The presence of V. parahaemolyticus in seafood may pose a risk for consumers, especially in countries where shellfish are eaten raw. In recent years, a significant increase of food poisoning caused by these bacteria has been also observed in Europe. Our results highlight the high level of V. parahaemolyticus contamination of seafood, along with the isolates being potentially pathogenic for humans. However, the first-line antimicrobials, such as tetracyclines and fluoroquinolones, remained highly effective against V. parahaemolyticus. The monitoring of antimicrobial resistance of isolates is important to ensure the high efficacy in the treatment of human infections. Most of V. parahaemolyticus strains possessed new sequence types (STs), which showed the high genetic diversity of the isolates tested.
KEYWORDS: Vibrio parahaemolyticus, virulence factors, antimicrobial resistance, MLST, PFGE, genetic diversity
ABSTRACT
Vibrio parahaemolyticus is a widespread bacterium in the marine environment and is responsible for gastroenteritis in humans. Foodborne infections are mainly associated with the consumption of contaminated raw or undercooked fish and shellfish. The aim of this study was to determine the antimicrobial resistance, virulence factors, and genetic profiles of V. parahaemolyticus isolates from seafood originating from different countries. A total of 104 (17.5%) isolates were recovered from 595 analyzed samples. The isolates were tested for the presence of the tdh and trh genes, involved in the pathogenesis of V. parahaemolyticus infections in humans, and these genes were detected in 3 (2.9%) and 11 (10.6%) isolates, respectively. The trh-positive isolates also possessed the ure gene, which is responsible for urease production. Moreover, the activity of protease A was identified in all V. parahaemolyticus strains. Antimicrobial resistance revealed that most isolates were resistant to ampicillin (75.0%) and streptomycin (68.3%), whereas all strains were sensitive to chloramphenicol and tetracyclines. Most of the isolates (55.8%) showed resistance against two classes of antimicrobials, mainly to ampicillin and streptomycin (46.2%). Only one isolate displayed a multiresistant pattern. Genotypic analysis of V. parahaemolyticus revealed a high degree of diversity among the isolates tested. The pulsed-field gel electrophoresis (PFGE) method distinguished 73 clonal groups, and the most numerous group consisted of 7 strains. Sequencing by the multilocus sequence typing (MLST) method showed 76 sequence types (STs), of which ST481 and ST1361 were most frequently identified. In addition, 51 (67.1%) new sequence types were discovered and added to the PubMLST international database.
IMPORTANCE The presence of V. parahaemolyticus in seafood may pose a risk for consumers, especially in countries where shellfish are eaten raw. In recent years, a significant increase of food poisoning caused by these bacteria has been also observed in Europe. Our results highlight the high level of V. parahaemolyticus contamination of seafood, along with the isolates being potentially pathogenic for humans. However, the first-line antimicrobials, such as tetracyclines and fluoroquinolones, remained highly effective against V. parahaemolyticus. The monitoring of antimicrobial resistance of isolates is important to ensure the high efficacy in the treatment of human infections. Most of V. parahaemolyticus strains possessed new sequence types (STs), which showed the high genetic diversity of the isolates tested.
INTRODUCTION
Vibrio parahaemolyticus is a marine microorganism recognized as an important cause of gastroenteritis in humans (1, 2). These bacteria naturally occur in warm seawaters along the coasts of many continents, and their highest level is found in the summer months. However, in recent years, increasing water temperature in seas and oceans has been observed as a result of global warming and may lead to the appearance of this pathogen in seawater, where it has not been previously present (2, 3). Human infection occurs usually after eating of contaminated food, especially raw or undercooked fish, crustaceans, and molluscs. Pathogenic V. parahaemolyticus bacteria mainly cause gastrointestinal disorders characterized by watery diarrhea, nausea, and abdominal cramps, but in severe cases, septicemia and generalized disease may develop, which sometimes lead to death (4). In Asian countries and the United States, V. parahaemolyticus has been one of the most common causes of food poisoning in humans (1, 5, 6). In Europe, the number of V. parahaemolyticus infections is growing year by year, mainly due to climate warming and changes in eating habits, i.e., increasing consumption of seafood (2).
Most of the V. parahaemolyticus strains isolated from the environment or food have no pathogenic potential, whereas clinical isolates usually possess virulence factors, such as thermostable direct hemolysin (TDH) and/or TDH-related hemolysin (TRH) (1). Both hemolysins, encoded by the tdh and trh genes, respectively, are the most important virulence markers associated with hemolytic, enterotoxic, and cytotoxic activities in the host cell (4, 7). Moreover, trh-positive V. parahaemolyticus strains almost always produce urease, which plays a significant role in the colonization of intestinal epithelial cells as well as in the activation of inflammatory cytokines (4, 8). However, V. parahaemolyticus strains that do not produce any hemolysins may possess other virulence factors important in gastroenteric infections (4, 9).
The vast majority of V. parahaemolyticus infections are self-limiting, and antimicrobial therapy is rarely required. The antimicrobials of choice for the treatment of severe infection cases are cephalosporins, tetracyclines, quinolones, and fluoroquinolones (10, 11). However, in recent years, many antimicrobial-resistant V. parahaemolyticus strains have emerged into the environment due to the excessive use of antibiotics and other chemotherapeutic agents in humans, agriculture, and aquaculture (10, 12). A determination of the antimicrobial resistance profile among V. parahaemolyticus strains may be used for monitoring changes in the sensitivity of the bacteria to antibiotics, especially those of the first-line choices in the treatment of human infections (13).
Several molecular typing methods have been developed for the differentiation of V. parahaemolyticus isolates, tracking the source of infection and detection of geographic distribution of virulent strains (14). Pulsed-field gel electrophoresis (PFGE) is a highly discriminatory technique used to determine the relatedness of clinical and environmental isolates and is still considered to be the gold standard for V. parahaemolyticus typing (15). Newer genotyping methods applied in epidemiological investigation, including multilocus sequence typing (MLST), are based on the sequencing of genes or whole genomes (14, 16). MLST allows the detection of slowly progressing sequence changes in the V. parahaemolyticus genome and may be used in monitoring the spread of pathogenic strains (14, 16).
The aim of the present study was to characterize V. parahaemolyticus isolates from seafood for the presence of virulence factors, antimicrobial resistance, and molecular diversity using PFGE and MLST methods.
RESULTS
Prevalence of V. parahaemolyticus.
A total 113 of 595 (19.0%) fish and shellfish samples tested were positive for V. parahaemolyticus as a result of biochemical identification, whereas PCR for the species-specific toxR and tlh genes confirmed the presence of these bacteria in 104 (92.0%) samples (Table 1). Most of the isolates were recovered from bivalve molluscs (92 out of 104 isolates [88.5%]), including clams (50 [54.4%]), mussels (25 [27.2%]), oysters (12 [13.0%]), and scallops (5 [5.4%]). The remaining 12 (11.5%) strains were obtained from fish. Regarding the geographical origin, V. parahaemolyticus was most commonly isolated from samples from the Netherlands (45 isolates [43.3%]) and Italy (34 [32.7%]), but some strains were also from Norway (13 [12.4%]), France (6 [5.8%]), and other countries (6 [5.8%]).
TABLE 1.
Sources of V. parahaemolyticus strains used in the present study
| Sample type tested | No. (%) of samples |
|
|---|---|---|
| Tested | Positive | |
| Shellfish | ||
| Clams | ||
| Manila clams | 122 | 50 |
| Razor clams | 24 | 0 |
| Hard clams | 7 | 0 |
| Cockle | 6 | 0 |
| Amandes | 4 | 0 |
| Mussels | 150 | 25 |
| Oysters | 129 | 12 |
| Scallops | 53 | 5 |
| Total | 495 (83.2) | 92 (15.5) |
| Fish | ||
| Perch | 37 | 5 |
| Cod | 16 | 0 |
| Salmon | 15 | 2 |
| Flounder | 13 | 1 |
| Herring | 10 | 2 |
| Ocean perch | 7 | 1 |
| Angler fish | 2 | 1 |
| Total | 100 (16.8) | 12 (2.0) |
| All samples | 595 (100.0) | 104 (17.5) |
Identification of virulence factors.
The tdh gene was identified only in 3 of 104 (2.9%) V. parahaemolyticus isolates analyzed, and all of them were recovered from Manila clams originating from Italy. The trh marker was detected in 11 (10.6%) isolates, of which 9 isolates were obtained from shellfish, especially from clams (7 strains) and fish (2 isolates). None of the tested isolates had both the tdh and trh genes. Moreover, all trh-positive strains possessed the ure gene and were able to produce urease in vitro. In addition, urease activity was revealed in one isolate from a fish that was trh negative. All V. parahaemolyticus isolates also showed protease A activity.
Antimicrobial resistance.
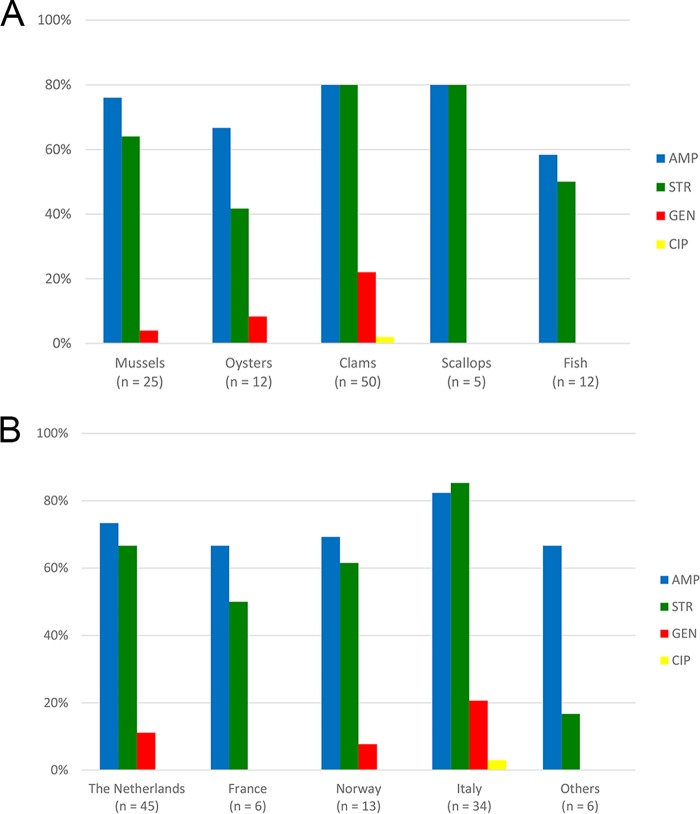
The antimicrobial resistance profiles of 64 V. parahaemolyticus isolates included in the present study were described previously (17). The analysis of the results for all 104 isolates revealed that 91 (87.5%) of them were resistant to at least one antimicrobial agent used in the study, and the remaining 13 (12.5%) strains were susceptible to all antimicrobials. The majority of the V. parahaemolyticus strains were resistant to ampicillin (78 [75.0%]) and streptomycin (71 [68.3%]), whereas resistance to gentamicin was found in 13 (12.5%) of the strains tested. On the other hand, only one isolate was resistant to ciprofloxacin, and all V. parahaemolyticus isolates were susceptible to tetracycline and chloramphenicol. Differences in antimicrobial resistance were observed among strains of various sources and geographical origins (Fig. 1). Most isolates resistant to ampicillin (40 [80.0%]) and streptomycin (40 [80.0%]) were recovered from clams. Ampicillin-resistant strains mainly originated from fish and shellfish from Italy (28 [82.3%]) and from the Netherlands (33 [73.3%]), whereas V. parahaemolyticus strains resistant to streptomycin were mainly recovered from Italian samples (29 [85.3%]). Isolates resistant to gentamicin were from mussels (1 isolate [4.0%]), oysters (1 isolate [8.3%]), and clams (11 [22.0%]), which originated from Italy, the Netherlands, and Norway. The only isolate resistant to ciprofloxacin was from Italian clams. It was noticed that the prevalences of streptomycin-resistant isolates from clams (40 out of 50 [80.0%]) and oysters (5 out of 12 [41.7%]) were statistically significant (P < 0.05). However, the analysis of the relationship between the country of origin and antimicrobial resistance did not reveal any differences (P > 0.05). Most of V. parahaemolyticus (58 [55.8%]) isolates showed resistance against two classes of antimicrobials, mainly to ampicillin and streptomycin (48 [46.2%]). Some strains were resistant to ampicillin, streptomycin, and gentamicin (10 [9.6%]). Only one isolate was resistant to three antimicrobial classes (ampicillin, streptomycin, and ciprofloxacin).
FIG 1.
Percentages of 104 V. parahaemolyticus isolates resistant to antimicrobials in relation to source (A) and origin (B) of samples. AMP, ampicillin; STR, streptomycin; GEN, gentamicin; CIP, ciprofloxacin.
MLST.
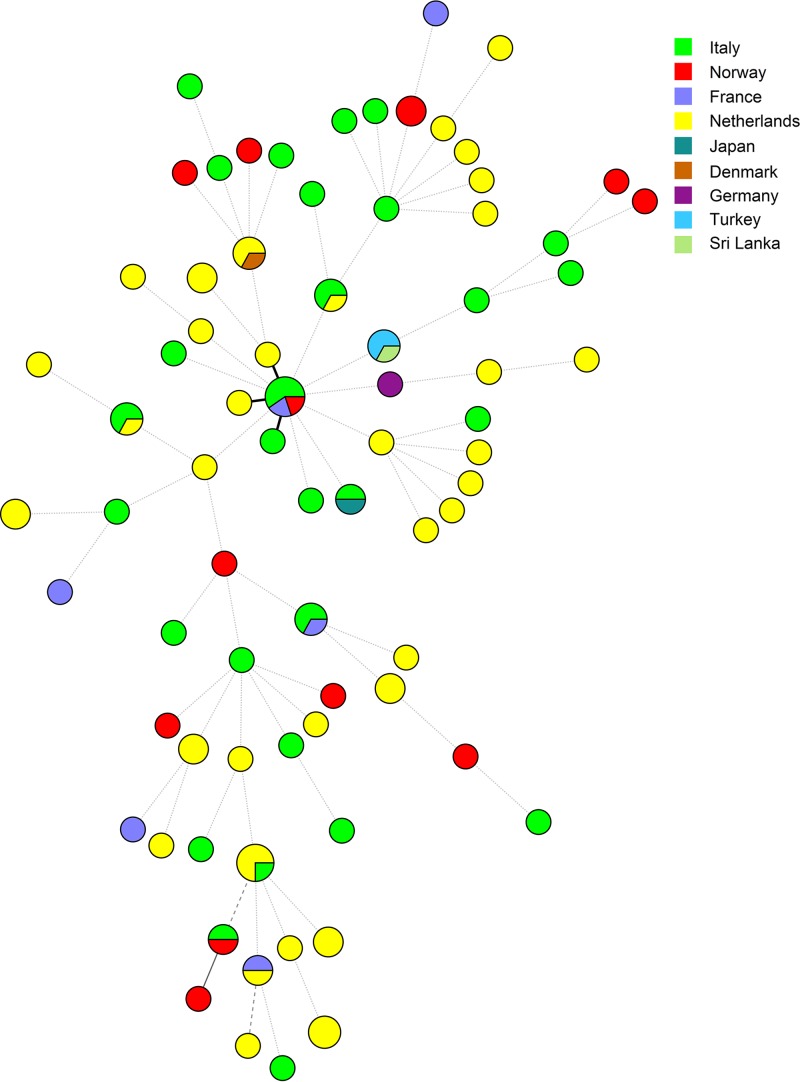
MLST analysis showed high molecular diversity among 104 V. parahaemolyticus isolates tested in the study. A total of 76 different sequence types (STs) were identified, including 17 STs with at least two isolates and 59 STs containing single strains. Furthermore, 51 (67.1%) novel STs were discovered and added to the international PubMLST/V. parahaemolyticus database (http://pubmlst.org/vparahaemolyticus) (Table 2). The two most frequent sequence types, ST481 and ST1361, possessed 5 and 4 isolates, respectively. Strains with ST481 were closely related (similarity above 80%) to the isolates of ST6 and ST1359 (difference only in the dtdS gene) and to the strain from ST1389 (which possessed a new allele for the dnaE gene). The minimum spanning tree based on the allele numbers and geographical origins of the isolates did not reveal a clear clustering connected with the origin of the strains (Fig. 2).
TABLE 2.
PFGE profiles in relation to sequence types and AMR patterns of V. parahaemolyticus strains originating from various sources and geographical areasa
| PFGE profile | No. of isolates | Source (no.) of isolatesb | Country (no.) of originc | AMR pattern (no.) | ST (no.)d |
|---|---|---|---|---|---|
| 1 | 2 | M (2) | NO (2) | AMP (1), AMP-STR (1) | ST1371 (2) |
| 2 | 1 | O | NL | AMP-STR-GEN | ST1407 |
| 3 | 2 | M (1), O (1) | FR (1), NL (1) | AMP (2) | ST1366 (2) |
| 4 | 1 | C | NL | AMP | ST1417 |
| 5 | 1 | M | NO | ST1388 | |
| 6 | 2 | C (1), S (1) | IT (1), NO (1) | AMP-STR (2) | ST1356 (1), ST1382 (1) |
| 7 | 1 | C | IT | AMP-STR | ST1368 |
| 8 | 1 | F | NL | AMP-STR | ST1385 |
| 9 | 1 | M | NO | AMP | ST1369 |
| 10 | 1 | M | NO | AMP-STR | ST1379 |
| 11 | 1 | M | NL | AMP-STR | ST1374 |
| 12 | 1 | C | IT | AMP-STR | ST162 |
| 13 | 1 | M | NL | AMP | ST57 |
| 14 | 1 | C | IT | AMP-STR | ST1380 |
| 15 | 1 | C | NL | AMP | ST1383 |
| 16 | 1 | O | NL | AMP-STR | ST1370 |
| 17 | 2 | O (2) | NL (2) | ST1411 (2) | |
| 18 | 1 | C | NL | AMP-STR-GEN | ST1419 |
| 19 | 1 | C | NL | AMP | ST445 |
| 20 | 1 | M | NO | STR-GEN | ST1320 |
| 21 | 3 | M (3) | NL (3) | AMP-STR (3) | ST1372 (3) |
| 22 | 2 | M (1), S (1) | IT (1), JP (1) | ST1386 (2) | |
| 23 | 1 | O | NL | AMP | ST49 |
| 24 | 2 | M (2) | DK (1), NL (1) | AMP-STR (2) | ST49 (2) |
| 25 | 1 | C | NL | AMP-STR | ST1375 |
| 26 | 1 | C | IT | AMP-STR | ST400 |
| 27 | 1 | F | NL | AMP-STR | ST1414 |
| 28 | 1 | O | NL | AMP | ST1373 |
| 29 | 2 | M (2) | NL (2) | STR (1), AMP-STR (1) | ST347 (2) |
| 30 | 1 | C | IT | AMP | ST1409 |
| 31 | 1 | C | FR | AMP | ST1363 |
| 32 | 2 | M (2) | NL (2) | AMP (1), AMP-STR (1) | ST445 (1), ST774 (1) |
| 33 | 2 | O (2) | NL (2) | AMP (1), STR (1) | ST1195 (2) |
| 34 | 1 | C | FR | AMP-STR | ST1159 |
| 35 | 1 | C | NL | AMP-STR | ST1285 |
| 36 | 1 | C | FR | ST1416 | |
| 37 | 2 | C (2) | NL (2) | ST810 (2) | |
| 38 | 1 | M | NL | AMP-STR | ST767 |
| 39 | 1 | F | DE | ST1413 | |
| 40 | 1 | F | NO | ST73 | |
| 41 | 1 | F | NL | AMP-STR | ST64 |
| 42 | 3 | C (3) | IT (2), NL (1) | AMP-STR (1), AMP-STR-GEN (2) | ST1358 (3) |
| 43 | 1 | C | IT | GEN | ST1387 |
| 44 | 1 | S | NL | AMP-STR | ST1381 |
| 45 | 1 | C | IT | AMP-STR | ST1410 |
| 46 | 1 | C | NL | AMP-STR | ST135 |
| 47 | 3 | F (3) | LK (1), TR (2) | AMP (3) | ST1390 (3) |
| 48 | 1 | F | NO | ST996 | |
| 49 | 1 | O | NL | AMP-STR | ST1406 |
| 50 | 1 | C | IT | AMP-STR-GEN | ST1415 |
| 51 | 3 | C (2), F (1) | FR (1), IT (2) | AMP-STR (1), AMP (2) | ST1362 (3) |
| 52 | 1 | C | IT | AMP-STR | ST1365 |
| 53 | 1 | C | IT | AMP-STR-CIP | ST1367 |
| 54 | 1 | C | IT | AMP-STR-GEN | ST1357 |
| 55 | 1 | C | IT | AMP-STR | ST1405 |
| 56 | 1 | C | IT | AMP-STR-GEN | ST1377 |
| 57 | 3 | C (1), F (2) | IT (2), NL (1) | AMP-STR (2), STR (1) | ST1384 (3) |
| 58 | 2 | C (2) | NL (2) | AMP-STR-GEN (2) | ST6 (1), ST1376 (1) |
| 59 | 1 | C | IT | AMP-STR | ST1412 |
| 60 | 7 | C (6), S (1) | FR (1), IT (4) NL (1), NO (1) | AMP-STR (3), AMP (2), STR (1), STR-GEN (1) | ST481 (5), ST1359 (1), ST1389 (1) |
| 61 | 2 | C (1), M (1) | IT (1), NO (1) | AMP-STR (2) | ST564 (1), ST1290 (1) |
| 62 | 1 | C | NL | AMP-STR | ST1418 |
| 63 | 1 | C | IT | AMP-STR | ST490 |
| 64 | 1 | O | NL | AMP | ST1404 |
| 65 | 1 | C | IT | AMP-STR-GEN | ST1408 |
| 66 | 1 | O | NL | AMP | ST1360 |
| 67 | 3 | C (1), M (2) | IT (1), NL (2) | AMP (2), AMP-STR (1) | ST1361 (3) |
| 68 | 1 | M | NL | ST1361 | |
| 69 | 1 | S | NO | AMP-STR | ST1267 |
| 70 | 2 | C (1), M (1) | IT (1), NO (1) | AMP-STR (2) | ST773 (2) |
| 71 | 1 | C | IT | AMP-STR | ST1355 |
| 72 | 1 | C | IT | ST1364 | |
| 73 | 1 | C | IT | AMP-STR | ST1262 |
AMR, antimicrobial resistance.
C, clams; O, oysters; M, mussels; S, scallops; F, fish.
NL, the Netherlands; IT, Italy; NO, Norway; FR, France; DK, Denmark; JP, Japan; DE, Germany; TR, Turkey; LK, Sri Lanka.
Novel sequence types (STs) are marked in bold.
FIG 2.
Minimum spanning tree of V. parahaemolyticus strains based on the allelic profiles. Colors indicate the geographical origins of isolates. Size of circles represents number of isolates with the same ST, and thickness of the branches indicates the degree of similarity among V. parahaemolyticus strains tested.
PFGE.
PFGE investigation showed 73 different molecular patterns, with 20 groups containing at least two isolates and the remaining 53 profiles covering only one strain (Table 2). The largest cluster group (PFGE 60) included 7 strains, mainly of clams (6 isolates) and scallops (1 isolate) originating from different countries. The majority of these isolates were classified to ST481 (5 strains) and were resistant to ampicillin and streptomycin. Most isolates with the same PFGE profile had similar ST and antibiotic resistance patterns and were mostly isolated from the same source. It was also found that V. parahaemolyticus isolates of the PFGE clonal groups 6, 32, 58, 60, and 61 possessed the same macrorestriction profiles but other STs. On the other hand, the isolates with the same sequence types ST49, ST445, and ST1361 had different PFGE profiles.
DISCUSSION
The present results represent a comprehensive study on the characterization of V. parahaemolyticus isolates toward the presence of virulence factors, antimicrobial resistance, and genetic relatedness.
The tdh and trh hemolysin markers, which play a significant role in the pathogenesis of infection in humans, were identified only in 3 (2.9%) and 11 (10.6%) of 104 isolates, respectively. In the study by Bilung et al. (18), the tdh gene was also detected at a lower level (3.2%) than the trh gene (17.7%). Similar results were also obtained by Ottaviani et al. (19). On the other hand, a higher prevalence of the tdh gene than the trh marker was observed in V. parahaemolyticus in Spain and in Thailand (20, 21). However, the distribution of tdh- and/or trh-positive strains may vary depending on the geographical origin, sample source, and the method of their detection. Furthermore, all trh-positive isolates identified in the present investigation possessed the ure gene, which is due to the presence of both genes in the same chromosomal pathogenicity island (8). Our results confirmed data reported by Sujeewa et al. (22) that not all isolates capable of producing urease were trh positive. Moreover, in the present investigation, another potential pathogenic factor, protease A, was found in 100% of the isolates tested, similar to the results in the study of Ottaviani et al. (9). This confirms the presence of other virulence factors, besides both hemolysins in V. parahaemolyticus, which play a significant role in pathogenicity.
In the current investigation, 75.0% and 68.3% of the isolates showed resistance to ampicillin and streptomycin, respectively, and these results were comparable to data obtained in other countries (10, 23, 24). The majority of V. parahaemolyticus strains resistant to both antibiotics were isolated from clams, mainly originating from Italy, which was similar to the findings in previous reports by Yu et al. (5), who found that 96.7% and 68.0% of V. parahaemolyticus strains of clam and oyster origin, respectively, were resistant to ampicillin and streptomycin. The highest resistance to ampicillin (100.0%) was previously observed among the isolates from Italian samples, whereas the percentage of streptomycin-resistant strains from this country was lower (32.2%) than that in the current study (23). The high resistance to β-lactams and aminoglycosides observed among V. parahaemolyticus strains may be due to extensive use of these antimicrobials in treatment, agriculture, and aquaculture during recent decades (10, 12). Moreover, in water environments, different microorganisms, including V. parahaemolyticus, are able to exchange their genetic determinants, which may also be the cause of increasing resistance to antibiotics (4, 23). Additionally, 46.2% of the isolates showed resistance to ampicillin (AMP) and streptomycin (STR), 9.6% to AMP, STR, and gentamicin (GEN), and only one isolate recovered from clams from Italy was resistant to ampicillin, streptomycin, and ciprofloxacin. In other investigations, most of the isolates (60 out of 76 [78.9%]) were resistant to two or more antimicrobial agents (25), whereas relatively low percentages (3.7% and 10.3%) of strains resistant to two and three classes of antimicrobials, respectively, were found in Italy (23). However, a comparison of data from various studies is often difficult due to different samples' origins, time and methods of isolate collection, and laboratory analysis. Furthermore, the present results confirmed those of previous reports from Europe, the United States, and Asian countries that V. parahaemolyticus strains were generally susceptible to antimicrobial agents used in the treatment of human infections (11, 21, 23). However, there is also a study showing a high prevalence of strains resistant to ciprofloxacin (46.9%), chloramphenicol (36.7%), and tetracycline (81.6%) (26).
Molecular typing revealed a high degree of diversity of the strains tested. The MLST method showed 76 STs, of which 51 (67.1%) STs were newly identified. Similar results were reported by Urmersbach et al. (27), where 130 V. parahaemolyticus strains obtained from a marine environment were classified into 82 STs, and 82.9% of them were described as new. Rahman et al. (28) also identified 63 unique STs in the isolates tested, and 49 (77.8%) STs were not described in the database. On the other hand, a lower percentage of isolates with new sequence types was also noted (29). Our findings and data from other countries indicated a high number of new alleles and STs in V. parahaemolyticus which have not been included into database. However, molecular information on the isolates collected in PubMLST are from clinical samples from the United States and Asia, whereas the isolates used in the present study were mainly originated from the European countries.
Based on the PFGE analysis, 73 clonal groups were identified among the isolates tested, with an overall similarity of 43.5%. Other authors indicated a closer genetic correlation between V. parahaemolyticus strains isolated in Europe or Japan, which were 63.4% and 71.0%, respectively (15, 30). However, there are also reports of a much lower level of similarity (28.4%) among isolates tested (31). The above-mentioned results are difficult to compare due to various sources of origin and different periods of V. parahaemolyticus isolation. It was also found that the largest PFGE 60 group, with 7 strains recovered mainly from clams in Italy, was resistant to ampicillin and streptomycin and belonged to ST481. This sequence type was previously identified in environmental isolates from Germany and Italy, as well as in clinical strains from China (27, 28) (http://pubmlst.org/vparahaemolyticus). In the present study, ST481 was closely related to new ST1359 and ST1389 detected in V. parahaemolyticus from clams originating from Italy and the Netherlands, respectively. Moreover, ST6 with the trh and ureR genes identified in the isolates from the Netherlands was genetically similar to strains of ST481. This sequence type was previously described in environmental isolates from Chile, Norway, and Italy (16, 32) (http://pubmlst.org/vparahaemolyticus). It may suggest a worldwide spread of some STs in V. parahaemolyticus, regardless of geographical location and environmental conditions.
In conclusion, the results of the present study indicate that V. parahaemolyticus may occur in seafood available in Poland, especially during warmer months. Some of these strains had pathogenic properties due to the presence of virulence factors. Most of the V. parahaemolyticus strains possessed new sequence types, which indicates a high diversity of the strains. Several isolates were resistant to ampicillin and streptomycin as well as to more than one class of antibiotics. Therefore, these bacteria may pose a serious threat to consumer health.
MATERIALS AND METHODS
Sources and identification of V. parahaemolyticus.
Vibrio parahaemolyticus strains were isolated during 2009 to 2015 from different species of raw shellfish (n = 495) and marine fish (n = 100) available on the Polish market but originating from various countries, such as the Netherlands (n = 277), Norway (n = 127), Italy (n = 91), France (n = 66), Turkey (n = 12), Poland (n = 10), Denmark (n = 4), Spain (n = 4), Germany (n = 2), and Sri Lanka (n = 2). All samples were analyzed according to the ISO 21872-1 standard (33) and then confirmed by PCRs for the species-specific tlh and toxR genes, as described previously (34, 35). Among all 104 V. parahaemolyticus isolates, 64 isolates were already described in a previous study on the prevalence and antimicrobial resistance (17). Detailed information on the number of V. parahaemolyticus isolates used in the current investigation is presented in Table 1.
Determination of virulence factors.
Vibrio parahaemolyticus isolates were investigated for the presence of the tdh, trh, and ureR genes using PCRs, as described previously (36, 37). The amplification products were visualized on 1.5% agarose gels (Sigma-Aldrich, USA) by staining with ethidium bromide (Sigma-Aldrich) and photographed using the Gel Doc 2000 documentation system (Bio-Rad, USA). Expression of the ure gene was determined by capacity of urease production on Christensen's urea agar with 1% NaCl (9). Protease A activity was examined on a nutrient agar with 1.5% skimmed milk and 1% NaCl (38).
Determination of antimicrobial resistance.
A broth microdilution method was used to establish the MICs of V. parahaemolyticus to antimicrobials selected according to the Clinical and Laboratory Standards Institute (CLSI) guideline (39), as described previously (17). The Sensititre custom susceptibility EUMVS2 plates (Trek Diagnostic Systems, UK) contained the following antibacterial agents (dilution range) were used: ampicillin (AMP, 0.5 to 32 mg/liter), ciprofloxacin (CIP, 0.03 to 8 mg/liter), streptomycin (STR, 2 to 128 mg/liter), gentamicin (GEN, 0.25 to 32 mg/liter), tetracycline (TET, 1 to 64 mg/liter), and chloramphenicol (CHL, 2 to 64 mg/liter). The cutoff values used for the interpretation of the MIC results were in accordance with the CLSI guideline (39), except streptomycin, for which the breakpoint has been described elsewhere (11, 24).
MLST analysis.
Multilocus sequence typing was performed by the sequence analysis of seven housekeeping genes (recA, gyrB, dnaE, dtdS, pntA, pyrC, and tnaA) according to the protocol available on the PubMLST website (http://pubmlst.org/vparahaemolyticus). The received nucleotide sequences for each locus were analyzed with BioNumerics software version 7.6 (Applied Maths, Belgium) and compared to the already-published sequences on the PubMLST website. Sequence types (STs) were determined on the basis of the obtained seven-digit allelic profiles. New alleles were submitted to the V. parahaemolyticus database in PubMLST. A phylogenetic tree was created using the unweighted pair group method using average linkages (UPGMA) and the minimum spanning tree (MST) methods (40, 41).
PFGE typing.
Pulsed-field gel electrophoresis was done according to the CDC PulseNet protocol (42). DNA was digested with 40 U of SfiI enzyme (Thermo Fisher Scientific, USA) at 50°C for 4 h. The separation of restriction fragments was performed in 1% SeaKem gold agarose (Lonza, USA) gels in 0.5× Tris-borate-EDTA (TBE) buffer (Sigma-Aldrich) using the CHEF-DR III system (Bio-Rad), with the following parameters: initial time, 10 s; final time, 35 s for 18 to 19 h at 6 V/cm and 14°C. The gels were stained in ethidium bromide (5 μg/ml; Sigma-Aldrich) for 15 min, and the DNA banding patterns were visualized with the Gel Doc 2000 system. Salmonella enterica serovar Braenderup H9812 was used as the molecular weight standard (42, 43). The macrorestriction profiles were analyzed by the BioNumerics software, and the dendrogram was generated by the UPGMA with the Dice correlation coefficient and a position tolerance of 1%. Clusters were defined on the basis of the 80% similarity cutoff.
Statistical analysis.
Statistical analysis of the results relating to antimicrobial resistance of V. parahaemolyticus by sample type or country of origin was carried out using Fisher's exact test for 2 by 2 contingency tables (Statistica, Krakow, Poland). P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
This study was supported financially by the Polish National Science Centre, decision DEC-2012/05/N/NZ7/00776, and by KNOW (Leading National Research Centre) Scientific Consortium Healthy Animal-Safe Food, decision of Ministry of Science and Higher Education no. 05-1/KNOW2/2015.
REFERENCES
- 1.Su Y-C, Liu C. 2007. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24:549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Baker-Austin C, Stockley L, Rangdale R, Martinez-Urtaza J. 2010. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ Microbiol Rep 2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Urtaza J, Bowers JC, Trinanes J, DePaola A. 2010. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res Int 43:1780–1790. doi: 10.1016/j.foodres.2010.04.001. [DOI] [Google Scholar]
- 4.Quadri F, Chowdhury NR, Takeda Y, Nair GB. 2005. Vibrio parahaemolyticus–seafood safety and associations with higher organisms, p 277–295. In Belkin S, Colwell RR (ed), Oceans and health: pathogens in the marine environment. Springer, New York, NY. [Google Scholar]
- 5.Yu WT, Jong KL, Lin YR, Tsai SE, Tey YH, Wong HC. 2013. Prevalence of Vibrio parahaemolyticus in oyster and clam culturing environments in Taiwan. Int J Food Microbiol 160:185–192. doi: 10.1016/j.ijfoodmicro.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC). 2016. National enteric disease surveillance: COVIS annual summary, 2014. Summary of human Vibrio cases reported to CDC, 2014. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/nationalsurveillance/pdfs/covis-annual-summary-2014-508c.pdf. [Google Scholar]
- 7.Letchumanan V, Chan KG, Lee LH. 2014. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol 5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida T, Suthienkul O, Park KS, Tang GQ, Yamamoto RK, Ishibashi M, Yamamoto K, Honda T. 1997. Evidence for genetic linkage of the ure and trh genes in Vibrio parahaemolyticus. J Med Microbiol 46:639–645. doi: 10.1099/00222615-46-8-639. [DOI] [PubMed] [Google Scholar]
- 9.Ottaviani D, Santarelli S, Bacchiocchi S, Masini L, Ghittino C, Bacchiocchi I. 2005. Presence of pathogenic Vibrio parahaemolyticus strains in mussels from the Adriatic Sea, Italy. Food Microbiol 22:585–590. doi: 10.1016/j.fm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Han F, Walker RD, Janes ME, Prinyawiwatkul W, Ge B. 2007. Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl Environ Microbiol 73:7096–7098. doi: 10.1128/AEM.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker-Austin C, McArthur JV, Tuckfield RC, Najarro M, Lindell AH, Gooch J, Stepanauskas R. 2008. Antibiotic resistance in the shellfish pathogen Vibrio parahaemolyticus isolated from the coastal water and sediment of Georgia and South Carolina, USA. J Food Prot 71:2552–2558. doi: 10.4315/0362-028X-71.12.2552. [DOI] [PubMed] [Google Scholar]
- 12.Cabello FC. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- 13.Elmahdi S, DaSilva LV, Parveen S. 2016. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol 57:128–134. doi: 10.1016/j.fm.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Xie Y, Xu J, Wang Q, Gu M, Yang J, Zhou M, Wang D, Shi C, Shi X. 2012. Molecular typing of Vibrio parahaemolyticus isolates from the middle-east coastline of China. Int J Food Microbiol 153:402–412. doi: 10.1016/j.ijfoodmicro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Suffredini E, Lopez-Joven C, Maddalena L, Croci L, Roque A. 2011. Pulsed-field gel electrophoresis and PCR characterization of environmental Vibrio parahaemolyticus strains of different origins. Appl Environ Microbiol 77:6301–6304. doi: 10.1128/AEM.00333-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, DePaola A. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol 190:2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopatek M, Wieczorek K, Osek J. 2015. Prevalence and antimicrobial resistance of Vibrio parahaemolyticus isolated from raw shellfish in Poland. J Food Prot 78:1029–1033. doi: 10.4315/0362-028X.JFP-14-437. [DOI] [PubMed] [Google Scholar]
- 18.Bilung LM, Radu S, Bahaman AR, Rahim RA, Napis S, Ling MWCV, Tanil GB, Nishibuchi M. 2005. Detection of Vibrio parahaemolyticus in cockle (Anadara granosa) by PCR. FEMS Microbiol Lett 252:85–88. doi: 10.1016/j.femsle.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 19.Ottaviani D, Leoni F, Rocchegiani E, Mioni R, Costa A, Virgilio S, Serracca L, Bove D, Canonico C, Di Cesare A, Masini L, Potenziani S, Caburlotto G, Ghidini V, Lleo MM. 2013. An extensive investigation into the prevalence and the genetic and serological diversity of toxigenic Vibrio parahaemolyticus in Italian marine coastal waters. Environ Microbiol 15:1377–1386. doi: 10.1111/j.1462-2920.2012.02839.x. [DOI] [PubMed] [Google Scholar]
- 20.Roque A, Lopez-Joven C, Lacuesta B, Elandaloussi L, Wagley S, Furones MD, Ruiz-Zarzuela I, de Blas I, Rangdale R, Gomez-Gil B. 2009. Detection and identification of tdh- and trh-positive Vibrio parahaemolyticus strains from four species of cultured bivalve molluscs on the Spanish Mediterranean coast. Appl Environ Microbiol 75:7574–7577. doi: 10.1128/AEM.00772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mala W, Alam M, Angkititrakul S, Wongwajana S, Lulitanond V, Huttayananont S, Kaewkes W, Faksri K, Chomvarin C. 2016. Serogroup, virulence, and molecular traits of Vibrio parahaemolyticus isolated from clinical and cockle sources in northeastern Thailand. Infect Genet Evol 39:212–218. doi: 10.1016/j.meegid.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Sujeewa AKW, Norrakiah AS, Laina M. 2009. Prevalence of toxic genes of Vibrio parahaemolyticus in shrimps (Penaeus monodon) and culture environment. Int Food Res J 16:89–95. [Google Scholar]
- 23.Ottaviani D, Leoni F, Televi G, Masini L, Santarelli S, Rocchegiani E, Susini F, Montagna C, Monno R, D'Annibale L, Manso E, Oliva M, Pazzani C. 2013. Extensive investigation of antimicrobial resistance in Vibrio parahaemolyticus from shellfish and clinical sources, Italy. Int J Antimicrob Agents 42:187–193. doi: 10.1016/j.ijantimicag.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Shaw KS, Rosenberg Goldstein RE, He X, Jacobs JM, Crump BC, Sapkota AR. 2014. Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS One 9:e89616. doi: 10.1371/journal.pone.0089616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daramola BA, Williams R, Dixon RA. 2009. In vitro antibiotic susceptibility of Vibrio parahaemolyticus from environmental sources in northern England. Int J Antimicrob Agents 34:499–500. doi: 10.1016/j.ijantimicag.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Noorlis A, Ghazali FM, Cheah YK, Tuan ZTC, Wong WC, Tunung R, Pui CF, Nishibuchi M, Nakaguchi Y, Son R. 2011. Antibiotic resistance and biosafety of Vibrio cholerae and Vibrio parahaemolyticus from freshwater fish at retail level. Int Food Res J 18:1523–1530. [Google Scholar]
- 27.Urmersbach S, Alter T, Koralage MSG, Sperling L, Gerdts G, Messelhäusser U, Huehn S. 2014. Population analysis of Vibrio parahaemolyticus originating from different geographical regions demonstrates a high genetic diversity. BMC Microbiol 14:59. doi: 10.1186/1471-2180-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman MS, Carraro R, Cardazzo B, Carraro L, Meneguolo DB, Martino ME, Andreani NA, Bordin P, Mioni R, Barco L, Novelli E, Balzan S, Fasolato L. 2017. Molecular typing of Vibrio parahaemolyticus strains isolated from mollusks in the North Adriatic Sea. Foodborne Pathog Dis 14:454–464. doi: 10.1089/fpd.2016.2263. [DOI] [PubMed] [Google Scholar]
- 29.Lüdeke CHM, Gonzalez-Escalona N, Fischer M, Jones JL. 2015. Examination of clinical and environmental Vibrio parahaemolyticus isolates by multi-locus sequence typing (MLST) and multiple-locus variable number tandem-repeat analysis (MLVA). Front Microbiol 6:564. doi: 10.3389/fmicb.2015.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Guan W, Alam MJ, Shen Z, Zhang S, Li L, Shinoda S, Shi L. 2009. Pulsed-field gel electrophoresis typing of multidrug-resistant Vibrio parahaemolyticus isolated from various sources of seafood. J Health Sci 55:783–789. doi: 10.1248/jhs.55.783. [DOI] [Google Scholar]
- 31.Ellingsen AB, Jorgensen H, Wagley S, Monshaugen M, Rorvik LM. 2008. Genetic diversity among Norwegian Vibrio parahaemolyticus. J Appl Microbiol 105:2195–2202. doi: 10.1111/j.1365-2672.2008.03964.x. [DOI] [PubMed] [Google Scholar]
- 32.Ellingsen AB, Olsen JS, Granum PE, Rorvik LM, González-Escalona N. 2013. Genetic characterization of trh positive Vibrio spp. isolated from Norway. Front Cell Infect Microbiol 3:107. doi: 10.3389/fcimb.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Organization for Standardization (ISO) 2017. Microbiology of the food chain. Horizontal method for the determination of Vibrio spp. Part 1: detection of potentially enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. ISO standard 21872-1:2017. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 34.Bej AK, Patterson DP, Brasher CW, Vickery MCL, Jones DD, Kaysner CA. 1999. Detection of total hemolysin Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh, and trh. J Microbiol Methods 36:215–225. doi: 10.1016/S0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 35.Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 37:1173–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y. 1992. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes 6:477–487. doi: 10.1016/0890-8508(92)90044-X. [DOI] [PubMed] [Google Scholar]
- 37.Parvathi A, Kumar HS, Bhanumathi A, Ishibashi M, Nishibuchi M, Karunasagar I, Karunasagar I. 2006. Molecular characterization of thermostable direct haemolysin-related haemolysin (TRH)-positive Vibrio parahaemolyticus from oysters in Mangalore, India. Environ Microbiol 8:997–1004. doi: 10.1111/j.1462-2920.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- 38.Iyer L, Vadivelu J, Puthucheary SD. 2000. Detection of virulence associated genes, haemolysin and protease amongst Vibrio cholerae isolated in Malaysia. Epidemiol Infect 125:27–34. doi: 10.1017/S0950268899004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute (CLSI). 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed CLSI document M45-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 40.Seneath PHA, Sokal RR. 1973. Numerical taxonomy. W. H. Freeman and Company, San Francisco, CA. [Google Scholar]
- 41.Salipante SJ, Hall BG. 2011. Inadequacies of minimum spanning trees in molecular epidemiology. J Clin Microbiol 49:3568–3575. doi: 10.1128/JCM.00919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention (CDC). 2013. Standard operating procedure for PulseNet PFGE of Vibrio cholerae and Vibrio parahaemolyticus. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/pulsenet/PDF/vibrio_pfge_protocol-508c.pdf.
- 43.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol 43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]