.
Bifidobacterial secretion systems identified between strains. (a) Distribution of the Sec- and Tat-dependent secretion machineries in the phylogenetic groups of B. longum. The phylogenomic tree was built based on core genome sequences shared among the bifidobacterial strains, using B. adolescentis ATCC 15703 as an outgroup. The phylogenomic tree was constructed by the neighbor-joining method. Bootstrap percentages above 50 are shown at node points, based on 1,000 replicates of the phylogenomic tree. (b) Genetic map between B. longum E18 and B. aesculapii DSM 26737 Tat-dependent secretion system loci. Each arrow indicates an ORF, whereas the length of the arrow is proportional to the length of the predicted ORF.
A complete Tat system composed of TatA, TatB, and TatC was identified in 22 B. longum genomes and in the chromosome of B. aesculapii DSM 26737 (Fig. 2). Recently, it has been demonstrated by means of a genomic identity approach that of the 84 analyzed B. longum genomes, 57 of them cluster together as members of B. longum subsp. longum (11). Interestingly, the 22 B. longum genomes possessing the Tat system were all members of the B. longum subsp. longum cluster (Fig. 2). Notably, a recent phylogenomic analysis attributed B. aesculapii DSM 26737 to the B. longum phylogenetic group (11). Furthermore, the genes flanking the Tat locus encode the same proteins in both species, i.e., a LacI family transcriptional regulator and a β-l-arabinofuranosidase that has been validated as a Tat-secreted protein of B. longum E18 (28). Altogether, these findings suggest that the Tat secretion system may have been inherited from a common ancestor of B. aesculapii and B. longum (Fig. 2).
Extracellular bifidobacterial glycobiome.
One of the key genetic features of bifidobacteria is their ability to utilize complex carbon sources (21, 34). Thus, classification according to the Carbohydrate-Active enZyme (CAZy) database (35) was performed to assess which secreted proteins possess the ability to enhance the carbohydrate-harvesting abilities and improve the associated fitness of the Bifidobacterium genus for the human gut environment. This analysis resulted in the identification of 4,397 secreted proteins possessing catalytic and carbohydrate-binding modules involved in the degradation and modification of carbohydrates, representing one-third of the collected secretome of the genus. Interestingly, the predicted secreted glycoside hydrolases (GHs) constitute 27% (representing 3,639 genes) of the total number of proteins in the BSD. Moreover, the most abundant families of secreted GHs were GH43 (20%), involved in the degradation of complex plant polysaccharides, such as (arabino)xylan, GH73 (12%), related to peptidoglycan hydrolysis, and GH13 (11%), a family with degradative abilities toward a wide range of carbohydrates, including plant-derived polysaccharides, such as starch and trehalose (Fig. 1).
When focusing on species-specific secretomes, bifidobacterial species that encode the highest percentages of extracellular GHs (from 30 to 40% of their secreted proteome) belong to the phylogenetic groups of Bifidobacterium pseudolongum and B. longum (11), while the members of the Bifidobacterium asteroides and Bifidobacterium bombi groups possess the lowest percentages of secreted GHs (Fig. 1 and Table 1). These findings may be correlated with the ecological niches in which the members of the Bifidobacterium asteroides and Bifidobacterium bombi groups are commonly found. In fact, members of the B. asteroides and B. bombi group are typical inhabitants of the hindgut of social insects; thus, their evolution has been driven by the availability of a limited range of carbon sources (36, 37). Nonetheless, their peculiar repertoire of GHs was recently found to be substantially different from other bifidobacterial species that evolved within the gut of mammals (21).
Further investigations were performed in order to associate extracellular GH families with the bifidobacterial taxa analyzed in the reconstructed bifidobacterial pan-secretome. Thus, the presence of GH families in each bifidobacterial species was used to construct a force-driven network (Fig. 3). Interestingly, four major clusters of bifidobacterial species were identified, each dominated by specific bifidobacterial species, i.e., B. animalis-B. adolescentis, B. longum, Bifidobacterium bifidum, and B. breve (Fig. 3). In this context, the B. animalis-B. adolescentis cluster was mainly characterized by the presence of GH3 family enzymes involved in the degradation of plant-derived polysaccharides, e.g., xylan, and GH13 shared with other members of the B. breve and B. longum clusters, which are predicted to degrade a wide range of carbohydrates, including starch and related substrates (Fig. 3). In contrast, the B. longum cluster was characterized by the presence of GH121 family enzymes represented by β-l-arabinobiosidase and GH43 involved in the degradation of complex plant polysaccharides, such as (arabino)xylan. Furthermore, GH101 family members were shared with the B. bifidum cluster, highlighting the presence of endo-α-N-acetylgalactosaminidase for the degradation of the O-glycosidic linkage of mucin-type glycoproteins. Remarkably, the B. bifidum cluster is constituted only by strains belonging to B. bifidum species, which differ from other bifidobacterial taxa by the presence of extracellular enzymes of the families GH95, GH84, GH110, GH89, GH29, GH20, and GH2 (Fig. 3). The presence of this unique repertoire of extracellular GHs, together with GH101 shared with B. longum cluster, is correlated with the capability of these strains to utilize HMOs and intestinal glycoconjugates, such as mucin (21, 38). Furthermore, another key GH family involved in the degradation of host glycans, i.e., the secreted GH33, which involves sialidases, was observed in the B. bifidum cluster as well as in the B. breve cluster. Finally, the fourth cluster represented by B. breve strains includes ubiquitous GHs, i.e., members of GH73, GH25, and GH23, involved in the hydrolysis of peptidoglycan.
FIG 3.
Network analysis based on the presence of extracellular GHs in each Bifidobacterium strain. Yellow circles represent the identified GHs, and their diameter is proportional to the number of strains that possess the enzyme. Each color denotes a different cluster, i.e., B. animalis-B. adolescentis, cyan; B. longum, purple; B. bifidum, orange; and B. breve, green. The bifidobacterial scientific names were synthetized with the first three characters of the species name, e.g., ADO, B. adolescentis; LON, B. longum; BIF, B. bifidum; and BRE, B. breve.
Secretome of bifidobacterial communities within the infant gut.
In order to explore the occurrence of bifidobacterial secreted proteins in the human gut, these genes were profiled in metagenomic and metatranscriptomic databases retrieved from the NCBI. In detail, we used data from a project that was aimed at exploring the microbial composition of the infant gut (BioProject PRJNA63661), and a project meant to evaluate vertical transmission of the microbiota from mothers to corresponding infants (BioProject PRJNA339914). Since a higher abundance of bifidobacteria was observed in the infant microbiota than in adults, we decided to use the infant-related data sets to allow a more in-depth analysis of the secreted proteins of this genus (39, 40).
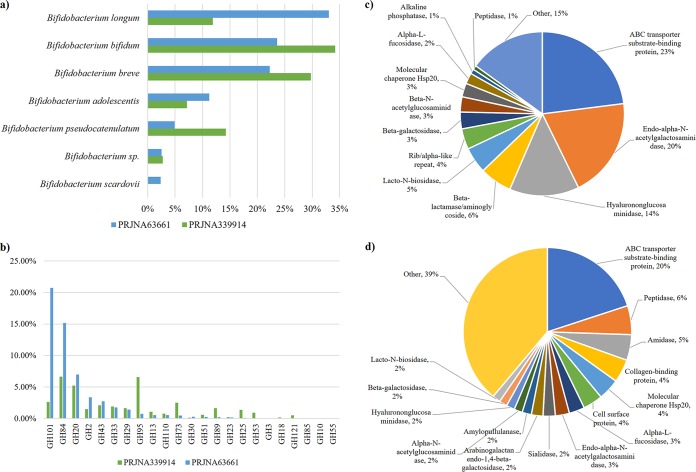
The metagenomic samples collected from healthy infants from the study with BioProject PRJNA63661 were screened for reads corresponding to genes included in the BSD database, unveiling dissimilar profiles between samples (Table S3). The number of metagenomic reads that belong to bifidobacterial secreted proteins ranged from 165,772 to none. As expected, the majority of the bifidobacterial extracellular proteins identified belong to B. longum, B. breve, and B. bifidum, i.e., bifidobacterial species that typically colonize the infant gut during early life (34).
In the same fashion, the analysis was performed on metatranscriptomic samples collected from four healthy infants from BioProject PRJNA63661. Notably, >50% of the reads that mapped on the BSD database correspond to ABC transporter substrate-binding proteins, as well as endo-α-N-acetylgalactosaminidases and hyalurononglucosaminidases (Fig. 4). Moreover, this analysis revealed that B. longum and then B. bifidum and B. breve, were the species with the highest expression of genes encoding extracellular proteins, i.e., 33%, 24%, and 22% of the identified reads, respectively (Fig. 4). Furthermore, these data highlight a high level of transcription of genes corresponding to glycosidic enzymes and related transporters for the uptake of simple sugars derived from the breakdown of complex metabolic substrates. Among extracellular GHs, we observed a predominance of GH101 family enzymes, corroborating the abundance of genes encoding endo-α-N-acetylgalactosaminidase in the genome of B. bifidum and B. longum species (see above) (Fig. 3). Moreover, we also identified GH enzymes belonging to the B. bifidum-associated GH84 and GH20 clusters (Fig. 4). While GH84 enzymes are associated with hyalurononglucosaminidase, GH20 reflects both hexosaminidase and lacto-N-biosidase. The substrates of these GHs are usually residues of mucin-type glycoproteins, as well as hyaluronic acid and lacto-N-biose, highlighting substantial metabolic activity directed to harvest host glycans by the identified bifidobacterial species.
FIG 4.
Metatranscriptomic profiling of bifidobacterial genes encoding secreted proteins within the infant gut. (a) Percentage of metatranscriptomic reads that match genes encoding secreted proteins based on bifidobacterial species. (b) Bifidobacterial extracellular GH distribution among the two data sets. (c and d) Functional distribution of the identified secretome in the PRJNA63661 data set (c) and the same distribution for PRJNA339914 samples (d).
A further metatranscriptomic data set (BioProject PRJNA339914) was employed in order to validate the above-described data. With respect to the PRJNA63661 data set, the two infant samples showed higher expression of genes encoding extracellular proteins belonging to B. bifidum (34%) and B. breve (30%) (Fig. 4). Regarding extracellular GH-specifying enzymes, a prevalence in GH84 and GH95, followed by GH20 and GH101 members, was noticed (Fig. 4). Accordingly, the same GHs were detected in data set PRJNA63661, with the exception of an increased family member of GH95 encoding fucosidases. Thus, within these samples, we observed a reduction in B. longum gene expression balanced by increased B. bifidum and B. breve gene transcription. Altogether, these data support previous findings about the cross-feeding activities established between B. bifidum and B. breve species (24, 32, 41).
Conclusions.
In this study, we reconstructed the pan-secretome of the genus Bifidobacterium through in silico analyses of 311 bifidobacterial strains belonging to all currently recognized bifidobacterial species. The most common extracellular proteins identified belong to solute-binding proteins of ABC transporter systems, amidases related to the peptidoglycan hydrolysis, glycosyl hydrolases of several families, and cell surface proteins, like pili and peptidases, such as d-alanyl–d-alanine carboxypeptidases. Interestingly, Bifidobacterium biavatii, followed by Bifidobacterium aesculapii, were predicted to encode the largest arsenal of secreted proteins among currently known members of the genus Bifidobacterium. Furthermore, a bifidobacterial secretion system screening allowed us to identify the Sec-dependent secretion machinery in the 311 genomes, while just 23 strains belonging to B. longum subsp. longum and B. aesculapii DSM 26737 also harbor the Tat protein translocation system. An extracellular glycobiome analysis allowed the identification of four bifidobacterial secretome clusters, represented by main species, i.e., B. animalis-B. adolescentis, B. longum, B. bifidum, and B. breve. This analysis revealed the presence of specific GH families in each cluster, unveiling that the B. bifidum cluster differs from that of other bifidobacterial (sub)species due to the presence of extracellular enzymes correlated with the ability of hydrolyze HMOs and intestinal glycoconjugates, such as mucin, i.e., families GH95, GH84, GH110, GH89, GH29, GH20, and GH2. Finally, the dissection of metatranscriptomic data sets derived from infant fecal samples allowed the transcriptional evaluation of bifidobacterial genes encoding extracellular proteins. This analysis showed that the majority of the bifidobacterial secreted proteins expressed in the infant gut belong to B. longum, B. bifidum, and B. breve species, representing ABC transporter substrate-binding proteins and GH enzymes, such as hyalurononglucosaminidase (GH84), endo-α-N-acetylgalactosaminidase (GH101), hexosaminidase and lacto-N-biosidase (GH20), and fucosidases (GH95), consistent with metabolic activities that involve the degradation of human milk oligosaccharides and/or mucin.
MATERIALS AND METHODS
Bifidobacterial genome sequences.
We retrieved complete and partial genome sequences of 311 bifidobacterial strains from the National Center for Biotechnology Information (NCBI) public database (Table S1). A single type strain for each bifidobacterial species was employed in our analyses in order to avoid genome content redundancy. Furthermore, genomes displaying insufficient quality, i.e., a high number of contigs or partial genome length, were discarded.
Proteome prediction and annotation.
Nucleotide genomic sequences obtained from the NCBI database were used as input for the MEGAnnotator pipeline in order to predict the protein-encoding open reading frames (ORFs) using the same methodology for each genome (42). While ORFs were predicted with Prodigal (43), gene annotation was defined by means of RAPSearch2 (Reduced Alphabet based Protein similarity Search) (44) in the nonredundant protein database from the NCBI. Furthermore, a hidden Markov model (HMM) search (http://hmmer.org/) of the manually curated Pfam-A protein family database was performed to identify domains between predicted ORFs (45). The results were inspected by Artemis (46), which was used for genome analyses and for manual editing where necessary. Functional annotation of each secreted protein was performed employing the eggNOG database (30).
Secretome and secretion systems prediction.
Several programs which are aimed at predicting extracellular and transmembrane proteins through signal peptide and transmembrane motif were employed based on a comparison performed by Caccia et al., i.e., PredSi, SignalP, Phobius, PredLipo, PredTat, LipoP, TMHMM, and PSORTb (47). These bioinformatic tools were selected due to their ability to analyze hundreds of proteins at the same iteration. A preliminary screening was performed using the proteome of B. bifidum PRL2010, followed by manual evaluation of the output to unveil which program results in a more accurate prediction (Table S2). Due to its multiple software implementation, PSORTb version 3.0.2 was shown to be the most accurate tool for the prediction of the bifidobacterial secretome (48). Nonetheless, PSORTb promotes specificity above sensitivity and allowed the collection of protein sequences with the lowest number of false positives compared to other tools (Table S2). Thus, the secreted proteins of the 311 sequenced bifidobacterial genomes were predicted using PSORTb version 3.0.2. Additionally, proteins whose putative cell localization was not clearly predicted by PSORTb were manually evaluated in order to increase the pool of secreted proteins of the genus.
In order to identify the secretion systems of each bifidobacterial strain, a bifidobacterial database based on secretion systems was built with the characterized genes of B. longum E18 (28). The RefSeq accession numbers of these genes are ESV34191.1, ESV34434.1, ESV33665.1, ESV33737.1, ESV32709.1, ESV32707.1 and ESV32708.1. The predicted secretion systems of the 311 bifidobacterial strains were obtained using BLAST, followed by manual evaluation of the results.
Glycobiome prediction.
The prediction of genes encoding extracellular enzymes possessing structurally related catalytic and carbohydrate-binding modules catalyzing hydrolysis, modification, or synthesis of glycoside bounds was performed using the CAZy database (35). A force-driven network was built using Gephi version 0.9.2 to highlight the correlation between GHs and bifidobacterial species.
Metagenomic and metatranscriptomic analyses.
Fecal metagenomic and metatranscriptomic data sets of infants were retrieved from two NCBI public databases, i.e., BioProjects PRJNA63661 and PRJNA339914. Each data set was filtered to obtain only high-quality reads (minimum mean quality score, 20; window size, 5; quality threshold, 25; minimum length, 80) using the fastq-mcf script (https://expressionanalysis.github.io/ea-utils/). The resulting reads were aligned against the human genome using the Burrows-Wheeler Aligner program (49) (BWA-MEM algorithm with trigger reseeding, 1.5; minimum seed length, 19; matching score, 1; mismatch penalty, 4; gap open penalty, 6; and gap extension penalty, 1) and further processed with the SAMtools software package (50) in order to remove human reads. Furthermore, the metatranscriptomic data sets were processed to remove rRNA-encompassing reads. Finally, the filtered reads were used to identify bifidobacterial secretome-associated reads within the data set for each sample by means of Bowtie 2 (51) through multiple-hit mapping and a “very sensitive” policy. The mapping was performed using a minimum score threshold function (–score-min C,-13,0) in order to limit reads of arbitrary length to two mismatches and retain those matches with at least 98% full-length identity. The software employed to calculate read counts corresponding to bifidobacterial genes was HTSeq (52), running in union mode.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the EU Joint Programming Initiative “A Healthy Diet for a Healthy Life” (JPI HDHL; http://www.healthydietforhealthylife.eu/) to D.V.S. (in conjunction with Science Fondation Ireland [SFI], grant 15/JP-HDHL/3280) and to M.V. (in conjunction with MIUR, Italy). D.V.S. is a member of APC Microbiome Ireland funded by the Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (grant SFI/12/RC/2273). The study is supported by Fondazione Cariparma, under the TeachInParma Project.
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. Part of this research was conducted using the High Performance Computing (HPC) facility of the University of Parma.
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00796-18.
REFERENCES
- 1.van Wely KH, Swaving J, Freudl R, Driessen AJ. 2001. Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol Rev 25:437–454. doi: 10.1111/j.1574-6976.2001.tb00586.x. [DOI] [PubMed] [Google Scholar]
- 2.Agarraberes FA, Dice JF. 2001. Protein translocation across membranes. Biochim Biophys Acta 1513:1–24. doi: 10.1016/S0304-4157(01)00005-3. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack MT, Schleiff E. 2010. The evolution of protein targeting and translocation systems. Biochim Biophys Acta 1803:1115–1130. doi: 10.1016/j.bbamcr.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Parachin NS, Mulder KC, Viana AA, Dias SC, Franco OL. 2012. Expression systems for heterologous production of antimicrobial peptides. Peptides 38:446–456. doi: 10.1016/j.peptides.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Gopal GJ, Kumar A. 2013. Strategies for the production of recombinant protein in Escherichia coli. Protein J 32:419–425. doi: 10.1007/s10930-013-9502-5. [DOI] [PubMed] [Google Scholar]
- 6.Rakoff-Nahoum S, Foster KR, Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, van Sinderen D, Ventura M. 2017. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11:2834–1847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, Mangifesta M, Gioiosa L, Palanza P, van Sinderen D, Ventura M. 2014. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol 80:6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, van Sinderen D, Ventura M. 2018. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci 75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sanchez B, Margolles A, van Sinderen D, Ventura M. 2016. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugli GA, Milani C, Duranti S, Mancabelli L, Mangifesta M, Turroni F, Viappiani A, van Sinderen D, Ventura M. 2018. Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Appl Environ Microbiol 84:e02249-17. doi: 10.1128/AEM.02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiri K, van Sinderen D, Ventura M. 2017. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milani C, Mangifesta M, Mancabelli L, Lugli GA, Mancino W, Viappiani A, Faccini A, van Sinderen D, Ventura M, Turroni F. 2017. The sortase-dependent fimbriome of the genus Bifidobacterium: extracellular structures with potential to modulate microbe-host dialogue. Appl Environ Microbiol 83:e01295-17. doi: 10.1128/AEM.01295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Martin P, Fernandez M, O'Connell-Motherway M, O'Connell KJ, Sauvageot N, Fitzgerald GF, MacSharry J, Zomer A, van Sinderen D. 2012. A conserved two-component signal transduction system controls the response to phosphate starvation in Bifidobacterium breve UCC2003. Appl Environ Microbiol 78:5258–5269. doi: 10.1128/AEM.00804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turroni F, Foroni E, O'Connell Motherway M, Bottacini F, Giubellini V, Zomer A, Ferrarini A, Delledonne M, Zhang Z, van Sinderen D, Ventura M. 2010. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol 76:3206–3219. doi: 10.1128/AEM.02938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrario C, Milani C, Mancabelli L, Lugli GA, Duranti S, Mangifesta M, Viappiani A, Turroni F, Margolles A, Ruas-Madiedo P, van Sinderen D, Ventura M. 2016. Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol Ecol 92:fiw056. doi: 10.1093/femsec/fiw056. [DOI] [PubMed] [Google Scholar]
- 19.Hidalgo-Cantabrana C, Sanchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. 2014. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol 80:9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, Ferretti P, Gorfer V, Ferrario C, Milani C, Mangifesta M, Anzalone R, Zolfo M, Viappiani A, Pasolli E, Bariletti I, Canto R, Clementi R, Cologna M, Crifo T, Cusumano G, Fedi S, Gottardi S, Innamorati C, Mase C, Postai D, Savoi D, Soffiati M, Tateo S, Pedrotti A, Segata N, van Sinderen D, Ventura M. 2017. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacConaill LE, Fitzgerald GF, Van Sinderen D. 2003. Investigation of protein export in Bifidobacterium breve UCC2003. Appl Environ Microbiol 69:6994–7001. doi: 10.1128/AEM.69.12.6994-7001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez B, Champomier-Verges MC, Anglade P, Baraige F, de Los Reyes-Gavilan CG, Margolles A, Zagorec M. 2008. A preliminary analysis of Bifidobacterium longum exported proteins by two-dimensional electrophoresis. J Mol Microbiol Biotechnol 14:74–79. doi: 10.1159/000106085. [DOI] [PubMed] [Google Scholar]
- 27.Sánchez B, Bressollier P, Urdaci MC. 2008. Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol Med Microbiol 54:1–17. doi: 10.1111/j.1574-695X.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- 28.Osswald A, Westermann C, Sun Z, Riedel CU. 2015. A phytase-based reporter system for identification of functional secretion signals in bifidobacteria. PLoS One 10:e0128802. doi: 10.1371/journal.pone.0128802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Theunissen D, Wels M, Siezen RJ. 2010. LAB-secretome: a genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of lactic acid bacteria. BMC Genomics 11:651. doi: 10.1186/1471-2164-11-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukjancenko O, Ussery DW, Wassenaar TM. 2012. Comparative genomics of Bifidobacterium, Lactobacillus and related probiotic genera. Microb Ecol 63:651–673. doi: 10.1007/s00248-011-9948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. 2017. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol 26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Robinson C, Bolhuis A. 2004. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694:135–147. doi: 10.1016/j.bbamcr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Killer J, Kopecny J, Mrazek J, Rada V, Benada O, Koppova I, Havlik J, Straka J. 2009. Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. Int J Syst Evol Microbiol 59:2020–2024. doi: 10.1099/ijs.0.002915-0. [DOI] [PubMed] [Google Scholar]
- 38.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sánchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 43.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, Tang H, Ye Y. 2012. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 47.Caccia D, Dugo M, Callari M, Bongarzone I. 2013. Bioinformatics tools for secretome analysis. Biochim Biophys Acta 1834:2442–2453. doi: 10.1016/j.bbapap.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 48.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.