Wood decay, driven primarily by wood-degrading basidiomycetes, is an essential component of global carbon cycles, and decay mechanisms are essential for understanding forest ecosystem function. These fungi efficiently consolidate pretreatment and saccharification of wood under mild conditions, making them promising templates for low-cost lignocellulose conversion. Species are categorized as ligninolytic white rots and polysaccharide-selective brown rots, with considerable undescribed variability in decay mechanism that may manifest in the sequential variation in protein secretion over the progression of decay. Here we resolved spatially a temporal progression of decay on intact wood wafers and compared secretome dynamics in two white and two brown rot fungi. We identified several universal mechanistic components among decay types, including early pectinolytic “pretreatment” and later-stage glycoside hydrolase-mediated saccharification. Interspecific comparisons also identified considerable mechanistic diversity within rot types, indicating that there are multiple avenues to facilitate white and brown rots.
KEYWORDS: manganese peroxidase, laccase, pectinase, proteomics
ABSTRACT
Wood-degrading fungi use a sequence of oxidative and hydrolytic mechanisms to loosen lignocellulose and then release and metabolize embedded sugars. These temporal sequences have recently been mapped at high resolution using directional growth on wood wafers, revealing previously obscured dynamics as fungi progressively colonize wood. Here, we applied secretomics in the same wafer design to track temporal trends on aspen decayed by fungi with distinct nutritional modes: two brown rot (BR) fungi (Postia placenta and Gloeophyllum trabeum) and two white rot (WR) fungi (Stereum hirsutum and Trametes versicolor). We matched secretomic data from three zones of decay (early, middle, and late) with enzyme activities in these zones, and we included measures of total protein and ergosterol as measures of fungal biomass. In line with previous transcriptomics data, the fungi tested showed an initial investment in pectinases and a delayed investment in glycoside hydrolases (GHs). Brown rot fungi also staggered the abundance of some oxidoreductases ahead of GHs to produce a familiar two-step mechanism. White rot fungi, however, showed late-stage investment in pectinases as well, unlike brown rot fungi. Ligninolytic enzyme activities and abundances were also different between the two white rot fungi. Specifically, S. hirsutum ligninolytic activity was delayed, which was explained almost entirely by the activity and abundance of five atypical manganese peroxidases, unlike more varied peroxidases and laccases in T. versicolor. These secretomic analyses support brown rot patterns generated via transcriptomics, they reveal distinct patterns among and within rot types, and they link spectral counts with activities to help functionalize these multistrain secretomic data.
IMPORTANCE Wood decay, driven primarily by wood-degrading basidiomycetes, is an essential component of global carbon cycles, and decay mechanisms are essential for understanding forest ecosystem function. These fungi efficiently consolidate pretreatment and saccharification of wood under mild conditions, making them promising templates for low-cost lignocellulose conversion. Species are categorized as ligninolytic white rots and polysaccharide-selective brown rots, with considerable undescribed variability in decay mechanism that may manifest in the sequential variation in protein secretion over the progression of decay. Here we resolved spatially a temporal progression of decay on intact wood wafers and compared secretome dynamics in two white and two brown rot fungi. We identified several universal mechanistic components among decay types, including early pectinolytic “pretreatment” and later-stage glycoside hydrolase-mediated saccharification. Interspecific comparisons also identified considerable mechanistic diversity within rot types, indicating that there are multiple avenues to facilitate white and brown rots.
INTRODUCTION
Wood-degrading basidiomycete fungi and their metabolic pathways for plant biomass decomposition are of biotechnological interest and are principal determinants of soil quality and CO2 evolution rates in nature (1, 2). Wood decomposition is a dynamic process where the decomposer species composition (3, 4) and the “signature” physiochemical modifications they make to wood substrates (5, 6) change over time, ultimately determining the fate of wood-bound carbon. Considerable interspecific variation in decay mechanisms is apparent among related species (7), and some variations may relate to time-dependent up- and downregulation patterns (8). Better resolution of temporal changes in basidiomycete wood decay mechanisms can therefore help identify processes that drive broader interspecific variations in the decay mechanisms of these important fungi.
Wood-degrading fungi are traditionally categorized as carbohydrate-selective brown rot (BR) fungi and lignin-degrading white rot (WR) fungi (9). As a general rule, WR fungi produce lignin-degrading peroxidases (10) and a greater diversity of glycoside hydrolase (GH) families (11) than BR fungi. Comparative genomics studies have, however, identified variable copy numbers for different classes of ligninolytic enzymes and other oxidases among species (11, 12). The white rot fungus Trametes versicolor (Polyporales), as an example relevant to the work we present here, is known to produce large amounts of laccase (13) and to possess in its genome lignin peroxidases (LiPs), versatile peroxidases (VPs), dye-decolorizing peroxidases (DyPs), and short manganese peroxidases (MnPs). On the other hand, the white rot fungus Stereum hirsutum (Russulales) encodes only “atypical” MnPs and laccases among its ligninolytic genes (11), a feature that does not apparently limit its ability to degrade lignin and that leaves S. hirsutum lumped with T. versicolor within a single rot “type.” This lumping into two rot type “bins,” however, is increasingly viewed as oversimplified, and genomic (14) and proteomic (15) studies indicate that rot types generally lie along a spectrum. Individual species within the BR and WR groups vary considerably in the enzymes they produce to degrade wood (7, 8).
These decay mechanisms are also dynamic over time, and this temporal variability is essential in how these mechanisms proceed. Temporal aspects of regulation are not evident from genomic DNA sequences and are likely controlled via gene networks that can be evaluated only via RNA transcripts rather than genomes. Using a recent transcriptomics study as an example for BR fungi, a directional colonization design using wood wafers showed that early stages of BR involve a unique set of differentially regulated oxidoreductases along with high pectinase expression (16). This temporal sequencing may loosen lignocellulose ahead of secretion of enzymes, providing better ingress for (hemi)cellulases, degrading pit membranes to improve cell-to-cell growth (17), and separating pectin from cellulose (18). This “pretreatment” would leave cellulose microfibrils more accessible to degradative enzymes that are upregulated after an initial delay (19).
Transcriptomics, however, is also limited in linking genes to their function, and gene products apparent in secretomes can provide more insight into the metabolites participating in lignocellulose deconstruction. These techniques are less quantitative than measuring expression levels but are inherently valuable “downstream” measurements posttranslation and can be coupled with enzyme activity assays in many cases to complement spectral counts of peptides. This approach has, for example, shown that WR fungi make early investments in peroxidase secretion (20) that are followed by an accumulation and increased activity of GHs, a pattern similar to that for BR fungi (8). These integrated secretome and specific-activity assessments have also been used to distinguish growth versus protein investments as a function of time among distinct wood-degrading clades within the BR fungi (8). This approach had not yet been coupled, however, with the higher-resolution temporal approaches to make interspecies comparisons within and among rot types.
In our study, growth characteristics and temporal dynamics among secretomes of four relevant WR and BR fungi were compared on thin aspen wood. Two BR and two WR fungi were grown on aspen wafers vertically, forcing a gradient of decay along the length of the wafer starting at the advancing hyphal front (16). Thin cross sections were taken along the advancing hyphal front to compare secretome characteristics over a progression of decay, tracking relative biomass investments and matching spectral counts via mass spectrometry with enzyme specific-activity assays. Our results contribute valuable secretome data sets that were generated using the high-resolution temporal sequence on solid wood and including four fungi commonly used as models in biochemical studies. Our results also confirm several key mechanistic patterns that were inferred previously from less-resolved time series or transcriptomic studies. Two novel insights also emerged from our results, as well, highlighting distinct temporal regulation of pectinases between BR and WR fungi and revealing two distinct peroxidase pathways in the WR fungi tested.
RESULTS
Secretome composition along hyphal fronts.
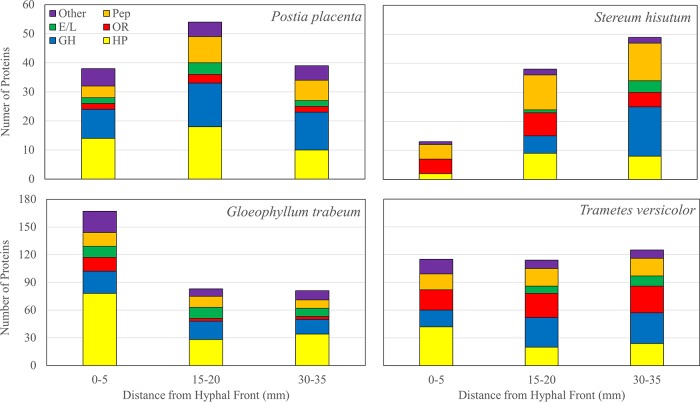
The secretome compositions of BR and WR fungi changed from undegraded to degraded wood sections (Fig. 1). The proportion of total proteins identified that were glycoside hydrolases (GHs) increased in all fungi from between 0 and 5 mm to between 30 and 35 mm from the hyphal front, ranging from 0 to 26.3% at 0 to 5 mm and from 19.8 to 33.3% at 30 to 35 mm (Fig. 1; see Table S1 in the supplemental material). The total number of GH spectra observed at each time point supports this trend, with GHs at least doubling, from between 0 and 5 to between 30 and 35 mm from the hyphal front in all species (Table 1). GH families 3 (GH 3), 5, 10, 28, and 79 were found among all fungi, and GH 30, 43, 53, and 131 were produced exclusively in T. versicolor extracts. GH 16 and GH 55 were identified exclusively in BR fungi in this study, while GHs 6 and 7 were found exclusively in WR fungi. Each fungus produced at least one GH 28 pectinase, some (PP 111730, GT 120615, and TV 125789) exclusively at 0 to 5 mm behind the hyphal front and one (SH 130790) only at 30 to 35 mm behind the hyphal front (see Tables S2 to S5 in the supplemental material). In addition to GH 28 proteins, both Gloeophyllum trabeum and T. versicolor produced GH 35 β-galactosidases, and the former also produced a carbohydrate esterase (CE) 8 pectin methylesterase (Tables S3 and S4).
FIG 1.
Total number of each of six protein categories identified in protein extracts of aspen wafer sections 0 to 5, 15 to 20, and 30 to 35 mm from the advancing hyphal front degraded by brown rot (Postia placenta and Gloeophyllum trabeum) and white rot (Stereum hirsutum and Trametes versicolor) fungi. HP, hypothetical protein; GH, glycoside hydrolase; OR, oxidoreductase; E/L; esterase/lipase, Pep, peptidase.
TABLE 1.
Total number of observations of proteins in each of six categories from degraded aspen wafer section extracts taken 0 to 5, 15 to 20, and 30 to 35 mm behind the advancing hyphal front of each fungusa
| Category | Total no. of observations for the following fungus and distance (mm): |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Postia placenta |
Gloeophyllum trabeum |
Trametes versicolor |
Stereum hirsutum |
|||||||||
| 0–5 | 15–20 | 30–35 | 0–5 | 15–20 | 30–35 | 0–5 | 15–20 | 30–35 | 0–5 | 15–20 | 30–35 | |
| Hypothetical protein | 191 | 323 | 481 | 5,515 | 1,234 | 422 | 789 | 610 | 354 | 5 | 113 | 169 |
| GH | 143 | 335 | 350 | 501 | 1,278 | 1,069 | 279 | 818 | 846 | 0 | 14 | 413 |
| OR | 16 | 63 | 31 | 1,002 | 115 | 8 | 952 | 510 | 365 | 47 | 296 | 159 |
| Esterase/lipase | 20 | 28 | 16 | 220 | 596 | 303 | 0 | 62 | 55 | 0 | 71 | 119 |
| Peptidase | 158 | 282 | 332 | 398 | 437 | 198 | 726 | 2,030 | 1,208 | 17 | 221 | 304 |
| Other | 52 | 62 | 68 | 947 | 68 | 30 | 477 | 148 | 110 | 25 | 87 | 73 |
| Total | 580 | 1,093 | 1,278 | 8,583 | 3,728 | 2,030 | 3,223 | 4,178 | 2,938 | 94 | 802 | 1,237 |
The number of observations (a proxy for abundance) was quantified from equal portions of protein from each section type among samples from each fungus. GH, glycoside hydrolase; OR, oxidoreductase.
Esterases and lipases constituted a smaller portion of the total number of proteins identified in all fungi (0 to 14.5%) and were found only in 0- to 5-mm extracts of BR fungi (5.3 to 7.2%) (Fig. 1). These trends are supported by the total number of esterase/lipase spectra identified 0 to 5 mm behind the hyphal front for BR (20 to 220) and WR (0) fungi (Table 1). All fungi except S. hirsutum produced carbohydrate esterase (CE) family 16 proteins, and CE 15 was found in extracts from all fungi except Postia placenta. CE 10 was found only in P. placenta extracts, CEs 4 and 8 were found only in G. trabeum extracts, and CE 12 was found only in S. hirsutum extracts (Tables S2 to S5).
Oxidoreductases (ORs) constituted a larger proportion of WR protein extracts than of BR protein extracts, constituting up to 23.2% and 36.9% of all identified proteins in T. versicolor and S. hirsutum, respectively, compared to 5.6% and 9.0% for P. placenta and G. trabeum, respectively (Fig. 1). This difference is driven primarily by MnPs and laccases found in S. hirsutum and T. versicolor and by LiPs and DyPs in extracts of the latter (Table 2). ORs were found in extracts of all three sections of brown rot fungi, but some secretion signal-containing ORs of unknown function were found exclusively in the 0- to 5-mm sections of G. trabeum (Gt 95549, Gt 130320, Gt 74773, and Gt 65654) extracts (see Table S7 in the supplemental material).
TABLE 2.
Laccases and peroxidases found in aspen wafer extracts and number of observations in protein extracts 0 to 5, 15 to 20, and 30 to 35 mm behind the advancing hyphal fronts of white rot fungia
| Fungus | Protein ID | Protein name | No. of observations at: |
||
|---|---|---|---|---|---|
| 0–5 mm | 15–20 mm | 30–35 mm | |||
| Trametes versicolor | 48870 | DyP1 | 174 | 57 | 33 |
| 48874 | DyP2 | 109 | 4 | 7 | |
| 146232 | Laccase 2 | 41 | 0 | 0 | |
| 138261 | Laccase 3 | 62 | 27 | 14 | |
| 115295 | Laccase 4 | 16 | 0 | 0 | |
| 43576 | LiP 1 | 8 | 42 | 41 | |
| 114944 | LiP 12 | 0 | 0 | 0 | |
| 43578 | LiP 2 | 6 | 34 | 66 | |
| 52333 | LiP 6 | 0 | 0 | 0 | |
| 134226 | LiP 9 | 12 | 69 | 34 | |
| 51375 | MnP 1, short | 53 | 1 | 1 | |
| 112835 | MnP 2, short | 2 | 5 | 4 | |
| 131080 | MnP 3, short | 11 | 0 | 0 | |
| 130496 | MnP 4, short | 211 | 38 | 30 | |
| 43477 | MnP 5, short | 0 | 3 | 0 | |
| 51455 | MnP 6, short | 53 | 10 | 5 | |
| 51457 | MnP 9, short | 0 | 0 | 0 | |
| Stereum hirsutum | 129431 | Laccase 5 | 1 | 3 | 0 |
| 142136 | MnP, atypical | 3 | 2 | 0 | |
| 134504 | MnP, atypical | 4 | 62 | 13 | |
| 161701 | MnP, atypical | 0 | 8 | 0 | |
| 134527 | MnP, atypical | 0 | 25 | 1 | |
| 171838 | MnP, atypical | 28 | 51 | 0 | |
The number of observations was quantified from equal portions of protein from each section type among samples from each fungus. MnP, manganese peroxidase; LiP, lignin peroxidase; DyP, dye-decolorizing peroxidase.
Enzyme activities.
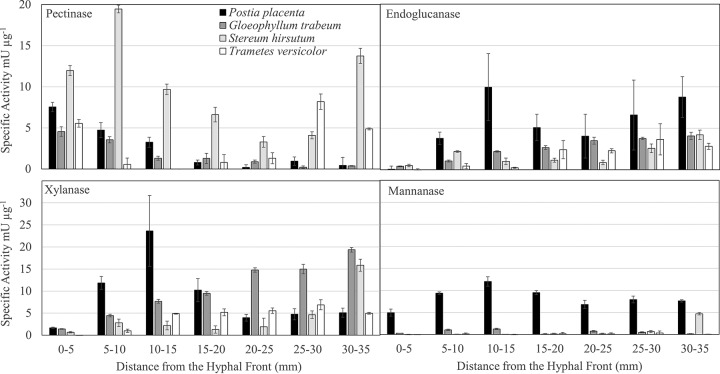
The specific activities of plant polysaccharide-degrading enzymes showed no consistent pattern along the advancing hyphal front among all fungi. BR fungi produced carbohydrate-active enzyme (CAZyme) specific activities equal to or greater than those of WR fungi (Fig. 2 and 3), despite secreting a reduced suite of GHs (Tables S2 to S5). Endoglucanase specific activity increased significantly (P < 0.05, Tukey's honestly significant difference [HSD] test) from between 0 and 5 mm to between 30 and 35 mm only in P. placenta, and a similar pattern was observed for xylanase specific activity in G. trabeum and S. hirsutum (Fig. 2). P. placenta xylanase specific activity increased above control levels only in sections 5 to 20 mm behind the hyphal front and then declined. P. placenta was also the only fungus to express mannanase specific activity above control levels in all sections (P < 0.05, Tukey's HSD) (Fig. 2), probably because of the high relative abundance of putative GH 5 endo-β-1,4-mannanase proteins in its secretome, which constituted 8.0 to 9.8% of all GH spectra (Table S2). Pectinase specific activity tended to be highest near the hyphal front (0 to 5 mm and 5 to 10 mm), although levels in WR fungi also reached similar levels in more-degraded wood (30 to 35 mm) (Fig. 2).
FIG 2.
Polysaccharide-degrading enzyme activities from aspen wafer section extracts along the advancing hyphal fronts of white and brown rot fungi. Error bars are standard deviations from three assays of one extract of a pool of 12 separate wafers.
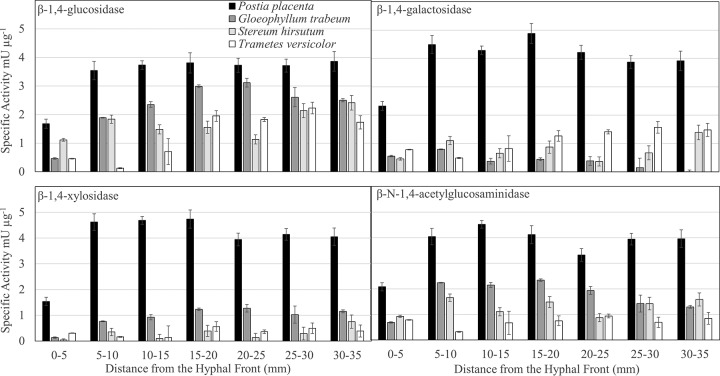
FIG 3.
Oligosaccharide- and disaccharide-degrading enzyme specific activities from aspen wafer section extracts along the advancing hyphal fronts of white and brown rot fungi. Error bars are standard deviations from three assays of one extract of a pool of 12 separate wafers.
Among the oligosaccharide- and disaccharide-degrading β-1,4-glucosidase (BGL), β-1,4-galactosidase (BGA), β-1,4-xylosidase (BXL), and β-1,4-N-acetylglucosaminidase (BNG) specific activities, BGA in G. trabeum was the only condition where activity was lower at 30 to 35 mm than at 0 to 5 mm (P = 0.025, Tukey's HSD) (Fig. 3). This was concurrent with the production of a GH 35 exclusively at the 0- to 5-mm wafer (Fig. 3; Table S3). BXL activity was generally low for all fungi except P. placenta and is likely a minor contributor to decay in all but this species (Fig. 3). Specific activity for BGL and BGA in T. versicolor extracts increased with distance from the hyphal front. Increases in BGL, BGA, BXL, and BNG specific activities in P. placenta extracts were seen from between 0 and 5 mm to between 5 and 10 mm, and then the levels remained stable along the length of the wafers (Fig. 3). In G. trabeum, BGL, BXL, and BNG specific activities increased from between 0 and 5 mm to between 5 and 10 mm and remained above the 0- to 5-mm levels along the length of the wafers. In S. hirsutum, a similar pattern was observed for BGL activity, but BNG activity fluctuated around the 0- to 5-mm levels throughout decay, and BXL levels were mostly not detectable (Fig. 3). Consistent with these trends, both WR fungi produced a putative GH 3 β-glucosidase exclusively after the earliest decay stage (Tables S4 and S5). Despite producing the highest oligosaccharide- and disaccharide-degrading enzyme activities, P. placenta produced only one GH 3 in all extracts, but this protein constituted 37.0 to 48.3% of all GH spectra in P. placenta extracts (Table S2), suggesting a high abundance.
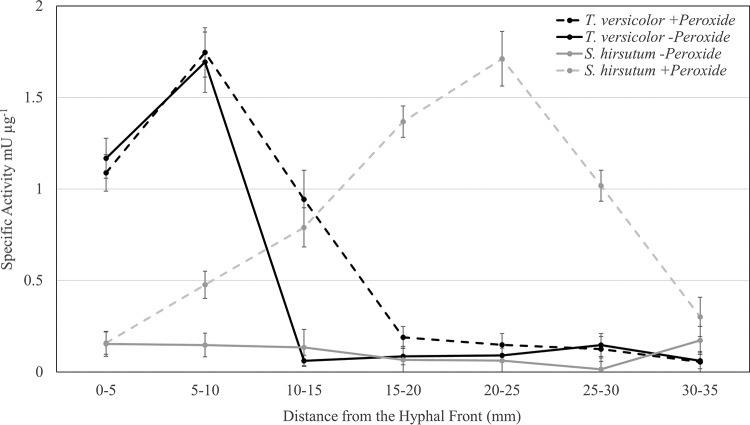
The most dramatic difference between BR and WR secretome activities was among ORs. Laccase and general peroxidase activities were measurable only in extracts from WR fungi. Laccase specific activity was above control levels only near the hyphal front (P = 0.033 for 0 to 5 mm and P = 0.045 for 5 to 10 mm) in S. hirsutum extracts but remained flat throughout decay, ranging from 0.17 to 0.01 mU μg−1 (Fig. 4). T. versicolor, on the other hand, produced 7 to 11 times greater laccase activity than S. hirsutum at 0 to 10 mm behind the hyphal front (1.17 to 1.69 mU μg−1) before the laccase activity dropped to levels not distinguishable from control levels in all other sections (0.14 to 0.06 mU μg−1) (Fig. 4). H2O2 and Mn2+ were added to laccase assay mixtures to determine the additive effect of peroxidases present in the secretome. Their addition drastically (3- to 65-fold) increased ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] oxidation in samples taken 5 to 30 mm behind the S. hirsutum hyphal front. In T. versicolor extracts, the difference in activity due to peroxide was less pronounced, and increases (15-fold) were seen only at 10 to 15 mm (Fig. 4) from the hyphal front.
FIG 4.
Laccase activities from aspen wafer extracts of sections taken along the advancing hyphal front of aspen wafers degraded by Trametes versicolor (black) and Stereum hirsutum (gray). Activities were measured with (dashed lines) and without (solid lines) the addition of hydrogen peroxide and Mn2+ to determine the ABTS oxidase activities of peroxidases in the extracts.
Growth rates and ergosterol analysis.
Fungal growth rates were comparable for P. placenta, G. trabeum, and S. hirsutum, advancing up the wafers at 2.7, 3.2, and 2.7 mm day−1, respectively, whereas T. versicolor grew notably faster at 5.3 mm day−1. Relatively slow growth by P. placenta and S. hirsutum did not result in increased ergosterol levels or protein secretion due to increased residence time at each 5-mm section (see Fig. S1 in the supplemental material).
DISCUSSION
This study reveals secretome changes on a scale of 1 to 2 days, which is comparable or higher resolution than in previous studies of secretome dynamics in wood decay fungi (15, 20). Early pectin metabolism (Fig. 2) and GH 28 secretion (see Tables S2 to S5 in the supplemental material) persisted across all rot types and species in this study despite the low (<2%) pectin content of aspen wood (21). This supports prior observations in our group of high pectinase expression seen in early stages of decay by Postia placenta (16), and this pattern suggests that early-stage pectinolysis may be necessary to convert wood lignocellulose enzymatically without harsh pretreatments. Consistent with previous studies (6, 8, 16, 20), increases in nonpectinolytic GH abundance (Fig. 1; Table 1) and most CAZyme specific activities (Fig. 2 and 3) increased as wood became more degraded. Our work suggests that this sequence is universal among WR and BR fungi and is likely a necessary “pretreatment” due to pectin's close association with (hemi)cellulose in the plant cell wall (18), where its removal allows GHs to access cell wall polysaccharides (19). The early dissolution of pectin-rich pit membranes (22) may also contribute to this pattern, as pits are an avenue of early wood colonization and exploration by decay fungi (17). However, later stages of WR decay also show heightened pectinase activity and GH 28 secretion, which may result from the dissolution of the lignin and pectin-rich middle lamellae seen in some types of WR (23).
The early stages of brown rot are characterized by hemicellulose removal (24), which requires the action of carbohydrate esterases (CEs) to remove branching groups such as acetyl groups on xylan in hardwoods (25). Early hemicellulose removal may explain the greater number of spectra attributed to CEs in brown rot secretomes in this study, particularly in early decay stages (Table 1), where hemicellulose is actively removed during brown rot. Peptidases were also major components of both white and brown rot secretomes in this study, particularly in S. hirsutum (Fig. 1). Peptidase secretion likely functions to recycle N recycling bound in the cell walls of wood-degrading basidiomycetes and partly explains their abundance despite low protein concentrations in wood (24). S. hirsutum is a secondary wood colonizer (26), and proteases are useful as a constitutive component of their secretomes for the utilization of cell wall proteins of predecessor mycelia in wood in conjunction chitinases (27).
It is well established that WR fungi secrete a more diverse suite of GHs than BR fungi (11, 14). However, the presence of a more diverse (hemi)cellulose-degrading arsenal in WR fungi (28) does not appear to drive increases in endoglucanase and hemicellulase specific activities compared to those of BR fungi (Fig. 2 and 3), and the difference must be driven by abundance or enzyme turnover. Protein quantitation using spectral counting indicates that the most abundant glycoside hydrolases in WR fungi were GH 6 and 7 proteins, together averaging 54.8% and 71.6% of all identified GH spectra at 15 to 20 mm and 30 to 35 mm behind the hyphal front, respectively (Tables S4 and S5). BR fungi produced greater amounts of GH 3 and 5 proteins, which together constituted 48.6% and 40.9% of all 15- to 20-mm and 30- to 35-mm GH spectra, respectively. This compared to 9.0% and 3.7% of all WR GH spectra at 15 to 20 and 30 to 35 mm, respectively. Indeed, BR fungi may compensate for losses in GH diversity by secreting their remaining GH suite in greater abundance than their WR counterparts. G. trabeum was a particularly heavy producer of GH 10 xylanases, and this family of enzymes constituted 34.1% and 51% of all GH spectra at 15 to 20 and 30 to 35 mm, respectively (Table S2). This pattern is supported by the high xylanase activity in G. trabeum extracts (Fig. 2) and suggests that different brown rot species may have different carbohydrate preferences, possibly leading to chemical variability in the decay substrate among species. It is also possible that the assay conditions used in this study did not measure the true catalytic potential of enzymes identified in the proteomics data. For example, T. versicolor secretes two putative β-xylosidases (Table S4), but virtually no β-xylosidase activity is detected (Fig. 3), and measurement of activities under different pH and temperature conditions could alter these results.
In addition to losses of several GH families, BR fungi are also known to lack lignin-degrading peroxidases (11) and phenol oxidase (laccase) activity (29). However, two flavin adenine dinucleotide (FAD)-binding domain-containing proteins secreted by G. trabeum (Gt 95549 and Gt 130320) (see Table S7 in the supplemental material) secreted early during decay may participate in an oxidative “pretreatment” phase of the BR decay mechanism (30). These BR ORs bear little homology to any functionally characterized proteins and merit further investigation. We cannot exclude the possibility that further brown rot ORs or other proteins escaped detection by our sampling and detection methods. The growth rates of the fungi tested here varied considerably, and ergosterol content along the wafers suggests that some of the species tested, especially T. versicolor, exhibit exploratory behavior at the hyphal front, leading to better resolution of decay stages than for the other species. This may have compounded secretome changes within discrete space in the slower-growing species, decreasing the likelihood that important, low-abundance proteins would be detected. In addition, proteins under 10 kDa were filtered out for this analysis, which would include things such as FeIII-reducing low-molecular-weight glycopeptides (31). Despite inclusion of a nonionic surfactant in our extraction buffer to reduce protein-substrate noncovalent bonding, it is likely that some degradative proteins also went undetected due to binding to the wood substrate.
WR fungi are unified as a group by the presence of lignin-degrading enzymes, including laccases and peroxidases, but this study identifies some of the mechanistic differences among WR species. Lignin-degrading OR activity in WR fungi followed different patterns in S. hirsutum and T. versicolor. Early secretion of ORs is known from other WR fungi (6, 20), but the types of peroxidases produced by the two WR fungi in this study differed. The only peroxidases secreted by S. hirsutum were 5 atypical MnPs, the only type encoded in its genome (11), while T. versicolor secreted 5 LiPs, 6 short MnPs (12), and 2 DyPs (32). Despite this disparity, H2O2-dependent ABTS oxidase activity in S. hirsutum extracts was greater and was produced over a larger time span (5 to 30 mm) (Fig. 4), supporting the functionality of atypical MnPs (33) and suggesting that they may be more effective than T. versicolor MnPs. Variation in oxidoreductases between S. hirsutum and T. versicolor could be one of the determinants of their ecological signatures. Both fungi are early secondary wood colonizers (34, 35), but precolonization of beech wood with either fungus produces disparate fungal successional patterns (4). This effect has been attributed to the production of biocidal secondary metabolites by S. hirsutum (36), but in light of this study, variations in decay mechanism and decay residue chemistry may also determine successor communities.
Overall, this study shows that the temporal changes in cultural characteristics and secretome composition during aspen wood decay are shared among BR and WR fungi. Early wood degradation likely requires hydrolysis of pectin, a barrier to access structural polysaccharides. Aside from pectinases, most plant-degrading GHs become more prominent as wood is more thoroughly degraded. Several ORs of BR fungi present in rotting wood remain uncharacterized, while WR oxidoreductases such as laccase and MnP are most active in early and intermediate decay stages, respectively. This work highlights some of the differences in the physiology of WR and BR fungi as well as interspecific variations in decay mechanisms, which may drive niche specialization and the ecological signatures of each species. We have also identified the importance of pectinolysis in early biological wood pretreatment, and pectin removal may be an important initiation step to facilitate fully enzymatic conversion of lignocellulose.
MATERIALS AND METHODS
Microcosm setup and harvest.
Two BR fungi (Postia placenta MAD-698 and Gloeophyllum trabeum ATCC 11539) and two WR fungi (Stereum hirsutum FP-91666 and Trametes versicolor A1-ATF of the UMN fungal collection) were cultivated on aspen wafers (60 by 25 by 3 mm) in modified ASTM standard soil block microcosms, as previously described (8, 16). Wafers with straight, horizontal hyphal fronts were harvested, and surface hyphae were removed and were cross-sectioned into seven 5-mm sections starting at the hyphal front and extending 35 mm behind it. Wafer sections were chopped into smaller pieces with sterilized razor blades prior to protein or ergosterol extractions. Growth rates were calculated as the total distance traveled by fungal hyphae divided by the total number of days of incubation less a 3-day (P. placenta, G. trabeum, and S. hirsutum) or 2-day (T. versicolor) lag period.
Protein extraction and biochemical assays.
Aspen wafer sections from equivalent distances from the advancing hyphal front (from between 0 and 5 mm to between 30 and 35 mm) degraded by each of the species mentioned above were pooled and chopped with sterile razor blades prior to extraction. Chopped sections were extracted with 80 ml of cold extraction buffer (0.5 M NaCl, 50 mM acetate, 0.05% Tween 80, pH 5.0) for 24 h at 4°C with gentle shaking. Extracts were filtered through polyester cloth, centrifuged at 4,000 × g for 30 min to remove solid particles, and filtered through sterile 0.2-μm polyvinylidene difluoride (PVDF) filters. Extracts were exchanged into 0.05 M citrate (pH 5.0) and concentrated using Vivaspin polyethersulfone (PES) 10-kDa-cutoff membranes prior to freezing at −20°C. Protein concentrations of extracts were determined using a Bio-Rad (Hercules, California, USA) protein assay kit.
Protein extracts of wafer section pools (12 wafers) from all seven separate sections along the hyphal front were assayed for various polysaccharide-degrading enzyme and ligninolytic oxidase activities. Cellulase and hemicellulase activities were measured by the dinitrosalicylic acid (DNS) method using solutions of 0.5% polygalacturonic acid (pectinase), 1.5% carboxymethyl cellulose (endoglucanase), 2% birchwood xylan (xylanase), and 0.5% locust bean gum (mannanase) (22). Triplicate portions of protein extracts were incubated with substrate solutions in 50 mM citrate (pH 5.0) at 50°C and quenched with DNS development solution. Reactions were developed at 90°C, and absorbance at 540 nm was measured and used to determine reducing sugars as galacturonic acid, glucose, xylose, and mannose reducing equivalents for pectinase, endoglucanase, xylanase, and mannanase activities, respectively. One unit was defined as the amount of enzyme required to produce 1 μmol of reducing sugar per minute. β-1,4-Glucosidase (BGL), β-1,4-galactosidase (BGA), β-1,4-xylosidase (BXL), and β-1,4-N-acetylglucosaminidase (BNG) activities were determined by measuring the release of 4-nitrophenol (4NP) from 4-nitrophenol-β-glucopyranoside, 4-nitrophenol-β-galactopyranoside, 4-nitrophenol-β-xylopyranoside, and 4-nitrophenol-N-acetyl-β-glucosamine, respectively. Reactions were carried out in 10 mM 4NP-substrate in 50 mM citrate (pH 5.0) at 50°C and were quenched with 2 volumes of 0.2 M Na2CO3. Absorbance at 400 nm was measured to quantify free 4NP, and 1 unit is defined as the amount of enzyme required to produce 1 μmol of free 4NP per minute under these conditions.
Protein extracts from WR fungi were also assayed for laccase and laccase + general peroxidase activity. Laccase and laccase + general peroxidase activities were measured by monitoring the oxidation of 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) at 420 nm (ε = 36,000 M−1 cm−1). Reactions were carried out in 5 mM ABTS for laccase and in 5 mM ABTS, 1 mM H2O2, and 1 mM MnCl2 for laccase plus general peroxidase. Both reactions were carried out at 20°C in 50 mM citrate buffer (pH 5.0), and one unit was defined as the amount of enzyme needed to oxidize 1 μmol of ABTS per minute under these conditions.
MS.
Purified protein extract from pools of 30 wafer sections from 0 to 5, 15 to 20, and 30 to 35 mm behind the advancing hyphal front for each fungus were used for liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics. Extracts were precipitated with trichloroacetic acid (TCA)-acetone, pelleted, and reconstituted in saturated guanidine-HCl. The protein concentration of each cellular extract was determined by bicinchoninic acid (BCA) assay (37). For each sample, processing replicates were performed; 50 μg of protein was aliquoted to low-retention Eppendorf tubes for downstream sample processing. All samples were incubated for 30 min at 60°C with tris(2-carboxyethyl)phosphine (TCEP) (Bond-Breaker; Thermo Fisher Scientific, Rockford, IL) to reduce disulfide bonds. Alkylation of cysteine residues was performed by treatment with 50 mM iodoacetamide, which was added from a 500 mM iodoacetamide–500 mM ammonium bicarbonate stock solution. After addition of iodoacetamide, all samples were incubated at room temperature in the dark for 40 min on a rocker. Each sample was then diluted to 0.9 M urea with 500 mM ammonium bicarbonate. To each 50-μg sample, 0.1 μg of MS-grade trypsin (Promega Corp., Madison, WI) was added and incubated overnight at 37°C. Peptides were extracted from each sample using solid-phase extraction with Discovery C18 50-mg resin columns (Supelco, St. Louis, MO). Each column was activated with 2 ml of methanol, followed by equilibration with 6 ml of 18-mΩ water. The sample was applied to the column, and then the column was washed with 8 ml of 50 mM ammonium bicarbonate. Peptides were eluted with two 0.9-ml washes of 80% acetonitrile. Samples were dried using a centrifugal concentrator (Thermo Scientific, Asheville, NC) and stored at −20°C until LC-MS analysis.
Peptides were solubilized in 150 μl solvent A (0.1% formic acid). For LC-MS analysis, 5 μl of sample was injected onto a Jupiter C18 resin reverse-phase column (3-μm particle size, 35-cm length, 75-μm inner diameter; Phenomenex, Torrance, CA). The peptides were eluted at 0.3 μl min−1 with an Agilent (Santa Clara, CA) 1200 high-performance liquid chromatograph (HPLC) with solutions of solvent A and 0.1% formic acid in acetonitrile (solvent B) using the following conditions: 0 to 30 min, isocratic at 100% solvent A; 30 to 32 min, linear gradient to 8% solvent B; 40 to 50 min, linear gradient to 12% solvent B; 50 to 105 min, linear gradient to 35% solvent B; 105 to 127 min, linear gradient to 60% solvent B; 127 to 130 min, linear gradient to 95% solvent B; and isocratic at 95% solvent B for 5 min. Eluted peptides were introduced into an Orbitrap XL mass spectrometer (Thermo Fisher, Waltham, MA) by electrospray ionization.
Spectra were collected in a data-dependent mode, with the five most intense ions in each survey scan selected for collisional induced dissociation in the five subsequent scans. Spectra were deconvoluted using the DeconMSn software (38) to more accurately assign parent ion mass and ion charge state and then were searched against predicted peptides derived from the fungal genome sequences via MS-GF+ (software used to analyze tandem mass spectra data [39]), using a 20-ppm parent ion mass tolerance in searches of tryptic peptides with a variable posttranslational modification of oxidized methionine. Protein databases were constructed in silico from genomes sequenced at the Joint Genome Institute (JGI). Genome sequences were obtained through the JGI Mycocosm database for Postia placenta (https://genome.jgi.doe.gov/Pospl1/Pospl1.home.html), Gloeophyllum trabeum (https://genome.jgi.doe.gov/Glotr1_1/Glotr1_1.home.html), Trametes versicolor (https://genome.jgi.doe.gov/Trave1/Trave1.home.html), and Stereum hirsutum (https://genome.jgi.doe.gov/Stehi1/Stehi1.home.html) (11, 40). A Q value cutoff (≤0.01) was utilized to obtain an ∼1% false-discovery rate (FDR) at each individual data set level, as assessed from a decoy identification search utilizing the reverse fungal genome sequence. Proteins with two or more spectra assigned to them were included in the analysis, and the entire list of proteins identified, including single observations, is listed in Data Set S1 in the supplemental material. Spectra observed in nondegraded aspen controls were subtracted from total spectral counts in degraded samples prior to analysis.
Ergosterol analysis.
Total ergosterol was measured as a biomarker for fungal biomass and was extracted from 5-mm sections taken along the advancing hyphal fronts of three separate aspen wafers for each fungus. Ergosterol was extracted using established methods (42) and measured by HPLC using a Phenomenex (Torrance, CA, USA) 4μ Hydro-RP 80a column by detection at 282 nm using previously described methods (43).
Data availability.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (41) partner repository, with the data set identifier PXD009480 (https://doi.org/10.6019/PXD009480).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the U.S. Department of Energy (DOE) Office of Science (Early Career grant DE-SC0004012 to J.S.S. from the Office of Biological and Ecological Research [BER] and BER grant DE-SC0012742 to J.S.S. and E.P.). This work was also funded by the National Science Foundation Graduate Research Fellowship Programs under grant 00039202 to G.N.P.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the DOE or NSF.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00159-18.
REFERENCES
- 1.Bardgett RD, Freeman C, Ostle NJ. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME J 2:805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- 2.A'Bear AD, Jones TH, Kandeler E, Boddy L. 2014. Interactive effects of temperature and soil moisture on fungal-mediated wood decomposition and extracellular enzyme activity. Soil Biol Biochem 70:151–158. doi: 10.1016/j.soilbio.2013.12.017. [DOI] [Google Scholar]
- 3.Rajala T, Peltoniemi M, Hantula J, Makipaa R, Pennanen T. 2011. RNA reveals a succession of active fungi during the decay of Norway spruce logs. Fungal Ecol 4:437–448. doi: 10.1016/j.funeco.2011.05.005. [DOI] [Google Scholar]
- 4.Hiscox J, Savoury M, Muller CT, Lindahl BD, Rogers HJ, Boddy L. 2015. Priority effects during fungal community establishment in beech wood. ISME J 9:2246–2260. doi: 10.1038/ismej.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Schilling JS. 2017. Role of carbon source in the shift from oxidative to hydrolytic wood decomposition by Postia placenta. Fungal Genet Biol 106:1–8. doi: 10.1016/j.fgb.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Kuuskeri J, Hakkinen M, Laine P, Smolander OP, Tamene F, Miettinen S, Nousiainen P, Kemell M, Auvinen P, Lundell T. 2016. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. Biotechnol Biofuels 9:192. doi: 10.1186/s13068-016-0608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDonald J, Suzuki H, Master ER. 2012. Expression and regulation of genes encoding lignocellulose-degrading activity in the genus Phanerochaete. Appl Microbiol Biotechnol 94:339–351. doi: 10.1007/s00253-012-3937-z. [DOI] [PubMed] [Google Scholar]
- 8.Presley GN, Schilling JS. 2017. Distinct growth and secretome strategies for two taxonomically divergent brown rot fungi. Appl Environ Microbiol 83:e02987-16. doi: 10.1128/AEM.02987-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatakka A, Hammel KE. 2010. Fungal biodegradation of lignocelluloses, p 319–340. In Hofrichter M. (ed), The mycota, vol 10, 2nd ed Industrial applications. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 10.Hofrichter M, Ullrich R, Pecyna MJ, Liers C, Lundell T. 2010. New and classic families of secreted fungal heme peroxidases. Appl Microbiol Biotechnol 87:871–897. doi: 10.1007/s00253-010-2633-0. [DOI] [PubMed] [Google Scholar]
- 11.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. 2012. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Duenas FJ, Lundell T, Floudas D, Nagy LG, Barrasa JM, Hibbett DS, Martinez AT. 2013. Lignin-degrading peroxidases in Polyporales: an evolutionary survey based on 10 sequenced genomes. Mycologia 105:1428–1444. doi: 10.3852/13-059. [DOI] [PubMed] [Google Scholar]
- 13.Bourbonnais R, Paice MG. 1990. Oxidation of non-phenolic substrates—an expanded role for laccase in lignin biodegradation. FEBS Lett 267:99–102. doi: 10.1016/0014-5793(90)80298-W. [DOI] [PubMed] [Google Scholar]
- 14.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N, Liu J, Yang J, Lin Y, Yang Y, Ji L, Li M, Yuan H. 2016. Comparative analysis of the secretomes of Schizophyllum commune and other wood-decay basidiomycetes during solid-state fermentation reveals its unique lignocellulose-degrading enzyme system. Biotechnol Biofuels 9:42. doi: 10.1186/s13068-016-0461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang JW, Presley GN, Hammel KE, Ryu JS, Menke JR, Figueroa M, Hu DH, Orr G, Schilling JS. 2016. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc Natl Acad Sci U S A 113:10968–10973. doi: 10.1073/pnas.1608454113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanchette RA. 1995. Degradation of the lignocellulose complex in wood. Can J Bot 73:S999–S1010. doi: 10.1139/b95-350. [DOI] [Google Scholar]
- 18.Dick-Perez M, Zhang YA, Hayes J, Salazar A, Zabotina OA, Hong M. 2011. Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50:989–1000. doi: 10.1021/bi101795q. [DOI] [PubMed] [Google Scholar]
- 19.Xiao C, Anderson CT. 2013. Roles of pectin in biomass yield and processing for biofuels. Front Plant Sci 4:67. doi: 10.3389/fpls.2013.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, Cullen D. 2014. Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose. Appl Environ Microbiol 80:2062–2070. doi: 10.1128/AEM.03652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willfor S, Pranovich A, Tamminen T, Puls J, Laine C, Suurnakki A, Saake B, Uotila K, Simolin H, Hemming J, Holmbom B. 2009. Carbohydrate analysis of plant materials with uronic acid-containing polysaccharides: a comparison between different hydrolysis and subsequent chromatographic analytical techniques. Ind Crops Prod 29:571–580. doi: 10.1016/j.indcrop.2008.11.003. [DOI] [Google Scholar]
- 22.Choat B, Cobb AR, Jansen S. 2008. Structure and function of bordered pits: new discoveries and impacts on whole-plant hydraulic function. New Phytol 177:608–625. doi: 10.1111/j.1469-8137.2007.02317.x. [DOI] [PubMed] [Google Scholar]
- 23.Blanchette RA. 1991. Delignification by wood-decay fungi. Annu Rev Phytopathol 29:381–398. doi: 10.1146/annurev.py.29.090191.002121. [DOI] [Google Scholar]
- 24.Cowling EB, Merrill W. 1966. Nitrogen in wood and its role in wood deterioration. Can J Bot 44:1539–1554. doi: 10.1139/b66-167. [DOI] [Google Scholar]
- 25.Puls J. 1997. Chemistry and biochemistry of hemicelluloses: relationship between hemicellulose structure and enzymes required for hydrolysis. Macromol Symp 120:183–196. doi: 10.1002/masy.19971200119. [DOI] [Google Scholar]
- 26.Hiscox J, Clarkson G, Savoury M, Powell G, Savva I, Lloyd M, Shipcott J, Choimes A, Cumbriu XA, Boddy L. 2016. Effects of pre-colonisation and temperature on interspecific fungal interactions in wood. Fungal Ecol 21:32–42. doi: 10.1016/j.funeco.2016.01.011. [DOI] [Google Scholar]
- 27.Lindahl BD, Finlay RD. 2006. Activities of chitinolytic enzymes during primary and secondary colonization of wood by basidiomycetous fungi. New Phytol 169:389–397. doi: 10.1111/j.1469-8137.2005.01581.x. [DOI] [PubMed] [Google Scholar]
- 28.Lafond M, Navarro D, Haon M, Couturier M, Berrin JG. 2012. Characterization of a broad-specificity beta-glucanase acting on beta-(1,3)-, beta-(1,4)-, and beta-(1,6)-glucans that defines a new glycoside hydrolase family. Appl Environ Microbiol 78:8540–8546. doi: 10.1128/AEM.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen E, Vejlby K. 1953. A new polyphenol oxidase test. Physiol Plant 6:533. doi: 10.1111/j.1399-3054.1953.tb08410.x. [DOI] [Google Scholar]
- 30.Kerem Z, Jensen KA, Hammel KE. 1999. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: evidence for an extracellular hydroquinone-driven fenton reaction. FEBS Lett 446:49–54. doi: 10.1016/S0014-5793(99)00180-5. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka H, Yoshida G, Baba Y, Matsumura K, Wasada H, Murata J, Agawa M, Itakura S, Enoki A. 2007. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol 128:500–511. doi: 10.1016/j.jbiotec.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Liers C, Bobeth C, Pecyna M, Ullrich R, Hofrichter M. 2010. DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl Microbiol Biotechnol 85:1869–1879. doi: 10.1007/s00253-009-2173-7. [DOI] [PubMed] [Google Scholar]
- 33.Hilden K, Makela MR, Steffen KT, Hofrichter M, Hatakka A, Archer DB, Lundell TK. 2014. Biochemical and molecular characterization of an atypical manganese peroxidase of the litter-decomposing fungus Agrocybe praecox. Fungal Genet Biol 72:131–136. doi: 10.1016/j.fgb.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Chapela IH, Boddy L. 1988. The fate of early fungal colonizers in beech branches decomposing on the forest floor. FEMS Microbiol Ecol 53:273–283. doi: 10.1111/j.1574-6968.1988.tb02674.x. [DOI] [Google Scholar]
- 35.Boddy L, Hiscox J. 2016. Fungal ecology: principles and mechanisms of colonization and competition by saprotrophic fungi. Microbiol Spectr 4. doi: 10.1128/microbiolspec.FUNK-0019-2016. [DOI] [PubMed] [Google Scholar]
- 36.Heilmann-Clausen J, Boddy L. 2005. Inhibition and stimulation effects in communities of wood decay fungi: exudates from colonized wood influence growth by other species. Microb Ecol 49:399–406. doi: 10.1007/s00248-004-0240-2. [DOI] [PubMed] [Google Scholar]
- 37.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 38.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. 2008. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics 24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S, Pevzner PA. 2014. MS-GF plus makes progress towards a universal database search tool for proteomics. Nat Commun 5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Wymelenberg AV, Gaskell J, Lindquist E, Sabat G, BonDurant SS, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavin JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kues U, Ramaiya P, Lucash S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, et al. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A 106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newell SY, Arsuffi TL, Fallon RD. 1988. Fundamental procedures for determining ergosterol content of decaying plant material by liquid chromatography. Appl Environ Microbiol 54:1876–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schilling JS, Jellison J. 2005. Oxalate regulation by two brown rot fungi decaying oxalate-amended and non-amended wood. Holzforschung 59:681–688. doi: 10.1515/HF.2005.109. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (41) partner repository, with the data set identifier PXD009480 (https://doi.org/10.6019/PXD009480).