We demonstrated that Variovorax boronicumulans CGMCC 4969 has two enzymatic systems—nitrilase and nitrile hydratase/amidase—that convert indole-3-acetonitrile (IAN) to the important plant hormone indole-3-acetic acid (IAA). The two IAA synthesis systems have very different regulatory mechanisms, affecting the IAA synthesis rate and duration. The nitrilase was induced by IAN, which was rapidly converted to IAA; subsequently, IAA was rapidly consumed for cell growth. The nitrile hydratase (NHase) and amidase system was constitutively expressed and slowly but continuously synthesized IAA. In addition to synthesizing IAA from IAN, CGMCC 4969 has a rapid IAA degradation system, which would be helpful for a host plant to eliminate redundant IAA. This study indicates that the plant growth-promoting rhizobacterium V. boronicumulans CGMCC 4969 has the potential to be used by host plants to regulate the IAA level.
KEYWORDS: indole-3-acetonitrile, biotransformation, indole-3-acetic acid, Variovorax boronicumulans
ABSTRACT
Variovorax is a metabolically diverse genus of plant growth-promoting rhizobacteria (PGPR) that engages in mutually beneficial interactions between plants and microbes. Unlike most PGPR, Variovorax cannot synthesize the phytohormone indole-3-acetic acid (IAA) via tryptophan. However, we found that Variovorax boronicumulans strain CGMCC 4969 can produce IAA using indole-3-acetonitrile (IAN) as the precursor. Thus, in the present study, the IAA synthesis mechanism of V. boronicumulans CGMCC 4969 was investigated. V. boronicumulans CGMCC 4969 metabolized IAN to IAA through both a nitrilase-dependent pathway and a nitrile hydratase (NHase) and amidase-dependent pathway. Cobalt enhanced the metabolic flux via the NHase/amidase, by which IAN was rapidly converted to indole-3-acetamide (IAM) and in turn to IAA. IAN stimulated metabolic flux via the nitrilase, by which IAN was rapidly converted to IAA. Subsequently, the IAA was degraded. V. boronicumulans CGMCC 4969 can use IAN as the sole carbon and nitrogen source for growth. Genome sequencing confirmed the IAA synthesis pathways. Gene cloning and overexpression in Escherichia coli indicated that NitA has nitrilase activity and IamA has amidase activity to respectively transform IAN and IAM to IAA. Interestingly, NitA showed a close genetic relationship with the nitrilase of the phytopathogen Pseudomonas syringae. Quantitative PCR analysis indicated that the NHase/amidase system is constitutively expressed, whereas the nitrilase is inducible. The present study helps our understanding of the versatile functions of Variovorax nitrile-converting enzymes that mediate IAA synthesis and the interactions between plants and these bacteria.
IMPORTANCE We demonstrated that Variovorax boronicumulans CGMCC 4969 has two enzymatic systems—nitrilase and nitrile hydratase/amidase—that convert indole-3-acetonitrile (IAN) to the important plant hormone indole-3-acetic acid (IAA). The two IAA synthesis systems have very different regulatory mechanisms, affecting the IAA synthesis rate and duration. The nitrilase was induced by IAN, which was rapidly converted to IAA; subsequently, IAA was rapidly consumed for cell growth. The nitrile hydratase (NHase) and amidase system was constitutively expressed and slowly but continuously synthesized IAA. In addition to synthesizing IAA from IAN, CGMCC 4969 has a rapid IAA degradation system, which would be helpful for a host plant to eliminate redundant IAA. This study indicates that the plant growth-promoting rhizobacterium V. boronicumulans CGMCC 4969 has the potential to be used by host plants to regulate the IAA level.
INTRODUCTION
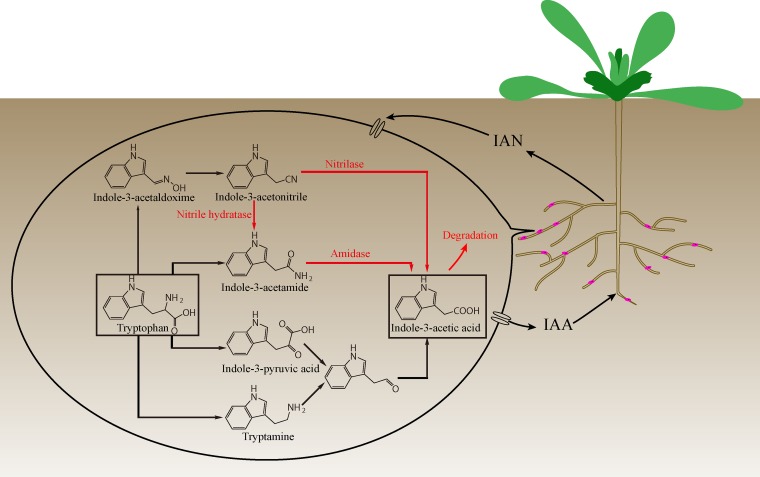
Plant growth-promoting rhizobacteria (PGPR) inhabit the rhizosphere and have a direct or indirect influence on the host plant. In the former case, plant growth is promoted either by providing nutrients or by producing phytohormones; in the latter case, plants are protected from acquiring infections (biotic stress) or aided to maintain healthy growth under environmental (abiotic) stress (1–3). The phytohormones produced by PGPR play an important role in regulating plant metabolic balance. Indole-3-acetic acid (IAA) is the most abundant and basic auxin natively occurring and functioning in plants, and it controls nearly every aspect of plant growth and development, such as cell division, elongation, fruit development, and senescence (4, 5). Bacterially produced IAA affects plant physiology and pathology, and it functions as a signaling molecule that participates in plant-microbe interactions (6). IAA production occurs in many plant-related bacteria. In the rhizosphere, >80% of bacteria are capable of synthesizing IAA (7). IAA synthesis may be tryptophan dependent or tryptophan independent. In the infrequently observed tryptophan-independent pathway, indole-3-glycerolphosphate or indole is regarded as the main precursor. However, no enzyme of this pathway has been characterized (8). Bacterial IAA synthesis mainly uses tryptophan as the precursor via four different pathways, classified according to their intermediates: indole-3-acetamide (IAM), indole-3-pyruvate, indole-3-acetonitrile (IAN), and tryptamine (9) (Fig. 1). The IAM and indole-3-pyruvate pathways are the two most common routes for IAA biosynthesis in bacteria, while the IAN pathway has been extensively studied in plants. Apart from synthesizing IAA, some PGPR, such as Pseudomonas, Arthrobacter, and Bradyrhizobium, can degrade IAA and use it as a carbon and nitrogen source (10, 11). Thus, IAA synthesis and degradation in PGPR play important roles in the regulation of IAA levels in host plants.
FIG 1.
Model of Variovorax boronicumulans CGMCC 4969-plant interactions based on analysis of whole-genome metabolic pathways and metabolic intermediates. The four common tryptophan-dependent indole-3-acetic acid (IAA) synthesis pathways are shown, and red indicates the pathways in CGMCC 4969.
The genus Variovorax belongs to the family Comamonadaceae and consists of metabolically diverse aerobic bacteria that possess extraordinary degradation abilities and can degrade a series of organic pollutants (12). They are also common plant symbionts found in the rhizosphere and used as model bacteria for the study of microbe-plant interactions (13, 14). We previously isolated Variovorax boronicumulans strain CGMCC 4969, which can degrade the neonicotinoid insecticides thiacloprid and acetamiprid to their corresponding amide metabolites (15, 16). V. boronicumulans CGMCC 4969 showed some plant growth-promoting traits, such as producing exopolymer substances, siderophores, ammonia, and hydrogen cyanide and secreting salicylate and 2, 3-dihydroxy benzoic acid (17). Those features can enhance the stress tolerance and disease resistance of the host plant and aid in nutrient availability and uptake. However, unlike most PGPR, V. boronicumulans CGMCC 4969 cannot produce IAA using tryptophan as a precursor. The same phenomenon was also observed in V. paradoxus strains S110, EPS, and B4. However, a few putative genes involved in the IAA transport system were found in the genomes of these Variovorax strains. Interestingly, we found that V. boronicumulans CGMCC 4969 produced IAA when IAN was used as the substrate. Thus, the IAA metabolic pathway in V. boronicumulans CGMCC 4969 and its synthesis mechanism from IAN were further explored.
Bacteria containing nitrile-converting enzymes are widely used in the production of high-value amides and carboxylic acid compounds, as well as for the bioremediation of nitrile-contaminated soil and water. However, a role for nitrile-converting enzymes in IAA synthesis has rarely been reported. IAA synthesis has mainly been reported in plant-associated bacteria, including phytopathogenic bacteria and PGPR. High levels of IAA produced by phytopathogenic bacteria such as Agrobacterium tumefaciens, Pseudomonas savastanoi, and Pseudomonas syringae pv. syringae cause necrotic lesions and gall tumor formation on host plants. The main IAA synthesis route in these bacteria is the IAM pathway (Fig. 1), in which tryptophan is converted to IAM by a tryptophan 2-monooxygenase (encoded by the tms-1 gene), and then IAM is converted to IAA by an IAM-specific hydrolase/amidase (encoded by tms-2) (9). The Agrobacterium tms-1 and tms-2 genes are not functional inside the bacterium but are transferred into plant cells where they are inserted into the plant chromosome and exert their pathogenic effect. The P. savastanoi tms-1 and tms-2 genes are only functional inside bacterium-induced plant tumors and depend on the continuous production of IAA by the infecting bacteria (18). Some PGPR, such as Rhizobium spp., Bradyrhizobium spp., and Pseudomonas fluorescens, cannot use this pathway because they lack tryptophan 2-monooxygenase (19, 20). However, they can synthesize IAA using nitrile hydrolase (NHase)/indoleacetamide hydrolase or nitrilase when IAN is provided as a substrate by the IAN pathway. In addition, the expression modes of the nitrile-converting enzymes are dissimilar, either inducible or constitutive. For example, Fusarium solani O1 nitrilase was induced in the presence of 2-cyanopyridine (21), but Klebsiella ozaenae nitrilase was constitutively expressed during the degradation of the herbicide bromoxynil (22). Many microorganisms harboring nitrilase or NHase and amidase have been isolated, but few have both enzymatic systems, especially acting on the same substrate (23). The expression modes of the nitrile-converting enzymes influencing IAA metabolic influx have rarely been reported. In the present study, the nitrile-converting enzymes involved in the IAA synthesis mechanism of the plant growth-promoting rhizobacterium V. boronicumulans CGMCC 4969 were characterized and their expression levels studied. This study helps to explain the versatile functions of nitrile-converting enzymes in IAA metabolism and the interactions between microbes and plants.
RESULTS AND DISCUSSION
Biotransformation of IAN by resting cells of V. boronicumulans CGMCC 4969 and metabolite identification.
A high-performance liquid chromatography (HPLC) analysis showed that resting cells of V. boronicumulans CGMCC 4969 transformed IAN into two polar metabolites, P1 and P2, with retention times of 4.18 and 5.42 min, respectively (Fig. 2A). Meanwhile, in controls without bacteria (Fig. 2B) or the IAN substrate (Fig. 2C), the corresponding peaks were not observed. Metabolites P1 and P2 showed the same retention times as the standards IAM and IAA, respectively. A liquid chromatography-mass spectroscopy (LC-MS) analysis indicated that the fragmentation patterns of P1 (Fig. 2D) and P2 (Fig. 2E) were the same as those observed for the standard compounds. Metabolite identification thus proved that V. boronicumulans CGMCC 4969 can synthesize IAM and IAA using IAN as the precursor.
FIG 2.
High-performance liquid chromatographs of the products of transformation of indole-3-acetonitrile (IAN) by resting cells of V. boronicumulans CGMCC 4969 and liquid chromatography-mass spectra of the metabolites. (A) Phosphate buffer containing 400 mg · liter−1 IAN and CGMCC 4969. (B) Phosphate buffer containing 400 mg · liter−1 IAN alone. (C) Phosphate buffer containing CGMCC 4969 alone. (D) Mass data for metabolite P1. (E) Mass data for metabolite P2.
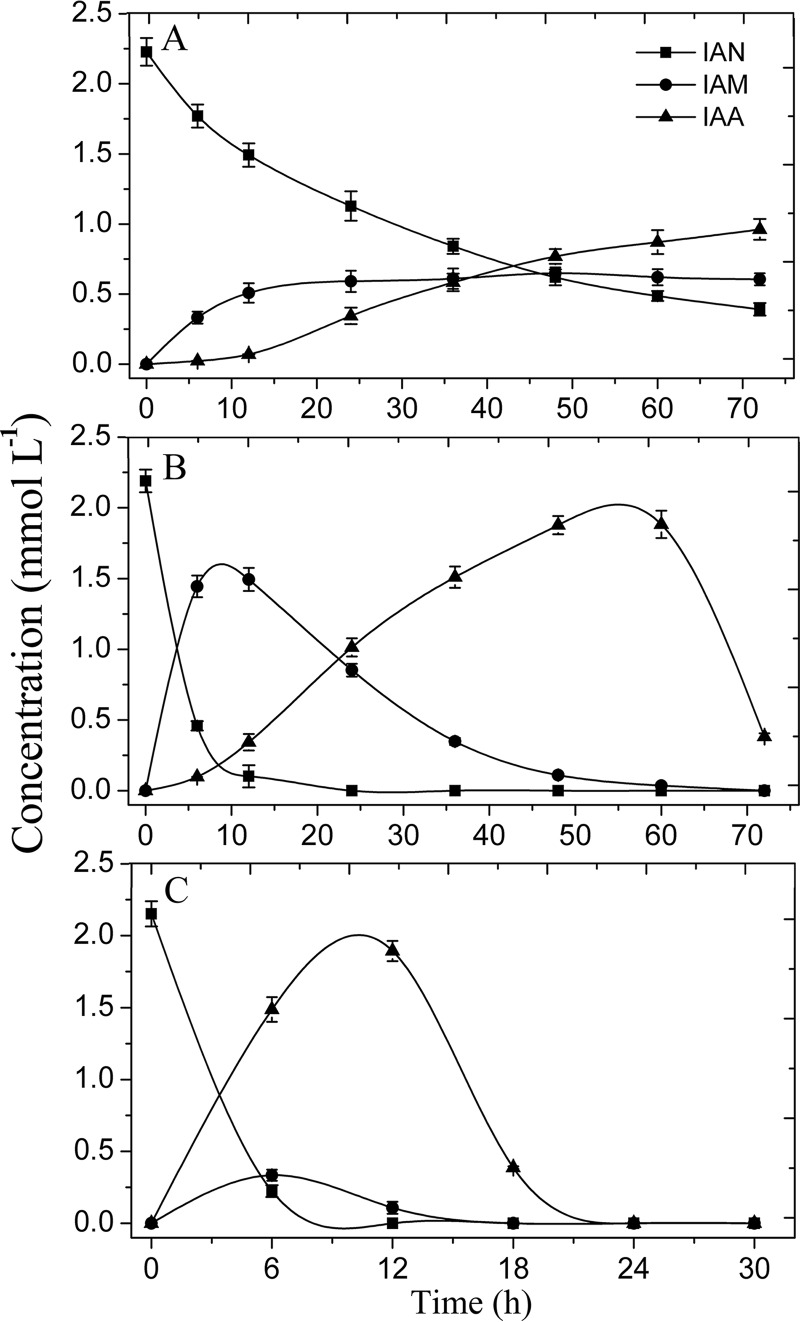
Resting cells of CGMCC 4969 transformed IAN to IAM, with a maximum content of 0.65 mmol · liter−1 at 48 h, while the IAA content gradually increased and reached 0.96 mmol · liter−1 at 72 h (Fig. 3A). The molar transformation rate (the total amounts of IAM and IAA divided by the consumed amount of IAN) at 72 h was 86.6%, which indicated that the synthesis of IAM and IAA was the main pathway of IAN metabolism in strain CGMCC 4969. CGMCC 4969 NHase is a cobalt-dependent metalloenzyme, and adding cobalt ions apparently enhanced its activity (15, 16). As Fig. 3B shows, when a final concentration of 0.1 mmol · liter−1 CoCl2 was added to the lysogeny broth (LB) used to preculture CGMCC 4969 cells, 1.44 mmol · liter−1 IAM was formed by the resting CGMCC 4969 cells in the initial 6 h (4.3-fold higher than in the control), while the IAA content was 0.1 mmol · liter−1 (Fig. 3B). IAM peaked at 12 h with a content of 1.49 mmol · liter−1, and the IAA content gradually increased to the maximum of 1.88 mmol · liter−1 by 60 h. Thus, the enhancement of the NHase/amidase pathway flux by cobalt supplementation accelerates the IAA synthesis rate from IAN by CGMCC 4969.
FIG 3.
Biotransformation of indole-3-acetonitrile (IAN) by resting cells of V. boronicumulans CGMCC 4969. (A) Resting cell transformation of IAN without adding cobalt ions and IAN to the culture medium. (B) Resting cell transformation of IAN after adding 0.1 mmol · liter−1 cobalt(II) ions to the culture medium. (C) Resting cell transformation of IAN after adding 100 mg · liter−1 IAN to the culture medium.
When IAN was added to the LB used for cell culture, the transformation of IAN by CGMCC 4969 resting cells was significantly different from that in the control that lacked IAN supplementation. The initial IAM formation after 6 h was 0.33 mmol · liter−1 (Fig. 3C), which was the same as the control value of 0.34 mmol · liter−1 (Fig. 3A), indicating that IAN addition into the preculture did not affect the NHase activity. However, IAA formation rapidly increased, to 1.49 mmol · liter−1 at 6 h, 64.6-fold higher than in the control (0.02 mmol · liter−1) (Fig. 3A). Thus, the additional IAN activated the nitrilase pathway, resulting in the increase in IAA synthesis by CGMCC 4969 resting cells.
Therefore, the biotransformation of IAN by resting cells revealed that there are two IAN metabolic pathways in V. boronicumulans CGMCC 4969 that act via (i) nitrilase and (ii) NHase and amidase. Cobalt can regulate the IAN metabolic flux via NHase/amidase, and IAN stimulated the metabolic flux via the nitrilase. Although both NHase/amidase and nitrilase pathways convert IAN to IAA, the rates of IAA synthesis and the accumulation of IAA are quite different. The nitrilase has a higher IAA synthesis rate than the NHase/amidase system and can quickly respond to synthesize large amounts of the phytohormone IAA. In the activated nitrilase pathway, when IAN and IAM were completely transformed, the IAA produced was rapidly degraded (from 12 to 24 h), while in the activated NHase/amidase pathway, the IAA content increased during this period.
Biotransformation of IAN by growing culture of V. boronicumulans CGMCC 4969.
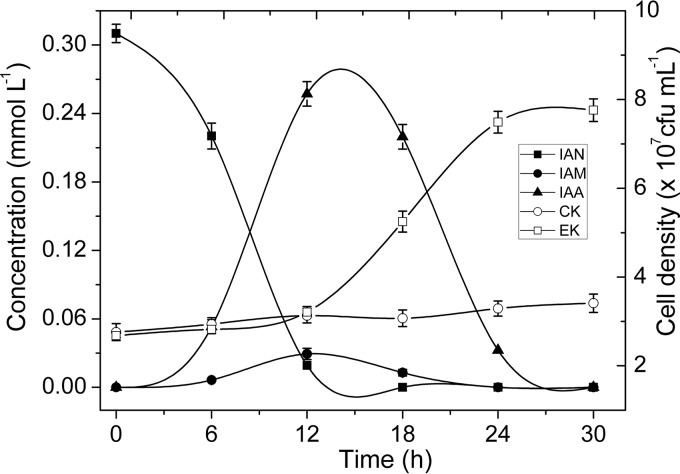
The metabolism of IAN in a growing culture of V. boronicumulans CGMCC 4969 was investigated by inoculating the strain into mineral salt medium (MSM) broth supplemented with 50 mg · liter−1 IAN. IAN was converted to IAM and IAA in the initial 12 h, and the cell numbers of CGMCC 4969 remained almost unchanged (Fig. 4). Subsequently, the IAA content decreased, and the cell numbers increased from 3.21 × 107 CFU · ml−1 at 12 h to 7.50 × 107 CFU · ml−1 at 24 h. After the IAA and IAM were consumed, the cell number did not increase further. Thus, IAN can be used as the sole carbon and nitrogen source for cell growth by CGMCC 4969. To further test the ability of CGMCC 4969 to degrade IAA, 50 mg · liter−1 IAA was added to MSM for transformation by growing cells. CGMCC 4969 completely degraded the IAA in 12 h, and no metabolites were detected by an HPLC analysis, confirming that CGMCC 4969 is capable of degrading IAA.
FIG 4.
Biotransformation of indole-3-acetonitrile (IAN) by growing cells of V. boronicumulans CGMCC 4969. CK (control), CGMCC 4969 cells inoculated in MSM; EK, CGMCC 4969 cells inoculated in MSM supplemented with 50 mg liter−1 IAN.
Thus, in addition to synthesizing IAA from IAN, CGMCC 4969 has a rapid IAA degradation system. IAA degradation-related genes have rarely been reported. Pseudomonas putida 1290 is a model microorganism for the study of the bacterial degradation of IAA, and an 8,994-bp genomic DNA fragment involving 10 genes (iacA to iacI) was identified as responsible for IAA mineralization (24, 25). However, this iac gene cluster was not found in the CGMCC 4969 genome. Several other pathways of IAA degradation have been proposed, such as the metabolism of IAA to anthranilic acid by the production of 2-formaminobenzoylacetic acid (26) or dioxindole-3-acetic acid (27) and the decarboxylation of IAA to skatole (which is further converted to catechol) (28). However, none of the relevant IAA metabolic intermediates of these pathways were observed by HPLC analysis in the present study. We hypothesize that CGMCC 4969 has a different IAA degradation pathway.
Genome features and bioinformatic analysis of the IAA metabolic pathway and nitrile-converting enzymes in V. boronicumulans CGMCC 4969.
V. boronicumulans CGMCC 4969 genomic DNA was isolated and sequenced. The genome of CGMCC 4969 consists of 7,137,898 bp in a single chromosome, with an average GC content of 68.54 mol%. The CGMCC 4969 genome contains 6,871 predicted genes, and biological roles were assigned to 6,471 predicted coding sequences on the basis of similarity searches. The IAA biosynthesis pathway of CGMCC 4969 was further investigated using a Kyoto Encyclopedia of Genes and Genomes metabolic pathway analysis. Bacteria synthesize IAA from tryptophan via four main pathways (Fig. 1). No homologous genes of the indole-3-pyruvic acid and tryptamine pathways were found in CGMCC 4969. Furthermore, genes for tryptophan 2-monooxygenase, which converts tryptophan to IAM in the IAM pathway, and the oxidoreductase that may function in converting tryptophan to indole-3-acetaldoxime in the indole acetaldoxime/IAN pathway were not detected in the genome. Those results were in accordance with our HPLC analysis, which showed that tryptophan could not be converted to IAA by CGMCC 4969.
The strain does have one NHase-coding gene cluster, two putative indoleacetamide hydrolase-coding genes, and two putative nitrilase-coding genes. The two putative indoleacetamide hydrolases and two putative nitrilases were named IamA, IamB, NitA and NitB. Their functions in the transformation of IAN to IAA were tested experimentally by overexpression in Escherichia coli.
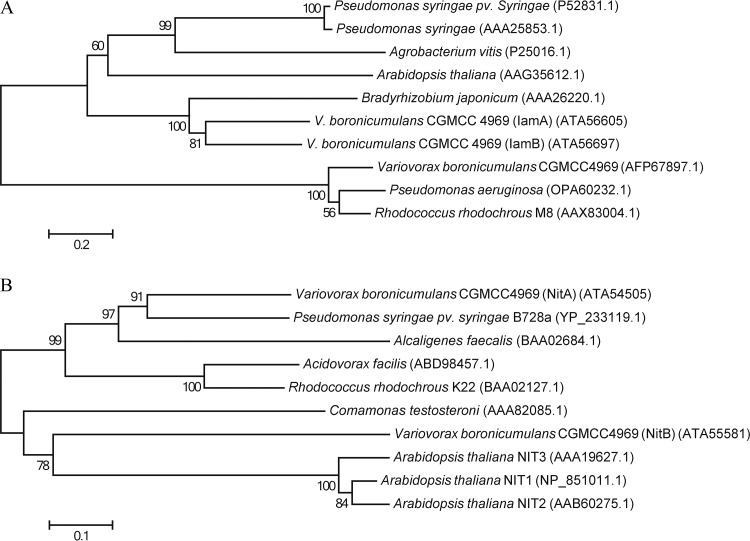
CGMCC 4969 NHase is a cobalt-type NHase, and it functions in the degradation of the neonicotinoid insecticides thiacloprid and acetamiprid (16). Amidases are classified into the amidase signature family and aliphatic amidases on the basis of their amino acid sequences (29). A multiple-sequence alignment analysis of the two putative indoleacetamide hydrolases showed that they contain a highly conserved GGSS motif and belong to the amidase signature family. A BLAST analysis showed that IamA had the highest identity (81%) to putative Cupriavidus necator indoleacetamide hydrolase, and IamB had the highest identity (59%) to putative Burkholderia sp. strain AU4i indoleacetamide hydrolase. The two putative CGMCC 4969 indoleacetamide hydrolases only share a 52.8% amino acid sequence identity with each other. In a phylogenetic analysis based on amino acid sequences, IamA and IamB clustered in a branch with indoleacetamide hydrolases from the PGPR Bradyrhizobium japonicum (30), independent from the phytopathogens Pseudomonas syringae pv. syringae (31) and Agrobacterium vitis (32), and also showed evolutionary divergence from the aliphatic amidases from Rhodococcus rhodochrous strain M8 (33) and P. aeruginosa (34) (Fig. 5A).
FIG 5.
Phylogenetic trees of indoleacetamide hydrolases and nitrilases. The phylogenetic trees were constructed on the basis of protein sequences by the neighbor-joining method, and the bootstrap percentages from 1,000 replicates are shown at the nodes. (A) Indoleacetamide hydrolase phylogenetic tree. (B) Nitrilase phylogenetic tree.
Nitrilases are frequently classified into one of three categories on the basis of substrate specificity: aliphatic nitrilases, which act primarily on aliphatic nitriles such as acrylonitrile; aromatic and heterocyclic nitrilases, which act primarily on aromatic or heterocyclic nitriles such as benzonitrile and cyanopyridine; and arylacetonitrilases, which act primarily on arylacetonitriles such as IAN and phenylacetonitrile (35). CGMCC 4969 NitA and NitB have a conserved Glu(49)-Lys(131)-Cys(165) and Glu(47)-Lys(132)-Cys(166) catalytic triads, respectively. NitB is a putative aliphatic nitrilase and showed the highest identity (81%) to putative Bordetella sp. strain FB-8 aliphatic nitrilase. Surprisingly, a phylogenetic analysis showed that NitB clustered in a branch with Arabidopsis thaliana nitrilases NIT1, NIT2, and NIT3 (36), independent from the aliphatic nitrilases from R. rhodochrous strain K22 (37) and Acidovorax facilis (38) (Fig. 5B). NitA putatively belongs to the arylacetonitrilases, whereas in a phylogenetic analysis, it clustered in a branch with nitrilase from the phytopathogen P. syringae pv. syringae B728a (39). The IAA secreted by phytopathogens acts as a virulence factor to subvert plant defense systems and facilitate invasion into plant tissues (9). CGMCC 4969 nitrilase showed a close relationship with nitrilases from phytopathogens, which may explain its beneficial traits for colonizing plant tissues.
Characterization of V. boronicumulans CGMCC 4969 nitrile-converting enzymes.
In V. boronicumulans CGMCC 4969, NHase activity for transforming IAN to IAM has been confirmed (16). The putative nitrilase-coding genes nitA and nitB and indoleacetamide hydrolase-coding genes iamA and iamB were overexpressed in E. coli Rosetta(DE3)/pLysS. The resting cell transformation of IAM in E. coli overexpressing indoleacetamide hydrolase genes indicated that the IamA transformed IAM to IAA, whereas the IamB did not. The resting cell transformation of IAN in E. coli overexpressing nitrilase genes indicated that the NitA showed the IAN-to-IAA transformation activity, whereas the NitB only transformed aliphatic nitriles. Subsequently, the NHase, IamA, and NitA were purified (see Fig. S1, lanes 3, 5, and 7, in the supplemental material) and their biochemical properties were characterized. The optimal temperature for the transformation of both IAN by NHase and IAM by IamA was 40°C, and for the transformation of IAN by NitA, it was 50°C (see Fig. S2, S3, and S4). The optimal pH for IAN transformation by both NHase and NitA was 7.0, and for the transformation of IAM by IamA, it was 8.0 to 9.0. The NHase, IamA, and NitA retained >60% residual activity for 5 h at 30°C but were unstable at 50°C, and >60% of the activity was lost at pH <4.0.
The Vmax of the NHase for IAN was 30.82 U · mg−1 and the Km was 2.87 mmol · liter−1 (Table 1). Although there have been many reports of the kinetic parameters of NHases toward aliphatic nitriles, aromatic nitriles, and heterocyclic nitriles, the parameters toward IAN have rarely been reported. Indoleacetamide hydrolase mainly exists in phytopathogens in the tryptophan 2-monooxygenase-dependent IAA synthesis pathway. Vega-Hernández et al. reported that NHase/indoleacetamide hydrolase catalyzed the conversion of IAN to IAA in Bradyrhizobium USDA110, and the specific activity of the conversion of IAM to IAA was only 0.8 U · mg−1 (20), two orders of magnitude lower than that of CGMCC 4969 IamA. CGMCC 4969 NitA has a Km value of 0.77 mmol · liter−1 for IAN, which is lower than that of Streptomyces sp. strain MTCC 7546 nitrilase (1.3 mmol · liter−1) (40). In addition, the Km value of the NHase is higher than that of NitA, which indicated that the nitrilase NitA has a higher affinity for IAN than the NHase.
TABLE 1.
Kinetic parameters of V. boronicumulans CGMCC 4969 nitrile-converting enzymesa
| Enzyme | Vmax (U · mg−1) | Km (mmol · liter−1) |
|---|---|---|
| NHase | 30.82 ± 1.64 | 2.87 ± 0.31 |
| IamA | 13.09 ± 1.05 | 0.52 ± 0.08 |
| NitA | 23.67 ± 2.51 | 0.77 ± 0.12 |
The kinetic parameters of the nitrile-converting enzymes were estimated over a range of substrate concentrations (0.1 to 6 mmol · liter−1) at 37°C in 50 mmol · liter−1 phosphate buffer at pH 7.5. Data shown are mean values ± standard deviations from three replicates.
The substrate spectra of NitA and IamA were assessed. NitA showed a higher specific activity for the arylacetonitrile IAN, phenylacetonitrile, and (R)-(+)-4-methylmandelonitrile but could not transform the aromatic and heterocyclic nitrile benzonitrile, bromoxynil, or 3-cyanopyridine (Table 2). NitA also exhibited comparatively low activity toward aliphatic nitriles, but interestingly, as the carbon chain increased in length from acetonitrile to hexanedinitrile, its activity increased. IamA showed strict substrate specificity and could only transform IAM and benzamide. The high activities of the nitrile-converting enzymes toward IAN or IAM indicate that the main role of the nitrile-converting enzymes in CGMCC 4969 is concerned with the synthesis of IAA.
TABLE 2.
Substrate specificities of purified V. boronicumulans CGMCC 4969 NitA and IamA
| Substrate | Relative activity (% [± SD])a |
|---|---|
| NitA | |
| IAN | 100.00 ± 0.62 |
| Phenylacetonitrile | 257.98 ± 2.41 |
| (R)-(+)-4-Methylmandelonitrile | 32.97 ± 1.26 |
| Acetonitrile | 0.39 ± 0.07 |
| Butyronitrile | 2.72 ± 1.61 |
| Isobutyronitrile | 0.44 ± 0.05 |
| Succinonitrile | 5.25 ± 0.13 |
| Hexanedinitrile | 23.14 ± 1.32 |
| 3-Cyanopyridine | ND |
| Benzonitrile | ND |
| Bromoxynil | ND |
| IamA | |
| IAM | 100.00 ± 2.13 |
| Benzamide | 64.89 ± 1.07 |
| Nicotinamide | ND |
| Phenylacetamide | ND |
| Acetamide | ND |
| Acrylamide | ND |
NitA and IamA activities were determined in standard assay conditions. The specific activities toward IAN (12.39 U · mg−1) and IAM (9.89 U · mg−1) were taken as 100%. SD, standard deviation; ND, not detected.
Transcription level analysis of nitrile-converting enzymes in V. boronicumulans CGMCC 4969.
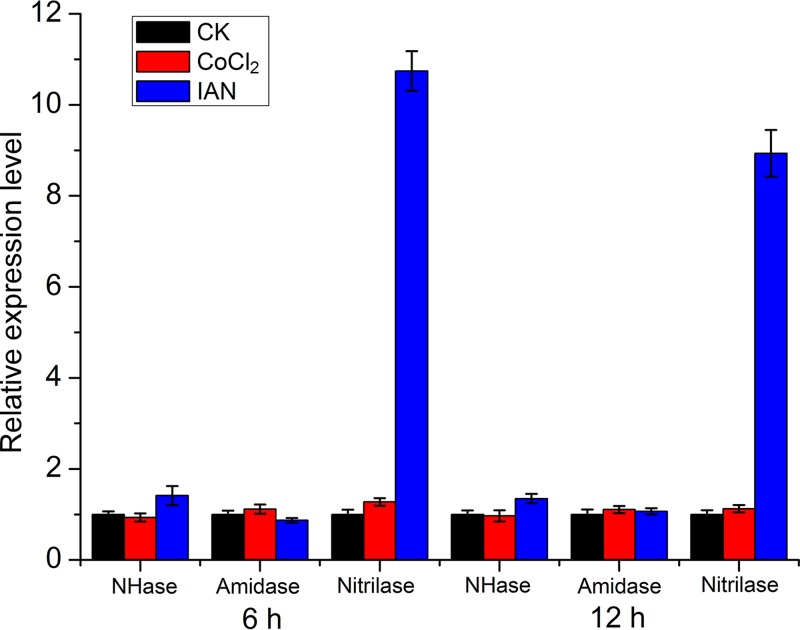
The addition of cobalt and IAN to the culture medium activated the CGMCC 4969 NHase/amidase and nitrilase pathways, respectively. Quantitative PCR (qPCR) was conducted to determine whether the expression of the nitrile-converting enzymes was influenced by the two different additives. When a final concentration of 0.1 mmol · liter−1 CoCl2 was added into the culture medium, the transcriptional levels of the NHase, IamA, and NitA showed no obvious changes; however, when a final concentration of 100 mg · liter−1 IAN was added to the culture medium, the transcriptional level of NitA increased by 10.7-fold at 6 h and 8.9-fold at 12 h compared with a culture in normal LB medium (Fig. 6).
FIG 6.
mRNA expression levels of genes encoding nitrile-converting enzymes in V. boronicumulans CGMCC 4969. CK (control), CGMCC 4969 cultured in LB medium; CoCl2, V. boronicumulans CGMCC 4969 cultured in LB medium supplemented with 0.1 mmol · liter−1 CoCl2; IAN, V. boronicumulans CGMCC 4969 cultured in LB medium supplemented with 100 mg · liter−1 IAN. “6 h” and “12 h” indicate the sampling times of V. boronicumulans CGMCC 4969. Error bars represent the standard deviations from three biological replicates.
Thus, the qPCR analysis showed that cobalt did not activate the NHase/amidase pathway by increasing the expression level of the NHase or IamA; it may function as a cofactor in the mature NHase, resulting in the improvement of NHase activity. However, IAN activated the nitrilase pathway by inducing the expression of the nitrilase. The qPCR results were in accordance with our findings in resting cells of CGMCC 4969 that the addition of 0.1 mmol · liter−1 CoCl2 enhanced the IAM content by 4.3-fold, while the addition of 100 mg/liter IAN enhanced the IAA content by 64.6-fold. Further analysis of the CGMCC 4969 genome showed that an AraC family transcriptional regulator (accession number ATA54506) is present in the downstream region of NitA and shows 27% sequence identity to the positive transcriptional regulator NitR of R. rhodochrous strain J1. R. rhodochrous J1 nitrilase was found to be required for isovaleronitrile-dependent induction, and the deletion of the central and 3′-terminal portion of the downstream nitR resulted in the complete loss of nitrilase activity (41).
Effect of V. boronicumulans CGMCC 4969 inoculation on the growth of Arabidopsis thaliana plantlets.
V. boronicumulans CGMCC 4969 is a plant growth-promoting rhizobium, and we investigated the effect of its inoculation on A. thaliana plantlets. After the inoculation of CGMCC 4969 onto the root of A. thaliana for 14 days, the main root length of A. thaliana plantlets increased 30%, the shoot weight increased 21%, and the total weight increased 24% compared with those of controls (Table 3). These results indicate that CGMCC 4969 promoted the growth of A. thaliana.
TABLE 3.
Influence of V. boronicumulans CGMCC 4969 on the growth of A. thaliana plantletsa
| Time (days) | Root length (cm) |
Shoot wt (mg) |
Total wt (mg) |
|||
|---|---|---|---|---|---|---|
| Control | Bacterial inoculation | Control | Bacterial inoculation | Control | Bacterial inoculation | |
| 0 | 3.3 ± 0.19 A | 3.2 ± 0.15 A | 11.07 ± 0.69 A | 10.80 ± 0.61 A | 11.68 ± 0.88 A | 11.36 ± 0.85 A |
| 14 | 10.87 ± 0.78 A | 14.13 ± 0.97 B | 41.07 ± 1.29 A | 49.88 ± 1.63 B | 43.65 ± 1.60 A | 53.98 ± 0.96 B |
Data indicate the means ± SDs from three biological replicates, and 12 A. thaliana plantlets were determined per replicate. Different uppercase letters within a column indicate significant difference (P ≤ 0.05) according to Welch's t test. Control, A. thaliana plantlets cultured independently without bacterial inoculation.
A. thaliana has four nitrilases, NIT1, NIT2, NIT3, and NIT4. NIT1, NIT2, and NIT3 are arylacetonitrilases and can transform IAN to IAA, which links them to auxin synthesis. NIT4 is widespread in the plant kingdom and is likely important in the cyanide detoxification pathway (36). Vorwerk et al. reported that A. thaliana NIT1, NIT2, and NIT3 have Km values toward the auxin precursor IAN of 11.1, 7.4, and 30.1 mmol · liter−1, respectively (42), which are much higher than the Km value of CGMCC 4969 NitA (0.77 mmol · liter−1), indicating that the CGMCC 4969 nitrilase has a higher affinity for IAN than the A. thaliana nitrilases. Additionally, the Vmax values toward IAN of A. thaliana NIT1, NIT2, and NIT3 were 640, 300, and 250 pkat/mg (38.4, 18, and 15 mU/mg), respectively, which are three orders of magnitude lower than that of CGMCC 4969 NitA. In long-term plant-microbe interactions, many plant-associated microbial nitrilases appear to have evolved to metabolize plant-derived nitriles or use plant nitriles as a carbon and nitrogen source (43). IAN contains a nitrile group, and some cruciferous plants, such as A. thaliana, are capable of synthesizing IAN (44). Thus, we speculate that when CGMCC 4969 colonizes the root of A. thaliana plantlets, its nitrilase may effectively compete with the plant's nitrilase for the common precursor IAN to produce IAA. Therefore, we hypothesize that Variovorax nitrile-converting enzymes may be involved in IAA production through the IAN pathway using plant-synthesized IAN as a substrate, leading to plant growth-promoting rhizobacterial Variovorax strains playing key roles in the regulation of auxin concentrations in the plant rhizosphere.
MATERIALS AND METHODS
Chemicals.
IAN, IAM, and IAA (98% purity) were purchased from Sigma-Aldrich (Shanghai, China). HPLC grade acetonitrile was purchased from Merck KGaA (Germany). All other reagents were of analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
Strains, plasmids, plants, and media.
The wild-type bacterium V. boronicumulans CGMCC 4969 was isolated from soil and deposited in the China General Microbiological Culture Collection Center (CGMCC; Beijing, China) under accession number 4969. E. coli Rosetta(DE3)/pLysS (Novagen) was used as the host strain for gene expression and stored in our laboratory The plasmid pET28a (Novagen) was used as the expression vector. The A. thaliana was Columbia wild type and purchased from Beijing Hua Yue Yang Biological Co. Ltd. (Beijing, China). LB medium and MSM were used for cell cultivation (45). Half-strength Murashige and Skoog (½MS) salt medium (Sigma-Aldrich, Sydney, Australia) was used for the cultivation of A. thaliana plantlets.
Biotransformation of IAN by V. boronicumulans CGMCC 4969.
To explore the metabolism of IAN in V. boronicumulans CGMCC 4969, the bacterium precultivated on an LB agar plate was inoculated in 100 ml of LB, incubated in a rotary shaker at 220 rpm for 24 h at 30°C, and harvested by centrifugation at 8,000 × g for 5 min, and then the cell pellet was washed twice with 50 mmol · liter−1 potassium phosphate buffer (pH 7.5). The cell sediment was resuspended in the same buffer with an IAN concentration of 400 mg · liter−1, and the cells were adjusted to an optical density at 600 nm of 5.0. These IAN transformation systems were defined as the standard resting cell transformations. Resting cell transformation broth, excluding IAN or cells, was used in negative controls. To further investigate the influence of IAN and cobalt on nitrile-converting enzyme activity, IAN and CoCl2 were added to LB to concentrations of 100 mg · liter−1 and 0.1 mmol · liter−1, respectively, to culture CGMCC 4969, and then the bacterium was prepared as resting cells to transform IAN in the aforementioned standard conditions. The transformation systems were incubated for the indicated times, and 0.5 ml of the transformation broths was sampled and centrifuged at 12,000 × g for 10 min. The supernatants were filtered through an organic-phase membrane with a 0.22-μm pore size and diluted 5-fold for HPLC analysis.
To determine whether V. boronicumulans CGMCC 4969 can use IAN as a carbon and nitrogen source for cell growth, MSM containing 50 mg · liter−1 IAN was used to cultivate CGMCC 4969. The culture broth was sampled at regular intervals to detect the cell growth and metabolites. The cell growth was monitored by counting the CFU on LB plates after 2 days of growth at 30°C. The metabolites of IAN transformation were detected by HPLC. MSM containing CGMCC 4969 alone was used as a control.
HPLC and LC-MS analyses.
An Agilent 1260 HPLC system equipped with a reverse-phase C18 precolumn (4.6 mm by 20 mm; Agilent Technologies) and an HC-C18 column (4.6 mm by 250 mm, 5-μm particle size; Agilent Technologies) was used to analyze the transformation of IAN and its metabolites. Elution was conducted at a 1-ml · min−1 flow rate in a mobile phase that contained water, acetonitrile, and 0.01% acetic acid (water-acetonitrile, 60:40). The signal was monitored at 230 nm using an Agilent G1314A UV detector. LC-MS was conducted using an Agilent 1290 infinity liquid chromatograph with a G1315B diode array detector and an Agilent 6460 triple quadrupole LC-MS system equipped with an electrospray ion source that was operated in positive ion mode.
Genome sequencing, assembly, gene annotation, and protein classification.
The genomic DNA of V. boronicumulans CGMCC 4969 was sequenced using a PacBio RS II platform and Illumina HiSeq 4000 platform at the Beijing Genomics Institute (BGI; Shenzhen, China). Four SMRT cell zero-mode waveguide arrays were used by the PacBio platform to generate the subread set. Draft genomic unitigs, which are uncontested groups of fragments, were assembled with the Celera Assembler against a high-quality corrected circular consensus sequence subread set.
Gene prediction was performed on the V. boronicumulans CGMCC 4969 genome assembly using glimmer3 (http://www.cbcb.umd.edu/software/glimmer/) with hidden Markov models. Tandem repeats annotation was obtained using Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html). For functional annotations, the best hit was abstracted using BLAST. Seven databases, Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups, nonredundant protein, Swiss-Prot, Gene Ontology, TrEMBL, and EggNOG, were used for general functional annotations.
Cloning and expressing recombinant indoleacetamide hydrolase and nitrilase in E. coli.
The hypothetical nitrile-converting enzyme-coding genes were cloned into plasmid pET28a and expressed in E. coli Rosetta(DE3)/pLysS. Genomic DNA was extracted using a bacterial genomic DNA extraction kit (TaKaRa, Dalian, China). Primers were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) and are listed in Table 4. Primer pair F1 and F2 was used for amplifying the indoleacetamide hydrolase-coding gene iamA, F3 and F4 were for amplifying the indoleacetamide hydrolase-coding gene iamB, F5 and F6 for amplifying the nitrilase-coding gene nitA, and F7 and F8 for amplifying the nitrilase-coding gene nitB. The PCR system and program used were as previously reported (46). The PCR products were analyzed by 1% agarose gel electrophoresis and ligated into pET28a according to the protocol of the ClonExpress II one-step cloning kit (Vazyme Biotech, Nanjing, China). The recombinant plasmids were verified by DNA sequencing performed by Springen Biotech (Nanjing, China). The transformation of the recombinant plasmids into competent E. coli Rosetta(DE3)/pLysS cells and the overexpression of the recombinant indoleacetamide hydrolase and nitrilase were conducted according to methods described in our previous report (47).
TABLE 4.
Primers used in this study
| Target | Primera | Sequence (5′→3′)b | Amplicon size (bp) |
|---|---|---|---|
| iamA | F1 | ACAGCAAATGGGTCGCGGATCCGA | 1,410 |
| ATTCATGACCGCATCCCCCGCCCT | |||
| F2 | ATCTCAGTGGTGGTGGTGGTGGTGC | ||
| TCGAGTCACTGCGCGACCAGCGGC | |||
| iamB | F3 | ACAGCAAATGGGTCGCGGATCCGAA | 1,401 |
| TTCATGCAACTCTGGCAACTGGAAGC | |||
| F4 | ATCTCAGTGGTGGTGGTGGTGGTGCT | ||
| CGAGTCAGGCGGGGTCGACCG | |||
| nitA | F5 | ACAGCAAATGGGTCGCGGATCCGAA | 1,005 |
| TTCATGCCGACCACCGTCCACC | |||
| F6 | ATCTCAGTGGTGGTGGTGGTGGTGCT | ||
| CGAGTCAGGCCACGACCGGCTC | |||
| nitB | F7 | ACAGCAAATGGGTCGCGGATCCGAAT | 1,047 |
| TCATGCTTGATCTGCCCAAGTTCAAGG | |||
| F8 | ATCTCAGTGGTGGTGGTGGTGGTGCTC | ||
| GAGTCATGGCGTGCCCTCCCCG | |||
| NHase | NHa-F | GCCAATACCGACGACCAGCAC | 172 |
| NHa-R | TGGTCTCGGGCAAGGTGGT | ||
| Indoleacetamide | Ind-F | ATCGCCGAACTCAACCCGC | 153 |
| Hydrolase | Ind-R | GTCGACGTTGACCTTGATGCTC | |
| Nitrilase | Nit-F | CGCTACCACGACAACTCGCTGAT | 111 |
| Nit-R | CGCCGCTTTCTCGCTGTAG | ||
| 16S | 16S-F | TACTGGGCGTAAAGCGTGCG | 174 |
| 16S-R | ATTGCCTTCGCCATCGGTGT |
Primers F1 to F8 were used for amplification of target genes. Primers NHa-F, NHa-R, Ind-F, Ind-R, Nit-F, Nit-R, 16S-F, and 16S-R were used for quantitative PCR analysis.
Underlined bases indicate the restriction enzyme cleavage sites of EcoRI (GAATTC) and XhoI (CTCGAG).
Purification and determination of nitrile-converting enzyme activity.
The purification of the recombinant His-tagged nitrile-converting enzymes was conducted by affinity chromatography, according to the instructions of the chromatography resin manufacturer (Novagen Inc., Madison, WI). The expression and purity of the recombinant proteins were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The standard enzyme assay was conducted by mixing 1 mmol · liter−1 IAN or 1 mmol · liter−1 IAM and an appropriate amount of enzyme in 50 mmol · liter−1 sodium phosphate buffer (pH 7.5). The reaction was performed at 37°C in a volume of 1 ml. NHase activity was analyzed by HPLC. NitA and IamA activities were assessed by quantifying the amount of ammonia released during the reaction according to the phenol-hypochlorite method (48). The optimal pH was determined by dissolving 1 mmol · liter−1 IAN in different 50 mmol · liter−1 sodium citrate (pH 4 to 6), phosphate (pH 6 to 8), and Tris-HCl (pH 8 to 10) buffers. The optimal temperature was determined by conducting the reaction from 20 to 60°C in 50 mmol · liter−1 sodium phosphate buffer (pH 7.5). The pH stability was determined by incubating the enzymes at 4°C for 12 h in buffers at different pH values and the residual activity was determined using the method described above. The thermal stability was assessed by incubating the enzymes at different temperatures, and the activity remaining was measured using the enzyme assays described above. The substrate specificities of IamA toward IAM, benzamide, nicotinamide, phenylacetamide, acetamide, acrylamide, and of NitA toward IAN, phenylacetonitrile, (R)-(+)-4-methylmandelonitrile, acetonitrile, butyronitrile, isobutyronitrile, succinonitrile, hexanedinitrile, 3-cyanopyridine, benzonitrile, and bromoxynil, were determined in the standard reaction system with 5 mmol · liter−1 substrate. The kinetic parameters of the nitrile-converting enzymes toward IAN and IAM were calculated by determining the initial velocity in the ranges 0.1 to 6 mmol · liter−1 IAN and IAM. The maximal reaction rate (Vmax) and apparent Michaelis-Menten constant (Km) were deduced from Eadie-Hofstee plots.
qPCR analysis of nitrile-converting enzyme genes in V. boronicumulans CGMCC 4969.
V. boronicumulans CGMCC 4969 cells were cultured in LB with or without 0.1 mmol · liter−1 CoCl2 and 100 mg liter−1 IAN and harvested after 6 and 12 h, respectively. The total RNAs were isolated using a spin column total RNA purification kit (Sangon Biotech, Shanghai, China) according to the manufacturer's instructions. The extracted total RNA (500 ng) was reverse transcribed using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa Biotech, Dalian, China). The synthesized cDNA was subjected to qPCR using the Applied Biosystems StepOne real-time PCR system (Carlsbad, CA, USA) and SYBR Premix Ex Taq II (Tli RNaseH Plus; TaKaRa Biotech). Specific primers were designed and are listed in Table 4. The reaction was performed in a 20-μl mixture containing 10 μl of SYBR Premix Ex Taq II (Tli RNaseH Plus, 2×), 0.8 μl of each primer (10 μmol · liter−1), 0.4 μl of ROX reference dye (50×), 2 μl of cDNA template, and 6 μl of sterilized deionized water. The thermal cycling conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. A melting curve was analyzed at the end of the qPCR to verify specific amplification. The 16S rRNA gene was used as a reference gene to normalize the amount of RNA in each sample (49). We conducted three biological replicates and each sample was measured in triplicates.
A. thaliana plantlet cultivation and V. boronicumulans CGMCC 4969 inoculation.
A. thaliana culture was conducted according to the revised method of Conn et al. (50). Seeds were surfaced sterilized by immersing in 75% (vol/vol) ethanol for 1 min, soaking in 6% sodium hypochlorite for 10 min, and thoroughly rinsing 10 times in sterile distilled water. Sterilized seeds were placed onto ½MS salt medium, and the plates were sealed with micropore tape and placed inversely at 4°C for 3 days to achieve stratification. Plants were then transferred to an illuminated incubator with a 16-h light/8-h dark cycle, a light density of 2,000 lx, and a temperature cycle of 25 and 20°C day/night. After incubating for 5 days, when two leaves appeared, the plants were transferred to new ½MS plates to continue growth under the conditions above.
A single bacterial colony of V. boronicumulans CGMCC 4969 grown on an LB agar plate was inoculated in a 250-ml flask containing 50 ml of LB and incubated in a rotary shaker at 220 rpm and 30°C for 24 h. Then, the bacterial cells were collected by centrifugation at 5,000 × g for 10 min, washed twice with sterile double-distilled water, and resuspended in double-distilled water with the concentration adjusted to 109 cells · ml−1. When the A. thaliana seedlings had grown for 7 days, 25 μl of bacterial suspension was uniformly sprayed onto the root of the plant, and then plant culture was continued. Other plantlets were sprayed with 25 μl sterile double-distilled water as a control. Fourteen days after inoculation, the A. thaliana plantlets were taken out, the surface water was blotted up, and the total fresh weight, shoot fresh weight, and the length of the main root were measured.
Accession number(s).
The sequence data for V. boronicumulans CGMCC 4969 (also known as strain J1) were submitted to the GenBank database with accession number CP023284. The GenBank accession numbers for IamA, IamB, NitA, and NitB are ATA56605, ATA56697, ATA54505, and ATA55581, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This research was financed by the National Science Foundation of China (grant number 31570104), the Top-notch Academic Programs Project (TAPP) of Jiangsu Higher Education Institutions, the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, and the Academic Natural Science Foundation of Jiangsu Province (grant number 14KJA180004).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00298-18.
REFERENCES
- 1.Bhattacharyya PN, Jha DK. 2012. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- 2.Goswami D, Thakker JN, Dhandhukia PC, Tejada Moral M. 2016. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric 2:1127500. doi: 10.1080/23311932.2015.1127500. [DOI] [Google Scholar]
- 3.Saharan BS. 2011. Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 2011:LSMR21. [Google Scholar]
- 4.McSteen P. 2010. Auxin and monocot development. Cold Spring Harb Perspect Biol 2:a001479. doi: 10.1101/cshperspect.a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossmann K. 2010. Auxin herbicides: current status of mechanism and mode of action. Pest Manag Sci 66:113–120. doi: 10.1002/ps.1860. [DOI] [PubMed] [Google Scholar]
- 6.Puyvelde SV, Cloots L, Engelen K, Das F, Marchal K, Vanderleyden J, Spaepen S. 2011. Transcriptome analysis of the rhizosphere bacterium Azospirillum brasilense reveals an extensive auxin response. Microb Ecol 61:723–728. doi: 10.1007/s00248-011-9819-6. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar D, Laha S. 2013. Production of phytohormone auxin (IAA) from soil born Rhizobium sp, isolated from different leguminous plant. Int J Appl Environ Sci 8:521–528. [Google Scholar]
- 8.Spaepen S, Vanderleyden J, Remans R. 2007. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 9.Duca D, Lorv J, Patten CL, Rose D, Glick BR. 2014. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek 106:85–125. doi: 10.1007/s10482-013-0095-y. [DOI] [PubMed] [Google Scholar]
- 10.Olesen MR, Jochimsen BU. 1996. Identification of enzymes involved in indole-3-acetic acid degradation. Plant Soil 186:143–149. [Google Scholar]
- 11.Leveau JHJ, Lindow SE. 2005. Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl Environ Microbiol 71:2365–2371. doi: 10.1128/AEM.71.5.2365-2371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satola B, Wübbeler JH, Steinbüchel A. 2013. Metabolic characteristics of the species Variovorax paradoxus. Appl Microbiol Biotechnol 97:541–560. doi: 10.1007/s00253-012-4585-z. [DOI] [PubMed] [Google Scholar]
- 13.Han JI, Choi HK, Lee SW, Orwin PM, Kim J, Laroe SL, Kim T, O'Neil J, Leadbetter JR, Sang YL. 2011. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J Bacteriol 193:1183–1190. doi: 10.1128/JB.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belimov AA, Dodd IC, Hontzeas N, Theobald JC, Safronova VI, Davies WJ. 2009. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol 181:413–423. doi: 10.1111/j.1469-8137.2008.02657.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HJ, Zhou QW, Zhou GC, Cao YM, Dai YJ, Ji WW, Shang GD, Yuan S. 2012. Biotransformation of the neonicotinoid insecticide thiacloprid by the bacterium Variovorax boronicumulans strain J1 and mediation of the major metabolic pathway by nitrile hydratase. J Agric Food Chem 60:153–159. doi: 10.1021/jf203232u. [DOI] [PubMed] [Google Scholar]
- 16.Sun SL, Yang WL, Guo JJ, Zhou YN, Rui X, Chen C, Ge F, Dai YJ. 2017. Biodegradation of the neonicotinoid insecticide acetamiprid in surface water by the bacterium Variovorax boronicumulans CGMCC 4969 and its enzymatic mechanism. RSC Adv 7:25387–25397. doi: 10.1039/C7RA01501A. [DOI] [Google Scholar]
- 17.Liu ZH, Cao YM, Zhou QW, Guo K, Ge F, Hou JY, Hu SY, Yuan S, Dai YJ. 2013. Acrylamide biodegradation ability and plant growth-promoting properties of Variovorax boronicumulans CGMCC 4969. Biodegradation 24:855–864. doi: 10.1007/s10532-013-9633-6. [DOI] [PubMed] [Google Scholar]
- 18.Follin A, Inzé D, Budar F, Genetello C, Montagu MV, Schell J. 1985. Genetic evidence that the tryptophan 2-mono-oxygenase gene of Pseudomonas savastonoi is functionally equivalent to one of the T-DNA genes involved in plant tumour formation by Agrobacterium tumefaciens. Mol Gen Genet 201:178–185. [Google Scholar]
- 19.Kiziak C, Conradt D, Stolz A, Mattes R, Klein J. 2005. Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151:3639–3648. doi: 10.1099/mic.0.28246-0. [DOI] [PubMed] [Google Scholar]
- 20.Vega-Hernández MC, León-Barrios M, Pérez-Galdona R. 2002. Indole-3-acetic acid production from indole-3-acetonitrile by strains of Bradyrhizobium. Soil Biol Biochem 34:665–668. doi: 10.1016/S0038-0717(01)00229-2. [DOI] [Google Scholar]
- 21.Vejvoda V, Kaplan O, Bezouška K, Pompach P, Sulc M, Cantarella M, Benada O, Uhnáková B, Rinágelová A, Lutz-Wahl S. 2008. Purification and characterization of a nitrilase from Fusarium solani O1. J Mol Catal B Enzym 50:99–106. doi: 10.1016/j.molcatb.2007.09.006. [DOI] [Google Scholar]
- 22.Stalker DM, Malyj LD, Mcbride KE. 1988. Purification and properties of a nitrilase specific for the herbicide bromoxynil and corresponding nucleotide sequence analysis of the bxn gene. J Biol Chem 263:6310–6314. [PubMed] [Google Scholar]
- 23.Fang S, An X, Liu H, Cheng Y, Hou N, Feng L, Huang X, Li C. 2015. Enzymatic degradation of aliphatic nitriles by Rhodococcus rhodochrous BX2, a versatile nitrile-degrading bacterium. Bioresour Technol 185:28–34. doi: 10.1016/j.biortech.2015.02.078. [DOI] [PubMed] [Google Scholar]
- 24.Leveau J, Gerards S. 2008. Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3-acetic acid. FEMS Microbiol Ecol 65:238–250. doi: 10.1111/j.1574-6941.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 25.Scott JC, Greenhut IV, Leveau JH. 2013. Functional characterization of the bacterial iac genes for degradation of the plant hormone indole-3-acetic acid. J Chem Ecol 39:942–951. doi: 10.1007/s10886-013-0324-x. [DOI] [PubMed] [Google Scholar]
- 26.Tsubokura S, Sakamoto Y, Ichihara K. 1961. The bacterial decomposition of indoleacetic acid. J Biochem 49:38–42. doi: 10.1093/oxfordjournals.jbchem.a127250. [DOI] [Google Scholar]
- 27.Jensen JB, Egsgaard H, Onckelen HV, Jochimsen BU. 1995. Catabolism of indole-3-acetic acid and 4- and 5-chloroindole-3-acetic acid in Bradyrhizobium japonicum. J Bacteriol 177:5762–5766. doi: 10.1128/jb.177.20.5762-5766.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deslandes B, Gariepy C, Houde A. 2001. Review of microbiological and biochemical effects of skatole on animal production. Livest Prod Sci 71:193–200. doi: 10.1016/S0301-6226(01)00189-0. [DOI] [Google Scholar]
- 29.Sharma M, Sharma NN, Bhalla TC. 2009. Amidases: versatile enzymes in nature. Rev Environ Sci Biotechnol 8:343–366. [Google Scholar]
- 30.Sekine M, Watanabe K, Syono K. 1989. Nucleotide sequence of a gene for indole-3-acetamide hydrolase from Bradyrhizobium japonicum. Nucleic Acids Res 17:6400. doi: 10.1093/nar/17.15.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazzola M, White FF. 1994. A mutation in the indole-3-acetic acid biosynthesis pathway of Pseudomonas syringae pv. syringae affects growth in Phaseolus vulgaris and syringomycin production. J Bacteriol 176:1374–1382. doi: 10.1128/jb.176.5.1374-1382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonnard G, Vincent F, Otten L. 1991. Sequence of Agrobacterium tumefaciens biotype III auxin genes. Plant Mol Biol 16:733–738. doi: 10.1007/BF00023438. [DOI] [PubMed] [Google Scholar]
- 33.Ryabchenko LE, Podchernyaev DA, Kotlova EK, Yanenko AS. 2006. Cloning the amidase gene from Rhodococcus rhodochrous M8 and its expression in Escherichia coli. Russ J Genet 42:886–892. doi: 10.1134/S1022795406080060. [DOI] [PubMed] [Google Scholar]
- 34.Lira F, García-León G, Oliver A, Martínez JL. 2017. Draft genome sequences of four Pseudomonas aeruginosa isolates obtained from patients with chronic obstructive pulmonary disease. Genome Announc 5:e00147-17. doi: 10.1128/genomeA.00147-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Reilly C, Turner PD. 2003. The nitrilase family of CN hydrolysing enzymes - a comparative study. J Appl Microbiol 95:1161–1174. doi: 10.1046/j.1365-2672.2003.02123.x. [DOI] [PubMed] [Google Scholar]
- 36.Howden AJ, Harrison CJ, Preston GM. 2009. A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J 57:243–253. doi: 10.1111/j.1365-313X.2008.03682.x. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi M, Yanaka N, Nagasawa T, Yamada H. 1992. Primary structure of an aliphatic nitrile-degrading enzyme, aliphatic nitrilase, from Rhodococcus rhodochrous K22 and expression of its gene and identification of its active site residue. Biochemistry 31:9000. doi: 10.1021/bi00152a042. [DOI] [PubMed] [Google Scholar]
- 38.Chauhan S, Wu S, Blumerman S, Fallon RD, Gavagan JE, Dicosimo R, Payne MS. 2003. Purification, cloning, sequencing and over-expression in Escherichia coli of a regioselective aliphatic nitrilase from Acidovorax facilis 72W. Appl Microbiol Biotechnol 61:118–122. doi: 10.1007/s00253-002-1192-4. [DOI] [PubMed] [Google Scholar]
- 39.Howden AJ, Rico A, Mentlak T, Miguet L, Preston GM. 2009. Pseudomonas syringae pv. syringae B728a hydrolyses indole-3-acetonitrile to the plant hormone indole-3-acetic acid. Mol Plant Pathol 10:857–865. doi: 10.1111/j.1364-3703.2009.00595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal A, Nigam VK. 2014. Nitrilase mediated conversion of indole-3-acetonitrile to indole-3-acetic acid. Biocatal Agric Biotechnol 3:351–357. [Google Scholar]
- 41.Komeda H, Hori Y, Kobayashi M, Shimizu S. 1996. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc Natl Acad Sci U S A. 93:10572–10577. doi: 10.1073/pnas.93.20.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Muller A, Weiler EW, Piotrowski M. 2001. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- 43.Howden AJM, Preston GM, Segura A, Preston G, Wit PD. 2009. Nitrilase enzymes and their role in plant-microbe interactions. Microb Biotechnol 2:441–451. doi: 10.1111/j.1751-7915.2009.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller A, Hillebrand H, Weiler EW. 1998. Indole-3-acetic acid is synthesized from l-tryptophan in roots of Arabidopsis thaliana. Planta 206:362–369. doi: 10.1007/s004250050411. [DOI] [PubMed] [Google Scholar]
- 45.Zhou GC, Wang Y, Zhai S, Ge F, Liu ZH, Dai YJ, Yuan S, Hou JY. 2013. Biodegradation of the neonicotinoid insecticide thiamethoxam by the nitrogen-fixing and plant-growth-promoting rhizobacterium Ensifer adhaerens strain TMX-23. Appl Microbiol Biotechnol 97:4065–4074. doi: 10.1007/s00253-012-4638-3. [DOI] [PubMed] [Google Scholar]
- 46.Sun SL, Lu TQ, Yang WL, Guo JJ, Rui X, Mao SY, Zhou LY, Dai YJ. 2016. Characterization of a versatile nitrile hydratase of the neonicotinoid thiacloprid-degrading bacterium Ensifer meliloti CGMCC 7333. RSC Adv 6:15501–15508. doi: 10.1039/C5RA27966F. [DOI] [Google Scholar]
- 47.Lu TQ, Mao SY, Sun SL, Yang WL, Ge F, Dai YJ. 2016. Regulation of hydroxylation and nitroreduction pathways during metabolism of the neonicotinoid insecticide imidacloprid by Pseudomonas putida. J Agric Food Chem 64:4866–4875. doi: 10.1021/acs.jafc.6b01376. [DOI] [PubMed] [Google Scholar]
- 48.Weatherburn MW. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 49.Iino T, Miyauchi K, Kasai D, Masai E, Fukuda M. 2013. Characterization of nitrate and nitrite utilization system in Rhodococcus jostii RHA1. J Biosci Bioeng 115:600–606. doi: 10.1016/j.jbiosc.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 50.Conn VM, Walker AR, Franco CM. 2008. Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol Plant Microbe Interact 21:208–218. doi: 10.1094/MPMI-21-2-0208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.