Glutarate is an important C5 linear-chain dicarboxylic acid, which is widely used in polyesters and polyamides such as nylon-4,5 and nylon-5,5 in the chemical industry. Glutarate is currently produced from the feedstocks derived from petroleum, specifically by oxidation of a mixture of cyclohexanone and cyclohexanol catalyzed by nitric acid. However, the chemical synthesis results in high pollution and dramatic greenhouse gas emission. Thus, the biological production of glutarate directly from the substrate is of great importance. Although there have been reports using Corynebacterium glutamicum to produce glutarate, it has serious limitations due to the limited lysine supply and long fermentation time. To solve this problem, a novel synthetic pathway was constructed in this study, and the highest glutarate titer was reported in Escherichia coli using a short fermentation time without lysine addition, making bio-based glutarate production much more feasible.
KEYWORDS: glutarate, Escherichia coli, high titer, reverse adipate degradation pathway, metabolic engineering
ABSTRACT
Glutarate is a linear-chain dicarboxylic acid with wide applications in the production of polyesters and polyamides such as nylon-4,5 and nylon-5,5. Previous studies focused on the biological production of glutarate from lysine with low yields and titers. Here, we report on glutarate production by Escherichia coli using a five-step reverse adipate degradation pathway (RADP) identified in Thermobifida fusca. By expressing the enzymes of RADP, the glutarate was detected by strain Bgl146 in shaken flasks. After fermentation optimization, the titer of glutarate by Bgl146 was increased to 4.7 ± 0.2 mM in shaken flasks. We further eliminated pathways for the major metabolites competing for carbon flux by CRISPR/Cas9 (ΔarcA, ΔldhA, ΔatoB, and ΔpflB). Moreover, the final strain Bgl4146 produced 36.5 ± 0.3 mM glutarate by fed-batch fermentation. These results constitute the highest glutarate titer reported in E. coli.
IMPORTANCE Glutarate is an important C5 linear-chain dicarboxylic acid, which is widely used in polyesters and polyamides such as nylon-4,5 and nylon-5,5 in the chemical industry. Glutarate is currently produced from the feedstocks derived from petroleum, specifically by oxidation of a mixture of cyclohexanone and cyclohexanol catalyzed by nitric acid. However, the chemical synthesis results in high pollution and dramatic greenhouse gas emission. Thus, the biological production of glutarate directly from the substrate is of great importance. Although there have been reports using Corynebacterium glutamicum to produce glutarate, it has serious limitations due to the limited lysine supply and long fermentation time. To solve this problem, a novel synthetic pathway was constructed in this study, and the highest glutarate titer was reported in Escherichia coli using a short fermentation time without lysine addition, making bio-based glutarate production much more feasible.
INTRODUCTION
Among the different dicarboxylic acids, glutarate is an important C5 linear-chain dicarboxylic acid that is widely used in polyesters and polyamides such as nylon-4,5 and nylon-5,5 in the chemical industry (1, 2). Glutarate is a petroleum derivative, specifically by oxidation of a mixture of cyclohexanone and cyclohexanol (KA oils) catalyzed by nitric acid (3–5). Thus, glutarate needs to be separated from a mixture of glutarate, adipate, and succinate (6). However, this chemical synthesis method could lead to high environmental pollution and greenhouse gas emissions (7). Therefore, there are great incentives to discover alternative approaches allowing for renewable and affordable glutarate production. The bio-based methods are attracting a lot of attention due to environment-friendly process and the feasibility of industrial application (8). There is thus a need for a bio-based method for glutarate production from renewable resources. Recently, many bacteria, such as Pseudomonas putida, could convert l-lysine to glutarate through the so-called aminovalerate (AMV) pathway (9–11). Thereafter, all the genes of the AMV pathway were transferred into Escherichia coli for heterologous expression, and the production of glutarate was 6.2 mM (1). In order to further increase glutarate production, lysine and α-ketoglutaric acid (α-KG) were added to the cells, and 12.9 mM glutarate could be synthesized by the recombinant E. coli WL3110 strain (12). Thus, the supply of lysine and α-KG has a key role in the production of glutarate by the AMV pathway. Researchers have begun to use Corynebacterium glutamicum (an l-lysine-overproducing bacterium) to produce glutarate (1, 13, 14). After heterologous overexpression of the first two enzymes of the AMV pathway, recombinant C. glutamicum could finally accumulate 90.2 mM glutarate by fed-batch fermentation (14). However, the highest titer of glutarate was obtained after 150 h, resulting in low productivity. All of these methods are dependent on AMV or lysine for production, and the biosynthetic pathway has serious limitations. In order to eliminate the lysine supply, recombinant E. coli BW25113 produced 2.3 mM glutarate on glucose (15). In order to improve the titers of glutarate, α-KG was added to the pathway, and CRISPRi was used to inhibit the essential genes in tricarboxylic acid (TCA) cycle, eventually producing 3.2 mM glutarate (15). Recently, Yu et al. constructed a functional biosynthetic pathway in E. coli, producing ∼0.1 mM glutarate with glucose and an extra 10 mM sodium glutamate (16, 17). Parthasarathy et al. (18) found that the production of adipate and glutarate shared the same pathway. Deng and Mao reported the existence of a native adipate synthesis pathway in the thermophilic actinobacterium Thermobifida fusca (19) and identified it as the reverse adipate degradation pathway (RADP) with five enzymes. We hypothesized that RADP could also convert acetyl coenzyme A (acetyl-CoA) and malonyl-CoA to glutarate from sugars. Subsequently, we constructed the T. fusca RADP in E. coli BL21(DE3) by expressing five enzymes for glutarate production. After a series of condition optimizations, the maximal glutarate titer was achieved in E. coli.
RESULTS
Assembling the glutarate biosynthetic pathway in E. coli.
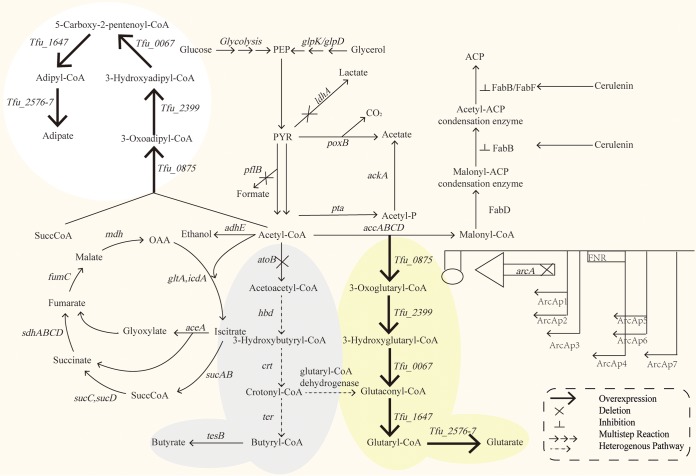
Parthasarathy et al. (18) proved that the production of adipate and glutarate shared the same pathway. Deng and Mao reported a native adipate synthesis pathway in T. fusca (19). Thus, we constructed the T. fusca RADP in E. coli BL21(DE3) by expressing five enzymes (Tfu_0875, Tfu_2399, Tfu_0067, Tfu_1647, and Tfu_2576-7) for glutarate production (Fig. 1). The glutarate was detected, and 0.5 ± 0.1 mM adipate was produced by the resulting strain (Bgl146) on 22.2 mM glucose in a 3-(N-morpholino)propanesulfonic acid (MOPS) culture in shaken flasks (Fig. 2a). Meanwhile, strain Bgl146 also produced many other organic acids, such as acetate, lactate, butyrate, and formate (Fig. 2a).
FIG 1.
A novel synthetic pathway for glutarate production in E. coli. The reverse adipate degradation pathway (RADP) includes five steps in Thermobifida fusca: β-ketothiolase (Tfu_0875), 3-hydroxyacyl-CoA dehydrogenase (Tfu_2399), 3-hydroxyadipyl-CoA dehydrogenase (Tfu_0067), 5-carboxy-2-pentenoyl-CoA reductase (Tfu_1647), and adipyl-CoA synthetase (Tfu_2576-7). The arcA, ldhA, atoB, sucD, and pflB genes were deleted using the CRISPR/Cas9 system. A rough arrow represents genes or enzymes subject to overexpression, “X” represents those deleted, and the “⊥” symbol represents suppression. Solid arrows indicate single steps, three arrows indicate multiple steps, and dashed arrows indicate that a step does not exist in E. coli. ldhA, l-lactate dehydrogenase; atoB, acetyl-CoA C-acetyltransferase; arcA, aerobic respiration control protein; pflB, formate C-acetyltransferase 1; TCA, tricarboxylic acid cycle; EMP, the glycolysis pathway; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PYR, pyruvate; SuccCoA, succinyl-CoA; ACP, acyl carrier protein.
FIG 2.
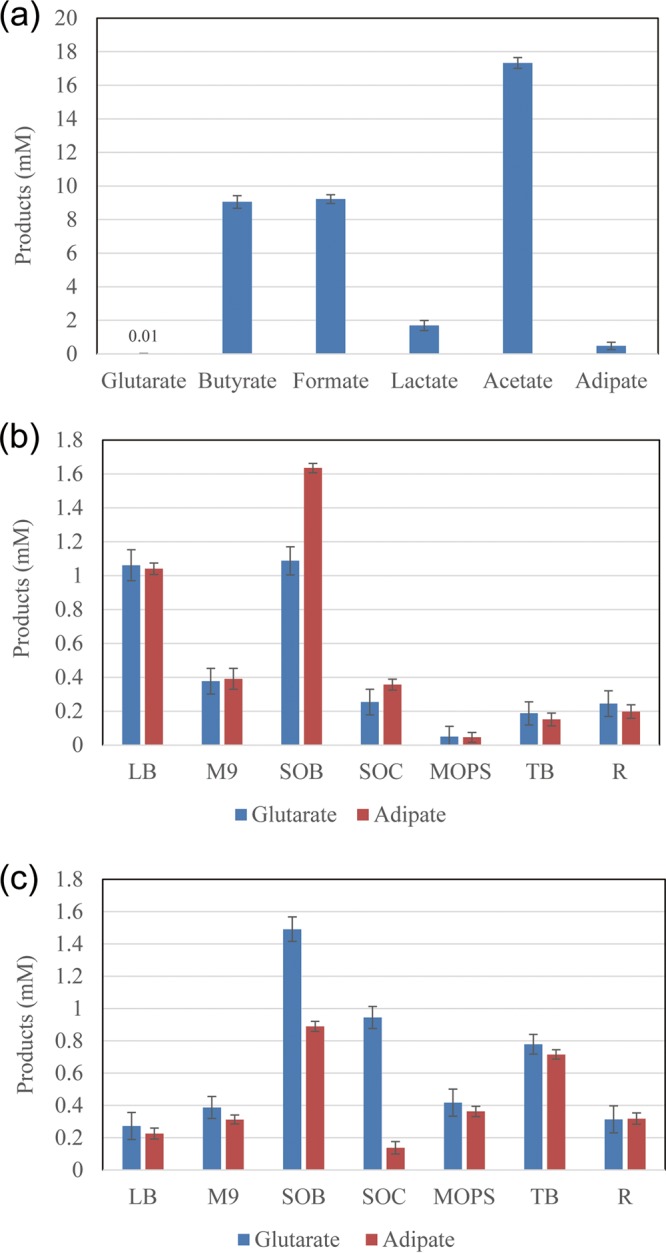
Characterizations of E. coli Bgl146. (a) Fermentation results of engineered strain Bgl146 on MOPS minimal medium. (b) Production of glutarate and adipate on M9, R, lysogeny broth (LB), superoptimal broth (SOB), superoptimal broth with catabolite repression (SOC), MOPS minimal medium (MOPS), and terrific broth (TB) media under aerobic conditions. (c) Fermentation results of glutarate and adipate on M9, R, LB, SOB, SOC, MOPS, and TB media under microaerobic conditions. E. coli Bgl146: BL21(DE3) harboring pAD1, pAD4, and pAD6. Error bars represent the SD from three independent assays.
Glutarate in the fermentation broth was then confirmed by quadrupole time-of-flight liquid chromatography/mass spectrometry (LCMS-Q-TOF) and gas chromatography-mass spectrometry (GC-MS). Glutarate, bis(trimethylsilyl) ester (derivatized glutarate) had a precursor ion/daughter ion of 129 → 75 (m/z) for quantification in the GC-MS (see Fig. S1 in the supplemental material). The ion peak [M-H]− at m/z 131 was responsible for the glutarate ion, in accordance with the standard glutarate in the LCMS-Q-TOF (see Fig. S2 in the supplemental material).
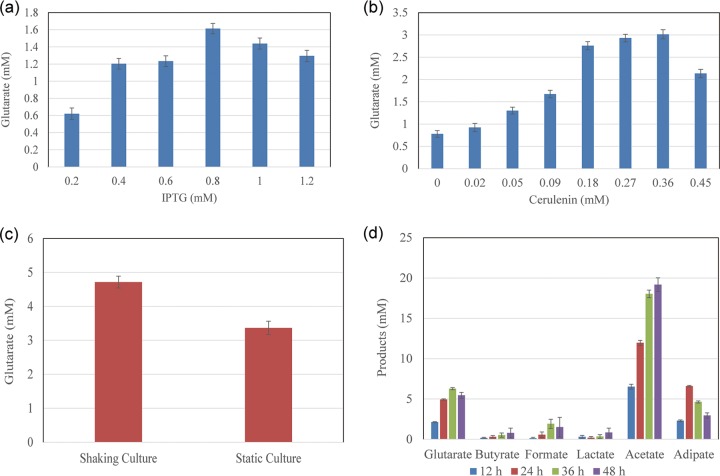
Although a feasible route to produce glutarate was demonstrated, the titer of glutarate was still very low, resulting from the low conversion rate from glucose to glutarate. To determine the optimal medium for glutarate production, seven different media, including M9, lysogeny broth (LB), superoptimal broth (SOB), superoptimal broth with catabolite repression (SOC), MOPS, R medium, and terrific broth (TB), were studied. Among these, the production of glutarate in SOB was up to 1.1 ± 0.1 mM, and that of adipate was about 1.6 ± 0.0 mM (Fig. 2b). The oxygen level has a vital effect on the production of adipate and glutarate (17). Subsequently, microaerobic conditions were explored, and SOB was still found to be the most suitable medium for the glutarate production (Fig. 2c). Meanwhile, 1.5 ± 0.1 mM glutarate and 0.9 ± 0.0 mM adipate were produced under microaerobic conditions. So, we finally decided to use SOB as the fermentation medium and to use microaerobic conditions for cultivation.
In order to further improve the titer, we explored the effect of the concentrations of the inducer IPTG (isopropyl-β-d-thiogalactopyranoside) on glutarate production for E. coli Bgl146. We evaluated six levels of IPTG induction and found that 0.8 mM IPTG was the most suitable one, with 1.6 ± 0.1 mM glutarate (Fig. 3a).
FIG 3.
Enhanced production of glutarate in E. coli. The strain Bgl146 was cultured on 22.2 mM glucose with following optimizations: the optimal concentration of IPTG in shaken flasks (a); the optimal concentration of cerulenin with 0.8 mM IPTG in shaken flasks (b); the fermentation in static and shaking culture (c). (d) Fermentation results for engineered strain E. coli Bgl4146 in shaken flasks on 22.2 mM glucose. E. coli Bgl4146: BL21(DE3) ΔarcA ΔldhA ΔatoB ΔpflB harboring pAD1, pAD4, and pAD6. Error bars represent the SD from three independent assays.
Great progress has been made to increase the concentration of malonyl-CoA by amplifying or deleting specific pathways (20, 21). However, intracellular malonyl-CoA is closely related to phosphoric acid and fatty acid syntheses, which are important for cell growth (22). In traditional gene deletion methods, the target products are synthesized using malonyl-CoA in the cell, resulting in slow cell growth or even cell death (23). Therefore, it is urgent to establish a micromodulation system in the cell to reach a balance between growth and metabolite production with the amount of malonyl-CoA. To increase the amount of malonyl-CoA available for glutarate production in this study, we tried a more effective method using cerulenin as an antibiotic to inhibit fatty acid synthesis pathway for accumulating of malonyl-CoA, which was the most effective in inhibiting the activities of FabB (3-oxoacyl-acyl carrier protein synthase I) and FabF (3-oxoacyl-acyl carrier protein synthase II) (24, 25). Then, we tested eight cerulenin concentrations (Fig. 3b), and 0.18 mM cerulenin was found to be the best concentration with 2.8 ± 0.1 mM glutarate production considering the high price of cerulenin (Fig. 3b).
Next, to explore the effect of a static culture (low dissolved oxygen) and a shaken-flask culture (higher dissolved oxygen) on the production of glutarate, strain Bgl146 was grown on 22.2 mM glucose in a static culture and a shaken flask with 0.18 mM cerulenin (26, 27) and 0.8 mM IPTG (28–30). The results showed that the titer of glutarate reached 4.7 ± 0.2 mM in the shaken flask, which was obviously higher than that of the static culture (Fig. 3c).
Strain engineering.
Through the optimization, we found that the production of glutarate was closely related to the oxygen levels and that the microaerobic condition was more beneficial for producing glutarate than the aerobic one (Fig. 2b and c). In the meantime, a shaken-flask culture was better than a static culture for the production of glutarate (Fig. 3c). Thus, the aerobic respiration control protein ArcA (31) drew our attention. The arcA gene was deleted to test its effect on the synthesis of glutarate. The engineered strains E. coli BL21, Bgl1, Bgl146, and Bgl1146 were cultured on 22.2 mM glucose under aerobic and microaerobic conditions. We found that in the microaerobic condition, BL21 and Bgl146 grew faster than the strains without the arcA gene (Bgl1 and Bgl1146) (see Fig. S3a in the supplemental material). However, this phenomenon did not occur under the aerobic condition (see Fig. S3b in the supplemental material). Moreover, the glutarate titers of Bgl1146 were slightly higher than those of Bgl146 under aerobic and microaerobic conditions (see Fig. S3c in the supplemental material). To increase the availability of acetyl-CoA, one of the precursors of glutarate, we deleted l-lactate dehydrogenase (ldhA), acetyl-CoA acetyltransferase (atoB), and formate C-acetyltransferase I (pflB) genes to test the effects of their knockouts on glutarate synthesis. The resulting strain Bgl4146 produced little lactate, butyrate, and formate (Fig. 3d), with the production of glutarate increased to 6.3 ± 0.1 mM from 22.2 mM glucose (Fig. 3d).
Production of glutarate in 5-liter bioreactor.
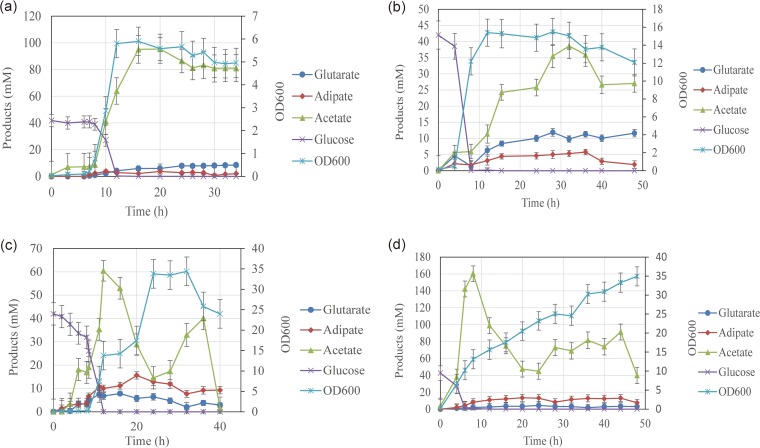
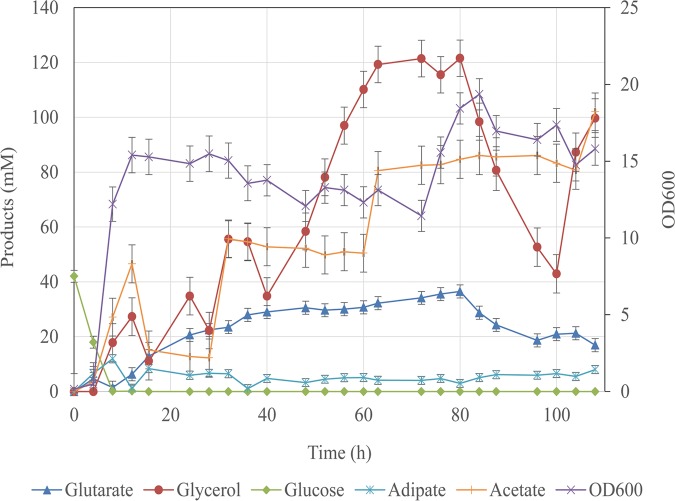
The Bgl4146 strain was cultured with 44.4 mM glucose in a 5-liter bioreactor with 0-, 0.5-, 1-, and 2-air volume/culture volume/min (vvm) aerations and 400-rpm agitation on SOB medium (Fig. 4). There was very little glutarate accumulation under a highly aerobic condition. Once the aeration was decreased to 0.5 vvm, the production was increased. However, when we decreased the aeration from 0.5 to 0 vvm, the titer of glutarate decreased slightly, indicating that less than 0.5-vvm aeration was not suitable for the production of glutarate. In Fig. 4, glutarate production at 0.5 vvm reached 11.9 ± 1.2 mM, and the titer of adipate reached 5.0 ± 1.2 mM. Finally, we chose 0.5-vvm aeration for fermentation. To achieve high titers of glutarate, fed-batch fermentation was used with glycerol as the major substrate with 0.5-vvm aeration and 400-rpm agitation. We observed a maximal glutarate titer of 36.5 ± 0.3 mM after an 80-h fermentation, with a maximal optical density at 600 nm (OD600) of 19.4 achieved after 84 h, representing a 3,650-fold increase compared to that of strain Bgl146 (Fig. 5). These results constitute the highest glutarate titer reported in E. coli.
FIG 4.
Effects of dissolved oxygen levels on glutarate production by strain Bgl4146. The maximal glutarate titers achieved by controlling dissolved oxygen levels are indicated. Strain Bgl4146 was cultured on 44.4 mM glucose with 0-vvm (a), 0.5-vvm (b), 1-vvm (c), or 2-vvm (d) aeration and 400-rpm agitation with 0.18 mM cerulenin and 0.8 mM IPTG. Error bars represent the SD from three independent assays.
FIG 5.
Fed-batch fermentation with strain Bgl4146. Substrate consumptions, metabolites, and cell growth during fed-batch fermentation are indicated. The highest titer was achieved in 5-liter bioreactor with a 0.5-vvm aeration rate and a 400-rpm agitation rate in SOB medium. Error bars represent the SD from three independent assays.
DISCUSSION
Glutarate is widely applied in producing polyesters and polyamides such as nylon-4,5 and nylon-5,5 in industry. We report here glutarate production by E. coli via a newly constructed biosynthetic pathway. The five-step RADP in T. fusca has been constructed in E. coli BL21(DE3). The glutarate was detected by the resulting strain (Bgl146) in shaken flasks. The results indicated that the engineered E. coli strain accumulated only small amounts of glutarate. The strategies used to increase glutarate production were as follows: (i) optimization of culture mediums, culture modes, inducers and inhibitors; (ii) deletion of arcA to test its effect on the titer of glutarate; (iii) deletion of ldhA to eliminate lactate production; (iv) deletion of atoB to eliminate butyrate production; and (v) deletion of pflB to eliminate formate production. Due to the low efficiency of homologous recombination in E. coli BL21(DE3) aided by the λ-Red system, the CRISPR/Cas9 system was used in this study to enable gene deletion (32). The final strain Bgl4146 produced 36.5 ± 0.3 mM glutarate by fed-batch fermentation. These results constitute the highest glutarate titer reported in E. coli.
Oxygen is essential for the synthesis of glutarate (17). The productions of glutarate and adipate were different under different dissolved oxygen levels. We found that the microaerobic condition was more beneficial to produce glutarate (Fig. 2b and c). The synthesis of glutarate started from malonyl-CoA and acetyl-CoA, while adipate started from succinyl-CoA and acetyl-CoA. In order to reduce the synthesis of adipate, the supply of succinyl-CoA should be decreased. There are mainly two routes for succinyl-CoA production: (i) via TCA and (ii) from oxaloacetate or malate to succinyl-CoA. Theoretically, under aerobic conditions succinyl-CoA should come from TCA, while under anaerobic conditions succinyl-CoA may come from oxaloacetate or malate to succinyl-CoA (33). In this study, all the fermentations were conducted aerobically or microaerobically; thus, the succinyl-CoA should mainly come from the TCA cycle. With the low oxygen level, the TCA cycle was significantly weakened, decreasing succinyl-CoA synthesis, while the synthesis of malonyl-CoA was not affected.
The intracellular malonyl-CoA directly associated with phospholipids and fatty acid synthesis (34, 35), which were important for the growth of cells (22). In the traditional gene knockout method, target product synthesis using intracellular malonyl-CoA yields slow cell growth and even cell death (23). Therefore, it was urgent to establish a micromodulation system in the cell to balance the amount of malonyl-CoA for growth and metabolite accumulations. In order to increase the amount of malonyl-CoA available for glutarate synthesis in this study, we tried a more effective method using cerulenin as an antibiotic to inhibit the fatty acid synthesis pathway for accumulating of malonyl-CoA, which was the most effective at inhibiting the activities of FabB (3-oxoacyl-acyl carrier protein synthase I) and FabF (3-oxoacyl-acyl carrier protein synthase II) (24, 25). Using cerulenin, 0.18 mM was the best concentration for producing 2.8 ± 0.1 mM glutarate (Fig. 3b); however, excessive cerulenin may cause bacteria producing the metabolic burden (36). This may also be the reason why the output could not be greatly improved.
Conclusions.
In this study, we constructed the glutarate synthesis pathway from T. fusca in E. coli BL21(DE3). We optimized the culture medium, culture mode, inducer, inhibitor, and rational deleted genes hindering glutarate production, resulting in a maximal glutarate titer of 36.5 ± 0.3 mM with glycerol as the major substrate. As we have seen, this constitutes the highest titer reported in E. coli (Table 1).
TABLE 1.
Comparison of cell factories for the production of glutarate
| Microorganism | Medium | Cultivation mode | Pathway | Titer (mM) | Additives | Source or reference(s) |
|---|---|---|---|---|---|---|
| Corynebacterium glutamicum | NAa | Fed-batch | AMV | 90.2 | No | 14 |
| Escherichia coli | MR | Fed-batch | AMV | 12.9 | Yes | 12 |
| M9 | Fed/shaken flask | α-Keto acid carbon chain extension and decarboxylation pathway | 3.2 | Yes | 15 | |
| MOPS | Shaken flask | A novel synthetic pathway | 0.1 | Yes | 16, 17 | |
| SOB | Fed-batch | RADP | 36.5 | No | This study |
NA, not available.
MATERIALS AND METHODS
Strains and plasmids.
The strains, plasmids, and primers used in this study are listed in Tables S1 and S2 in the supplemental material. Tfu_0875 was synthesized and ligated into plasmid pUC57-1 by Genewiz (Suzhou, China) and amplified by the primers pUC57-1F and pUC57-1R. The pRSFDuet-1 plasmid and the PCR product were digested, and Tfu_0875 was ligated into pRSFDuet-1 using the T4 DNA ligase. The ligation product was introduced into E. coli JM109 for screening by colony PCR and Sanger sequencing by the primers veri-pRSF-0875F and veri-pRSF-0875R. The final plasmid was named pRSF-Tfu_0875. Tfu_2399 was amplified by the primers pUC57-2F (BglII) and pUC57-2R (KpnI) and ligated into pRSF-Tfu_0875, which was digested with BglII and KpnI to form pAD1. Plasmids pAD6 also was constructed according to the same protocol as described previously. Tfu_0067 and Tfu_1647 were amplified by the primers pUC57-3R and pUC57-3F and the primers pUC57-4R and pUC57-4F derived from pUC57-3 and pUC57-4, respectively. pTrc99A was linearized by NcoI and HindIII restriction digestion, and primers pAD4-0067F and pAD4-1647R contained 39-nucleotide sequences homologous to the upstream and downstream regions of pTrc99A at the NcoI and HindIII sites, with the two genes linked by a ribosome binding site (RBS) (AAGAGGTATATATTA). Plasmid pAD4 was constructed using Gibson assembly methods (37) and transformed into E. coli JM109 competent cells. The remaining plasmids (parcA, pldhA, patoB, psucD, and ppflB) developed in this study were constructed according to the protocol described above and are described in Table S1 in the supplemental material. The synthesized genes are shown in Table S3 in the supplemental material.
Genetic work.
Gene deletions were achieved with CRISPR/Cas9 method (32). The strain harboring pCas was grown at 30°C to an OD600 ranging from 0.2 to 0.4 and subsequently induced by 10 mM arabinose at 30°C for 60 min. The parcA plasmid was used as the template to obtain the arcA gene upstream and downstream 500 bp from E. coli. The templates and pTargetF were introduced into E. coli by electroporation. The pTargetF harboring sgRNA (constitutive expression) targeted the gene to be deleted. After the deletion, IPTG was used to induce the expression of sgRNA, which cut the targeting pTargetF using Cas9, and the pCas was eliminated at 42°C and 200 rpm for 12 h. By the same method, the genes arcA, ldhA, atoB, and pflB were deleted in E. coli BL21(DE3).
Glutarate fermentation.
Recombinant strains Bgl146 with desirable plasmids (pAD-1, pAD-4, and pAD-6) were inoculated into LB medium and cultivated at 37°C. The cells were then grown at 30°C with an initial OD600 of 0.8 and a shaking speed of 200 rpm. The different selective media were M9, LB, SOB, SOC, MOPS, TB, and R medium (the medium recipes are provided in the supplemental material). After inoculation, the flasks were immediately transferred to the microaerobic environment and controlled by the fermentation bung (38) or aerobic environment. All genes from organisms other than E. coli were codon optimized for E. coli by Genewiz (Suzhou, China). The production of glutarate by E. coli Bgl4146 in a 5-liter bioreactor (Baoxing, Shanghai, China) was conducted at 37°C for culturing cells and at 30°C for inducing gene expression with 44.4 mM glucose, using 0-, 0.5-, 1-, and 2-vvm aeration and 400-rpm agitation with 0.18 mM cerulenin and 0.8 mM IPTG. For the fed-batch fermentation, a 5-liter bioreactor was used for the final strain (Bgl4146), and the initial culturing temperature was 37°C. During the first stage, 2% of the working volume seed culture was inoculated to the bioreactor, and fermentation was initiated with 44.44 mM glucose. Then, 0.18 mM cerulenin and 0.8 mM IPTG was added once the OD600 reached 0.8; after this, the culturing temperature was decreased to 30°C. During the second stage, the desired amount of glycerol was added to the bioreactor. The pH was kept at 7.0 during the fermentation process.
Metabolite analysis.
The concentrations of glucose, glycerol, and glutarate were measured with an HPLC system (Rigol, Suzhou, China) using an HPX-87H ion-exclusion column (Bio-Rad, Hercules, CA), a refractive index, and an UV detector. The mobile phase was 30 mM H2SO4 at 0.3 ml/min at 42°C, and glutarate was detected at 210 nm (39, 40). The analytical methods to determine the concentrations of glutarate in the fermentation broth by GC-MS were essentially the same for adipate quantification, as described earlier (39, 41). To confirm the biosynthesis of glutarate, the organic acids were also analyzed by a Waters MALDI SYNAPT Q-TOF MS equipped with a Waters Acquity UPLC CSH C18 column (2.1 by 100 mm) and a Waters Acquity PDA detector (200 to 400 nm) (42). The mobile phase consisted of 0.1% formic acid (A) and 100% acetonitrile (B) with a 0.3-ml/min flow velocity at 45°C with the gradient elution program (see Table S4 in the supplemental material).
Data analysis.
All experimental data were measured in triplicate, and error bars represent the standard deviations (SD).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Natural Science Foundation of Jiangsu Province (BK20150136), the National Natural Science Foundation of China (31500070), the Fundamental Research Funds for the Central Universities (JUSRP51705A), the Open Foundation of Jiangsu Key Laboratory for Biomass-Based Energy and Enzyme Technology (BEETKA1801), and the Distinguished Professor Project of Jiangsu Province.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00814-18.
REFERENCES
- 1.Adkins J, Jordan J, Nielsen DR. 2013. Engineering Escherichia coli for renewable production of the 5-carbon polyamide building-blocks 5-aminovalerate and glutarate. Biotechnol Bioeng 110:1726–1734. doi: 10.1002/bit.24828. [DOI] [PubMed] [Google Scholar]
- 2.Bermúdez M, León S, Alemán C, Muñoz-Guerra S. 2000. Comparison of lamellar crystal structure and morphology of nylon 46 and nylon 5. Polymer (Guildf) 41:8961–8973. doi: 10.1016/S0032-3861(00)00239-1. [DOI] [Google Scholar]
- 3.Polen T, Spelberg M, Bott M. 2013. Toward biotechnological production of adipic acid and precursors from biorenewables. J Biotechnol 167:75–84. doi: 10.1016/j.jbiotec.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Sato K, Aoki M, Noyori R. 1998. A “green” route to adipic acid: direct oxidation of cyclohexenes with 30 percent hydrogen peroxide. Science 281:1646–1647. doi: 10.1126/science.281.5383.1646. [DOI] [PubMed] [Google Scholar]
- 5.Niu W, Draths KM, Frost JW. 2002. Benzene-free synthesis of adipic acid. Biotechnol Prog 18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- 6.Nishikido J, Tamura N, Fukuoka Y. March 1979. Method for obtaining glutaric acid, succinic acid, and adipic acid from an acid mixture comprising them. US patent US4146730. [Google Scholar]
- 7.US Environmental Protection Agency. 2011. Inventory of U.S. greenhouse gas emissions and sinks: 1990-2009. Fed Regist 33:273–279. https://www.mendeley.com/research-papers/inventory-greenhouse-gas-emissions-sinks-19902009/. [Google Scholar]
- 8.Tsuge Y, Kawaguchi H, Sasaki K, Kondo A. 2016. Engineering cell factories for producing building block chemicals for bio-polymer synthesis. Microb Cell Fact 15:19. doi: 10.1186/s12934-016-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL. 2005. Multiple and interconnected pathways for l-lysine catabolism in Pseudomonas putida KT2440. J Bacteriol 187:7500–7510. doi: 10.1128/JB.187.21.7500-7510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fothergill JC, Guest JR. 1977. Catabolism of l-lysine by Pseudomonas aeruginosa. J Gen Microbiol 99:139–155. doi: 10.1099/00221287-99-1-139. [DOI] [PubMed] [Google Scholar]
- 11.Revelles O, Wittich RM, Ramos JL. 2007. Identification of the initial steps in d-lysine catabolism in Pseudomonas putida. J Bacteriol 189:2787–2792. doi: 10.1128/JB.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park SJ, Kim EY, Noh W, Park HM, Oh YH, Lee SH, Song BK, Jegal J, Lee SY. 2013. Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. Metab Eng 16:42–47. doi: 10.1016/j.ymben.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Rohles CM, Giesselmann G, Kohlstedt M, Wittmann C, Becker J. 2016. Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb Cell Fact 15:154. doi: 10.1186/s12934-016-0553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin JH, Park SH, Oh YH, Choi JW, Lee MH, Cho JS, Jeong KJ, Joo JC, Yu J, Si JP. 2016. Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid. Microb Cell Fact 15:174. doi: 10.1186/s12934-016-0566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Wu Y, Sun X, Yuan Q, Yan Y. 2017. De novo biosynthesis of glutarate via α-keto acid carbon chain extension and decarboxylation pathway in Escherichia coli. ACS Synth Biol 6:10. doi: 10.1021/acssynbio.6b00323. [DOI] [PubMed] [Google Scholar]
- 16.Yu JL, Xia XX, Zhong JJ, Qian ZG. 2017. A novel synthetic pathway for glutarate production in recombinant Escherichia coli. Process Biochem 59:167–171. doi: 10.1016/j.procbio.2017.06.026. [DOI] [Google Scholar]
- 17.Yu JL, Xia XX, Zhong JJ, Qian ZG. 2017. Enhanced production of glutarate by using anaerobic-aerobic shift cultivation and an anaerobically inducible promoter in an engineered Escherichia coli. Process Biochem 62:53–58. doi: 10.1016/j.procbio.2017.09.001. [DOI] [Google Scholar]
- 18.Parthasarathy A, Pierik AJ, Kahnt J, Zelder O, Buckel W. 2011. Substrate specificity of 2-hydroxyglutaryl-CoA dehydratase from Clostridium symbiosum: toward a bio-based production of adipic acid. Biochemistry 50:3540–3550. doi: 10.1021/bi1020056. [DOI] [PubMed] [Google Scholar]
- 19.Deng Y, Mao Y. 2015. Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J Appl Microbiol 119:1057–1063. doi: 10.1111/jam.12905. [DOI] [PubMed] [Google Scholar]
- 20.Fowler ZL, Gikandi WW, Koffas MA. 2009. Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production. Appl Environ Microbiol 75:5831–5839. doi: 10.1128/AEM.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA. 2011. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng 13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Rathnasingh C, Raj SM, Lee Y, Catherine C, Ashok S, Park S. 2012. Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J Biotechnol 157:633–640. doi: 10.1016/j.jbiotec.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Wu J, Yu O, Du G, Zhou J, Chen J. 2014. Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from l-tyrosine in Escherichia coli. Appl Environ Microbiol 80:7283–7292. doi: 10.1128/AEM.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers JK, Church GM. 2016. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci U S A 113:2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heath RJ, Rock CO. 1995. Regulation of malonyl-CoA metabolism by acyl-acyl carrier protein and beta-ketoacyl-acyl carrier protein synthases in Escherichia coli. J Biol Chem 270:15531–15538. doi: 10.1074/jbc.270.26.15531. [DOI] [PubMed] [Google Scholar]
- 26.Omura S. 1980. Cerulenin. Methods Enzymol 72:520–532. doi: 10.1016/S0076-6879(81)72041-X. [DOI] [PubMed] [Google Scholar]
- 27.Omura S. 1976. The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol Rev 40:681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marbach A, Bettenbrock K. 2012. lac operon induction in Escherichia coli: systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J Biotechnol 157:82–88. doi: 10.1016/j.jbiotec.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Hansen LH, Knudsen S, Sørensen SJ. 1998. The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens. Curr Microbiol 36:341–347. doi: 10.1007/s002849900320. [DOI] [PubMed] [Google Scholar]
- 30.Kosinski MJ, Rinas U, Bailey JE. 1992. Isopropyl-β-d-thiogalactopyranoside influences the metabolism of Escherichia coli. Appl Microbiol Biotechnol 36:782–784. doi: 10.1007/BF00172194. [DOI] [Google Scholar]
- 31.Park DM, Sohail AM, Ansari AZ, Robert L, Kiley PJ. 2013. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet 9:e1003839. doi: 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. 2015. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q, Huang B, Wu H, Li Z, Ye Q. 2017. Efficient anaerobic production of succinate from glycerol in engineered Escherichia coli by using dual carbon sources and limiting oxygen supply in preceding aerobic culture. Bioresour Technol 231:75–84. doi: 10.1016/j.biortech.2017.01.051. [DOI] [PubMed] [Google Scholar]
- 34.Magnuson K, Jackowski S, Rock CO, Cronan JE Jr. 1993. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev 57:522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu J, Yu O, Du G, Zhou J, Chen J. 2014. Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from l-tyrosine in Escherichia coli. Appl Environ Microbiol 23:7283–7292. doi: 10.1128/AEM.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zha W, Rubin-Pitel SB, Shao Z, Zhao H. 2009. Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab Eng 11:192–198. doi: 10.1016/j.ymben.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 38.Nastasia M. February 2001. Fermentation bung device and method. US patent US6183982. [Google Scholar]
- 39.Cheong S, Clomburg JM, Gonzalez R. 2016. Energy- and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative claisen condensation reactions. Nat Biotechnol 34:556–561. doi: 10.1038/nbt.3505. [DOI] [PubMed] [Google Scholar]
- 40.Babu T, Yun EJ, Kim S, Kim DH, Liu KH, Kim SR, Kim KH. 2015. Engineering Escherichia coli for the production of adipic acid through the reversed β-oxidation pathway. Process Biochem 50:2066–2071. doi: 10.1016/j.procbio.2015.09.018. [DOI] [Google Scholar]
- 41.Yu JL, Xia XX, Zhong JJ, Qian ZG. 2014. Direct biosynthesis of adipic acid from a synthetic pathway in recombinant Escherichia coli. Biotechnol Bioeng 111:2580–2586. doi: 10.1002/bit.25293. [DOI] [PubMed] [Google Scholar]
- 42.Li LL, Chang M, Tao GJ, Wang XS, Liu Y, Liu RJ, Jin QZ, Wang XG. 2016. Analysis of phospholipids in Schizochytrium sp. S31 by using UPLC-Q-TOF-MS. Anal Methods 8:763–770. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.