Abstract
Ripening affects the nutritional contents and quality of fleshy fruits, and it plays an important role during the process of fruit development. Studies have demonstrated that ubiquitin-conjugating (UBC or E2) genes can regulate fruit ripening, but the characterization of UBCs in pear is not well documented. The recently published genome-wide sequences of Pyrus bretschneideri and Pyrus communis have allowed a comprehensive analysis of this important gene family in pear. Using bioinformatics approaches, we identified 83 (PbrUBCs) and 84 (PcpUBCs) genes from P. bretschneideri and P. communis, respectively, which were divided into 13 subfamilies. In total, 198 PbrUBC paralogous, 215 PcpUBC paralogous, and 129 orthologous gene pairs were detected. Some paralogous gene pairs were found to be distributed on the same chromosome, suggesting that these paralogs may be caused by tandem duplications. The expression patterns of most UBC genes were divergent between Pyrus bretschneideri and Pyrus communis during pear fruit development. Remarkably, the transcriptome data showed that UBC genes might play a more important role in fruit ripening for further study. This is the first report on the systematic analysis of two Pyrus UBC gene families, and these data will help further study the role of UBC genes in fruit development and ripening, as well as contribute to the functional verification of UBC genes in pear.
Keywords: UBC, Pyrus, expression, gene pairs
1. Introduction
Ubiquitination is an essential cellular process for eukaryotes [1]. The ubiquitin proteasome pathway involves many aspects of eukaryotic cell regulation because of its ability to degrade intracellular proteins [2,3]. Ubiquitin conjugation is a multistep reaction mediated by the action of three enzymes, including E1s (ubiquitin-activating enzymes), E2s (ubiquitin-conjugating enzymes), and E3s (ubiquitin ligases). E2s act in the middle step of the protein ubiquitination pathway [4]. Previous reports suggested that the E2 family members have a certain degree of expansion during evolution, for example, more ancestral eukaryotes such as algae have fewer E2 enzymes (< or =20) than certain plants and animals (>40) [5]. The Saccharomyces cerevisiae genome encodes 13 UBC proteins [6]; 19, 18, and 12 UBC proteins are identified in the algae Chlamydomonas reinhardtii, Micromonas sp. RCC299, and Ostreococcus, respectively; 20 in Caenorhabditis elegans [7]; and 75, 74, 52, 48, 34, and 37 UBC proteins in Zea mays, Musa nana, Solanum lycopersicum, Oryza sativa, Carica papaya, and A. thaliana, respectively [1,8,9,10,11,12].
Researchers have studied UBC genes in some plants, indicating that these genes are involved in tolerance against biotic and abiotic stresses, and plant growth and development [1,8,9,10,11,12]. For example, the A. thaliana UBC1 and AtUBC2 participate in the activation of the flowering suppressor FLC gene and inhibit flowering [13]. Overexpression of the AtUBC32 gene in A. thaliana reduced the sensitivity of plants to salt stress [14]. Overexpression of Vigna radiata UBC1 (VrUBC1), Arachis hypogaea UBC2 (AhUBC2), or Glycine max UBC2 (GmUBC2) in A. thaliana can enhance plant drought resistance [15,16,17]. The A. thaliana UBC13 (AtUBC13) has been implicated in iron deficiency responses and epidermal cell differentiation [18,19]. Additionally, AtUBC21 (AtPEX4) has been shown to be specific for ubiquitination in peroxisome maintenance [20]. In Z. mays, 16 and 48 ZmUBCs were significantly up-regulated in response to drought and salt stress [10], respectively. Similarly, in O. sativa, 14 OsUBC genes were differentially expressed under salt and drought stress [21].
Pear (Pyrus spp.) is one of the leading cultivated fruit trees which are widely grown in temperate regions, and its fleshy fruits play an important role in human health and nutrition [22,23]. The pear is the third largest temperate fruit tree after apple (Malus domestica) and grape (Vitis vinifera). Previously published manuscripts have carried out some studies on the mechanisms related to fruit ripening, as the fruit ripening is an important and complex process. The UBC genes play an important role in the fruit ripening process. In S. lycopersicum, Wang et al. (2014) found that SlUBC32 was down-regulated in the S. lycopersicum rin mutant and up-regulated during fruit ripening, indicating that this gene plays a key role in the regulation of fruit ripening [11]. In M. nana, Dong et al. (2016) identified that the expressions of 32 MaUBCs were increased or decreased during different ripening stages [9]. In C. papaya, Jue et al. (2017) suggested that 13 and two CpUBCs were up-regulated and down-regulated during one and two ripening stages, respectively [12]. However, the UBC genes involved in fruit ripening in two Pyrus species (P. bretschneideri and P. communis) have not yet been identified. To further understand the function of UBC genes, we carried out a systematic analysis of two Pyrus species, including phylogenetic relationships, sequence characteristics, chromosomal locations, and the expression differences during fruit development. These results highlight the role of UBC genes in pear fruit development and provide important information for further exploring the functional differences between two Pyrus species.
2. Materials and Methods
2.1. Sequence Retrieval and Identification of UBC Genes
To identify the UBC proteins in two Pyrus species (i.e., P. bretschneideri and P. communis), we used HMMER v3.1b2 obtained from the HMMER website (http://www.hmmer.org/download.html) [24]. The HMM profile of the UBC domain (PF00179) was downloaded from Pfam 31.0 (http://pfam.xfam.org/) [25]. Using HMMER v3.1b2 software, an HMM search was carried out for P. bretschneideri [23] and P. communis [26] genomes with a significance e-value of 0.001. SMART [27], Pfam [25], and INTERPRO [28] were used to confirm the presence of the UBC domain. Information on PbrUBCs and PcpUBCs, including intron and exon numbers, chromosomal locations, and coding sequences (CDS), was obtained from the GigdDB (http://gigadb.org/) and GDR databases (https://www.rosaceae.org/) [29], respectively.
2.2. Phylogenetic Analysis and Gene Duplication
The MUSCLE program was used to perform the alignments of all UBC amino acid sequences using default parameters [30]. ModelFinder was used to detect the best substitution model of these alignment sequences. The phylogenetic tree was generated using full-length sequences by the Maximum Likelihood (ML) method with 1000 bootstrap replications and the VT + G4 model implemented in IQ-TREE software obtained from IQ-TREE (http://www.iqtree.org/) [31]. The FigTree software (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the ML tree. We have utilized the MCscanX software for the analysis of gene duplication events [32], and the “add_ka_and_ks_to_collinearity.pl” script for the analysis of the non-synonymous (Ka)/synonymous (Ks) substitution ratio.
2.3. Gene Structure and Motif Analysis
The GFF files of P. bretschneideri and P. communis were downloaded from GigdDB (http://gigadb.org/) and GDR databases (https://www.rosaceae.org/), respectively. The TBtools software was used to plot the map of the exon-intron structure [33]. The MEME online tool was used to search for the conservative motif of both PbrUBC and PcpUBC proteins, with the maximum width of 200 amino acids and a limit of 20 motifs, and other default parameters [34].
2.4. Expression Analysis
To further understand the expression of UBC genes in both P. bretschneideri and P. communis, we downloaded the RNA-Seq data from the public NCBI database. The sample details and accession numbers for the above data are presented in the availability of data and materials section. The FASTX-toolkit was used to remove the low-quality base-calls (Q < 20) of raw reads [35]. The TopHat2 software was used to map the clean reads to the reference genome with default parameters [36], and the Cufflinks software was used to assemble and calculate the expression FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) values [37]. The R software was used to plot the heatmap of these UBC genes.
2.5. Expression Correlation of Orthologous UBC Genes in Two Pyrus Species
Using RNA-Seq data, we obtained the expression profiles of orthologous UBC genes. Then, we estimated the similarity between the expression patterns of the orthologous gene pair by using Pearson’s correlation coefficient (r). The degree of expression diversity was confirmed by significant values (r) based on previous studies [38,39]. In general, r > 0.5, 0.3 < r < 0.5, and r < 0.3 suggest non-divergence, ongoing divergent, and divergent, respectively [38,39].
2.6. Availability of Data and Materials
P. bretschneideri fruit developmental stage 1 (Fruit_stage1: 15DAB), Accession: SRX1595645; P. bretschneideri Fruit_stage2 (30DAB), Accession: SRX1595646; P. bretschneideri Fruit_stage3 (55DAB), Accession: SRX1595647; P. bretschneideri Fruit_stage4 (85 DAB), Accession: SRX1595648; P. bretschneideri Fruit_stage5 (115 DAB), Accession: SRX1595650; P. bretschneideri Fruit_stage6 (mature stage), Accession: SRX1595651; P. bretschneideri Fruit_stage7 (fruit senescence stage), Accession: SRX1595652; P. communis Fruit_stage1 (15DAB), Accession: SRX1595636; P. communis Fruit_stage2 (30DAB), Accession: SRX1595637; P. communis Fruit_stage3 (55DAB), Accession: SRX1595638; P. communis Fruit_stage4 (85 DAB), Accession: SRX1595639; P. communis Fruit_stage5 (115 DAB), Accession: SRX1595640; P. communis Fruit_stage6 (mature stage), Accession: SRX1595641; P. communis Fruit_stage7 (fruit senescence stage), Accession: SRX1595644. P. bretschneideri drought-tolerant for 0 h, Accession: SRX4110141; P. bretschneideri drought-tolerant for 1 h, Accession: SRX4110140; P. bretschneideri drought-tolerant for 3 h, Accession: SRX4110143; P. bretschneideri drought-tolerant for 6 h, Accession: SRX4110142; P. bretschneideri recovery for 24 h, Accession: SRX4110139.
3. Results
3.1. Identification of UBC Genes in Two Pyrus Species
The protein of P. bretschneideri and P. communis was downloaded from the GigdDB (http://gigadb.org/) and GDR database (https://www.rosaceae.org/), respectively. To identify the potential UBC protein family in two Pyrus species, we obtained the UBC domain from the Pfam database and generated the HMM profile in the HMMER 3.0 package. A total of 85 and 94 putative UBC proteins were identified by searching the generated HMM profile with the E-value of 0.001 against the P. bretschneideri and P. communis protein sequence database, respectively. Further scanning of these UBC proteins for the UBC domain was conducted by a motif scan using the INTERPRO, SMART, and Pfam database, and found some UBC proteins not contained in the UBC domain. Finally, we identified 83 and 84 putative UBC proteins in the P. bretschneideri and P. communis genome, named PbrUBC01-83 and PcpUBC01-84 according to their order on the chromosomes, respectively. The information of these PbrUBC and PcpUBC genes, such as chromosome location, gene identifier, and protein length (aa), is shown in Table 1.
Table 1.
The detailed information of UBC family members in both P. bretschneideri and P. communis.
| Gene Name | Gene Identifier | Chromosme | 5′ End | 3′ End | Protein Size (aa) |
|---|---|---|---|---|---|
| PcpUBC01 | PCP011547.1 | chr2 | 908379 | 915288 | 378 |
| PcpUBC02 | PCP008190.1 | chr2 | 2303235 | 2304455 | 406 |
| PcpUBC03 | PCP026226.1 | chr2 | 9163625 | 9165622 | 161 |
| PcpUBC04 | PCP025062.1 | chr2 | 11398030 | 11400462 | 160 |
| PcpUBC05 | PCP029820.1 | chr2 | 11983749 | 11985588 | 154 |
| PcpUBC06.1 | PCP020092.1 | chr3 | 547762 | 556678 | 986 |
| PcpUBC06.2 | PCP041697.1 | chr3 | 551276 | 556678 | 387 |
| PcpUBC07 | PCP000885.1 | chr3 | 3236140 | 3247151 | 841 |
| PcpUBC08 | PCP013747.1 | chr3 | 4266095 | 4267481 | 148 |
| PcpUBC09 | PCP031109.1 | chr3 | 6092388 | 6096394 | 192 |
| PcpUBC10 | PCP029016.1 | chr3 | 9480497 | 9483572 | 146 |
| PcpUBC11 | PCP029037.1 | chr3 | 11156660 | 11161769 | 769 |
| PcpUBC12 | PCP008664.1 | chr3 | 16102272 | 16105426 | 277 |
| PcpUBC13.1 | PCP006596.1 | chr4 | 3944944 | 3952328 | 304 |
| PcpUBC13.2 | PCP033076.1 | chr4 | 3944944 | 3946278 | 148 |
| PcpUBC14 | PCP032557.1 | chr4 | 13065481 | 13065759 | 92 |
| PcpUBC15 | PCP004584.1 | chr5 | 4531710 | 4535205 | 920 |
| PcpUBC16 | PCP000343.1 | chr5 | 6602829 | 6603942 | 120 |
| PcpUBC17 | PCP000325.1 | chr5 | 6785831 | 6791593 | 1148 |
| PcpUBC18 | PCP013561.1 | chr6 | 7061585 | 7063890 | 183 |
| PcpUBC19.1 | PCP003994.1 | chr6 | 9051182 | 9054610 | 490 |
| PcpUBC19.2 | PCP038680.1 | chr6 | 9053326 | 9054610 | 152 |
| PcpUBC20 | PCP006677.1 | chr6 | 10001882 | 10012807 | 890 |
| PcpUBC21 | PCP025848.1 | chr7 | 1250213 | 1253437 | 172 |
| PcpUBC22 | PCP025852.1 | chr7 | 1277835 | 1279055 | 189 |
| PcpUBC23 | PCP027463.1 | chr7 | 2394123 | 2396438 | 148 |
| PcpUBC24 | PCP039252.1 | chr7 | 5395929 | 5399115 | 181 |
| PcpUBC25 | PCP041398.1 | chr7 | 9425098 | 9426267 | 78 |
| PcpUBC26 | PCP023269.1 | chr7 | 14434633 | 14435892 | 419 |
| PcpUBC27 | PCP018282.1 | chr8 | 526845 | 534958 | 439 |
| PcpUBC28 | PCP024977.1 | chr8 | 2320747 | 2321922 | 189 |
| PcpUBC29 | PCP014622.1 | chr8 | 5359408 | 5371794 | 989 |
| PcpUBC30 | PCP013024.1 | chr8 | 12182295 | 12185615 | 188 |
| PcpUBC31.1 | PCP025013.1 | chr9 | 5181126 | 5187890 | 423 |
| PcpUBC31.2 | PCP042586.1 | chr9 | 5181126 | 5183531 | 195 |
| PcpUBC32.1 | PCP029211.1 | chr9 | 5697502 | 5704326 | 356 |
| PcpUBC32.2 | PCP037499.1 | chr9 | 5697502 | 5700526 | 237 |
| PcpUBC33 | PCP022975.1 | chr10 | 4876099 | 4877045 | 137 |
| PcpUBC34 | PCP006043.1 | chr10 | 13347113 | 13347565 | 150 |
| PcpUBC35 | PCP019803.1 | chr10 | 17163601 | 17167148 | 921 |
| PcpUBC36 | PCP037747.1 | chr11 | 5179080 | 5182742 | 153 |
| PcpUBC37 | PCP003934.1 | chr11 | 5615653 | 5620782 | 686 |
| PcpUBC38 | PCP014945.1 | chr11 | 12048835 | 12052442 | 227 |
| PcpUBC39 | PCP007359.1 | chr11 | 13280818 | 13282231 | 148 |
| PcpUBC40 | PCP041407.1 | chr12 | 2420398 | 2421551 | 148 |
| PcpUBC41 | PCP043134.1 | chr12 | 15435377 | 15437609 | 146 |
| PcpUBC42 | PCP003304.1 | chr13 | 3769096 | 3771160 | 152 |
| PcpUBC43 | PCP028885.1 | chr13 | 7446155 | 7448170 | 373 |
| PcpUBC44 | PCP010847.1 | chr14 | 1973506 | 1975549 | 195 |
| PcpUBC45 | PCP000493.1 | chr14 | 3899314 | 3905194 | 754 |
| PcpUBC46 | PCP006717.1 | chr14 | 4144989 | 4159895 | 1373 |
| PcpUBC47 | PCP033096.1 | chr14 | 4155864 | 4157211 | 152 |
| PcpUBC48 | PCP017973.1 | chr14 | 6488805 | 6490257 | 144 |
| PcpUBC49 | PCP031605.1 | chr14 | 8511513 | 8512945 | 265 |
| PcpUBC50.1 | PCP001680.1 | chr14 | 12856983 | 12862739 | 444 |
| PcpUBC50.2 | PCP032125.1 | chr14 | 12860593 | 12862739 | 146 |
| PcpUBC51 | PCP022610.1 | chr15 | 2571489 | 2577399 | 467 |
| PcpUBC52 | PCP032902.1 | chr15 | 3995850 | 3996286 | 78 |
| PcpUBC53 | PCP013613.1 | chr15 | 5801442 | 5811234 | 828 |
| PcpUBC54 | PCP005846.1 | chr15 | 15546784 | 15549513 | 160 |
| PcpUBC55 | PCP020991.1 | chr15 | 18866853 | 18872463 | 319 |
| PcpUBC56 | PCP014540.1 | chr15 | 21362811 | 21364686 | 161 |
| PcpUBC57 | PCP006806.1 | chr16 | 1486195 | 1494553 | 341 |
| PcpUBC58 | PCP026158.1 | chr16 | 4290275 | 4292359 | 152 |
| PcpUBC59 | PCP021281.1 | chr16 | 5293770 | 5294891 | 373 |
| PcpUBC60 | PCP021299.1 | chr16 | 5419315 | 5421158 | 307 |
| PcpUBC61 | PCP010786.1 | chr17 | 6829215 | 6832764 | 267 |
| PcpUBC62 | PCP042470.1 | chr17 | 15393404 | 15402997 | 596 |
| PcpUBC63 | PCP012277.1 | chr17 | 17062013 | 17063062 | 349 |
| PcpUBC64 | PCP007143.1 | chr17 | 17728444 | 17733945 | 621 |
| PcpUBC65 | PCP018399.1 | scaffold00612 | 73468 | 83219 | 651 |
| PcpUBC66 | PCP021641.1 | scaffold00634 | 76924 | 79227 | 148 |
| PcpUBC67 | PCP007398.1 | scaffold00805 | 133398 | 134716 | 174 |
| PcpUBC68 | PCP018521.1 | scaffold00852 | 13601 | 20410 | 278 |
| PcpUBC69 | PCP004264.1 | scaffold00983 | 16646 | 19316 | 148 |
| PcpUBC70 | PCP021954.1 | scaffold01394 | 34467 | 36817 | 191 |
| PcpUBC71 | PCP007629.1 | scaffold01465 | 20581 | 22904 | 291 |
| PcpUBC72 | PCP002761.1 | scaffold01522 | 35381 | 36867 | 160 |
| PcpUBC73 | PCP020361.1 | scaffold01593 | 68663 | 70923 | 191 |
| PcpUBC74 | PCP045045.1 | scaffold01774 | 29298 | 33191 | 754 |
| PcpUBC75 | PCP001255.1 | scaffold01881 | 39525 | 44855 | 1147 |
| PcpUBC76 | PCP004531.1 | scaffold01923 | 15838 | 21766 | 337 |
| PcpUBC77 | PCP028469.1 | scaffold02358 | 32352 | 36077 | 463 |
| PcpUBC78 | PCP022164.1 | scaffold02454 | 7209 | 11865 | 389 |
| PcpUBC79 | PCP012568.1 | scaffold02548 | 27443 | 29342 | 178 |
| PcpUBC80 | PCP043259.1 | scaffold04878 | 3046 | 6910 | 134 |
| PcpUBC81 | PCP001444.1 | scaffold05041 | 9798 | 11719 | 183 |
| PcpUBC82 | PCP022346.1 | scaffold17014 | 829 | 2135 | 180 |
| PcpUBC83 | PCP040042.1 | scaffold23907 | 470 | 1254 | 125 |
| PcpUBC84 | PCP011188.1 | scaffold27287 | 92 | 1717 | 232 |
| PbrUBC01 | Pbr021045.1 | Chr1 | 3279060 | 3282148 | 149 |
| PbrUBC02 | Pbr022046.1 | Chr1 | 5355491 | 5357926 | 192 |
| PbrUBC03 | Pbr018716.1 | Chr1 | 9129424 | 9132884 | 149 |
| PbrUBC04 | Pbr013632.1 | Chr1 | 9364933 | 9367901 | 149 |
| PbrUBC05 | Pbr029889.2 | Chr2 | 11205065 | 11207039 | 195 |
| PbrUBC06 | Pbr025178.1 | Chr2 | 13123810 | 13126801 | 161 |
| PbrUBC07 | Pbr022865.1 | Chr2 | 15059120 | 15062337 | 184 |
| PbrUBC08 | Pbr022866.1 | Chr2 | 15070696 | 15073441 | 182 |
| PbrUBC09 | Pbr040498.1 | Chr2 | 15603251 | 15605586 | 162 |
| PbrUBC10 | Pbr024232.2 | Chr3 | 7098302 | 7101956 | 160 |
| PbrUBC11 | Pbr027637.1 | Chr3 | 9231735 | 9233562 | 181 |
| PbrUBC12 | Pbr023139.1 | Chr3 | 17780615 | 17782004 | 149 |
| PbrUBC13 | Pbr000740.1 | Chr3 | 19467052 | 19469759 | 170 |
| PbrUBC14 | Pbr013150.2 | Chr3 | 22108425 | 22111475 | 149 |
| PbrUBC15 | Pbr034016.1 | Chr3 | 25287937 | 25291916 | 168 |
| PbrUBC16 | Pbr030934.3 | Chr4 | 12531057 | 12533684 | 161 |
| PbrUBC17 | Pbr027417.1 | Chr5 | 12936909 | 12946872 | 201 |
| PbrUBC18 | Pbr027395.1 | Chr5 | 13135775 | 13142057 | 1149 |
| PbrUBC19 | Pbr000361.1 | Chr5 | 25903450 | 25906904 | 853 |
| PbrUBC20 | Pbr011471.1 | Chr6 | 1539205 | 1541448 | 153 |
| PbrUBC21 | Pbr009129.1 | Chr6 | 7334609 | 7336195 | 77 |
| PbrUBC22 | Pbr014124.1 | Chr6 | 9284322 | 9290606 | 296 |
| PbrUBC23 | Pbr018194.1 | Chr6 | 13523851 | 13526524 | 184 |
| PbrUBC24 | Pbr040529.1 | Chr6 | 16795923 | 16797553 | 181 |
| PbrUBC25 | Pbr032353.1 | Chr7 | 10867139 | 10869902 | 224 |
| PbrUBC26 | Pbr006183.1 | Chr8 | 15645575 | 15647047 | 190 |
| PbrUBC27 | Pbr004154.1 | Chr8 | 4690292 | 4691740 | 129 |
| PbrUBC28 | Pbr032653.1 | Chr9 | 4250106 | 4253822 | 461 |
| PbrUBC29 | Pbr032645.1 | Chr9 | 4153561 | 4154900 | 279 |
| PbrUBC30 | Pbr031810.1 | Chr10 | 268749 | 273571 | 458 |
| PbrUBC31 | Pbr016259.1 | Chr10 | 4489977 | 4494846 | 922 |
| PbrUBC32 | Pbr009080.1 | Chr10 | 10152737 | 10153189 | 151 |
| PbrUBC33 | Pbr009081.1 | Chr10 | 10158759 | 10159211 | 151 |
| PbrUBC34 | Pbr020740.1 | Chr10 | 17324391 | 17325467 | 149 |
| PbrUBC35 | Pbr020719.1 | Chr10 | 17573570 | 17574635 | 149 |
| PbrUBC36 | Pbr020703.1 | Chr10 | 17785964 | 17790523 | 891 |
| PbrUBC37 | Pbr038220.3 | Chr11 | 4285552 | 4289283 | 190 |
| PbrUBC38 | Pbr038323.1 | Chr11 | 5463176 | 5468616 | 589 |
| PbrUBC39 | Pbr017901.1 | Chr11 | 11843306 | 11844937 | 181 |
| PbrUBC40 | Pbr031559.1 | Chr11 | 13000377 | 13002724 | 149 |
| PbrUBC41 | Pbr041320.1 | Chr11 | 21287679 | 21289091 | 152 |
| PbrUBC42 | Pbr017298.1 | Chr11 | 24728725 | 24731067 | 149 |
| PbrUBC43 | Pbr028474.1 | Chr12 | 194166 | 195789 | 149 |
| PbrUBC44 | Pbr016440.1 | Chr12 | 3192332 | 3192601 | 90 |
| PbrUBC45 | Pbr039044.1 | Chr12 | 10209302 | 10211625 | 309 |
| PbrUBC46 | Pbr015391.1 | Chr12 | 19750784 | 19753479 | 147 |
| PbrUBC47 | Pbr010810.1 | Chr13 | 289004 | 291284 | 149 |
| PbrUBC48 | Pbr011958.2 | Chr13 | 9713060 | 9715379 | 231 |
| PbrUBC49 | Pbr010372.1 | Chr14 | 2425473 | 2427613 | 147 |
| PbrUBC50 | Pbr010424.1 | Chr14 | 2933078 | 2935217 | 147 |
| PbrUBC51 | Pbr038166.1 | Chr14 | 7236972 | 7240490 | 273 |
| PbrUBC52 | Pbr026720.1 | Chr14 | 8739052 | 8742374 | 169 |
| PbrUBC53 | Pbr027115.1 | Chr14 | 12937009 | 12939516 | 196 |
| PbrUBC54 | Pbr005908.1 | Chr15 | 2962026 | 2964448 | 162 |
| PbrUBC55 | Pbr009224.1 | Chr15 | 4281248 | 4283960 | 158 |
| PbrUBC56 | Pbr019673.1 | Chr15 | 7696112 | 7697156 | 100 |
| PbrUBC57 | Pbr016945.1 | Chr15 | 14705287 | 14708021 | 524 |
| PbrUBC58 | Pbr017248.1 | Chr15 | 19933740 | 19935641 | 141 |
| PbrUBC59 | Pbr017249.1 | Chr15 | 19937558 | 19939328 | 182 |
| PbrUBC60 | Pbr015294.2 | Chr15 | 23696990 | 23705032 | 371 |
| PbrUBC61 | Pbr024308.1 | Chr15 | 24667752 | 24669854 | 153 |
| PbrUBC62 | Pbr024286.1 | Chr15 | 24961413 | 24963994 | 190 |
| PbrUBC63 | Pbr024279.1 | Chr15 | 25164896 | 25167005 | 178 |
| PbrUBC64 | Pbr017425.1 | Chr15 | 26387149 | 26392677 | 702 |
| PbrUBC65 | Pbr040652.1 | Chr15 | 36951735 | 36952300 | 112 |
| PbrUBC66 | Pbr020836.2 | Chr15 | 42195161 | 42198738 | 228 |
| PbrUBC67 | Pbr012108.1 | Chr16 | 3304638 | 3306350 | 202 |
| PbrUBC68 | Pbr013690.1 | Chr16 | 9644295 | 9646021 | 188 |
| PbrUBC69 | Pbr022472.1 | Chr17 | 2772442 | 2779627 | 468 |
| PbrUBC70 | Pbr026816.1 | Chr17 | 3678331 | 3681687 | 454 |
| PbrUBC71 | Pbr034051.1 | Chr17 | 5335721 | 5339074 | 454 |
| PbrUBC72 | Pbr008641.1 | Chr17 | 6164722 | 6165771 | 350 |
| PbrUBC73 | Pbr040232.1 | Chr17 | 20655869 | 20656618 | 106 |
| PbrUBC74 | Pbr003941.1 | scaffold1182.0 | 19987 | 21731 | 175 |
| PbrUBC75 | Pbr005003.1 | scaffold1241.0 | 889 | 1709 | 133 |
| PbrUBC76 | Pbr005004.1 | scaffold1241.0 | 11548 | 13331 | 182 |
| PbrUBC77 | Pbr006049.1 | scaffold1301.0 | 22029 | 24744 | 149 |
| PbrUBC78 | Pbr009005.1 | scaffold1564.0 | 5159 | 7986 | 149 |
| PbrUBC79 | Pbr028213.1 | scaffold467.0 | 324186 | 326069 | 190 |
| PbrUBC80 | Pbr028219.1 | scaffold467.0 | 348935 | 352574 | 172 |
| PbrUBC81 | Pbr032413.2 | scaffold581.0.1 | 30262 | 34773 | 126 |
| PbrUBC82 | Pbr034367.1 | scaffold640.0 | 26565 | 30347 | 154 |
| PbrUBC83 | Pbr042566.1 | scaffold992.0 | 77372 | 83394 | 147 |
Note: Red logo represents tandem duplication.
3.2. Phylogenetic Analysis of UBC Genes in Two Pyrus Species
To gain insight into the evolutionary relationships of UBC genes in two Pyrus species, we built an ML tree with all PbrUBCs and PcpUBCs sequences using IQ-TREE software with the VT+G4 model, and investigated the gene structures of PbrUBC and PcpUBC genes based on the GFF3 annotation files. Phylogenetic analysis revealed that these PbrUBCs and PcpUBCs could be clustered into 13 subfamilies (Figure 1), using A. thaliana UBC genes as a template [1]. Subfamily H had 45 Pyrus UBC members and was the largest clade of all subfamilies, which represented 26.01% of the total Pyrus UBC genes. However, subfamily M and subfamily G only contained three and two Pyrus UBC members, respectively. We also found that the distribution of Pyrus UBC members was uneven in some subfamilies, suggesting that they had undergone dynamic changes from the common ancestor. Based on the phylogenetic analysis, we found that the UBC members from these Pyrus species presented a higher similarity with each other, which was consistent with their (i.e., P. bretschneideri and P. communis) evolutionary relationship. Additionally, we also detected the orthologous gene pairs between P. bretschneideri and P. communis. Finally, 129 orthologous gene pairs were found in these Pyrus species (Figure 2 and Table S1). This orthologous analysis supported the evolutionary relationships and the classification of subfamilies of UBC genes between the P. bretschneideri and P. communis genome.
Figure 1.
Phylogenetic tree of UBC genes from P. bretschneideri, P. communis, and A. thaliana. The phylogenetic tree was built using IQ-TREE software, and HsUfc1 from Homo sapiens was used as the out-group. According to published articles, the ML tree could be divided into 13 subfamilies (A–M).
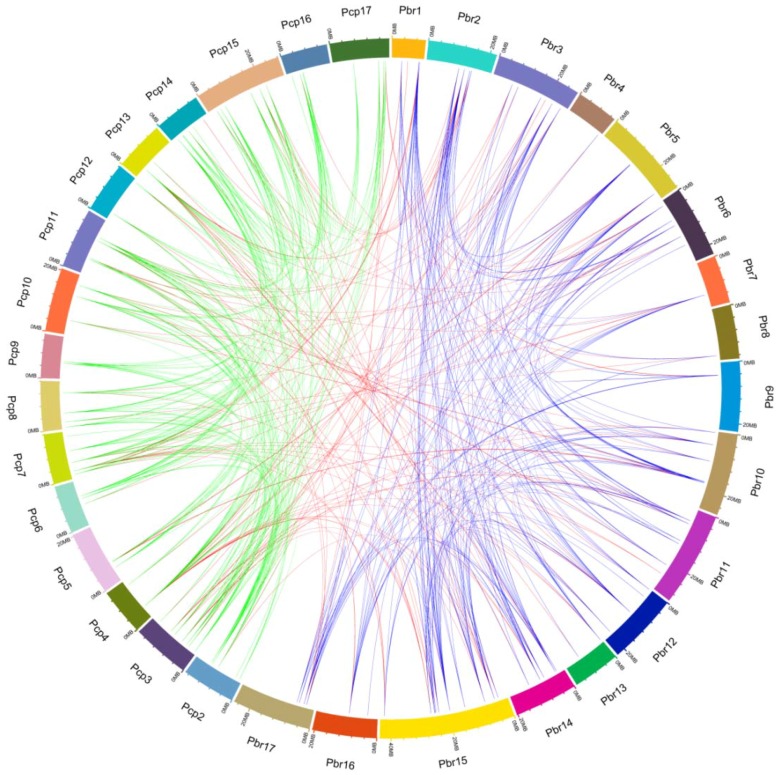
Figure 2.
Microsynteny of UBC genes across P. bretschneideri and P. communis. The outermost scale represents the megabases (Mb). The P. communis and P. bretschneideri chromosomes are labeled Pcp and Pbr, and are represented by different color boxes, respectively. Blue, green, and red lines represent the P. bretschneideri paralogous gene pairs, P. communis paralogous gene pairs, and orthologous gene pairs, respectively.
Observation of the gene structure in these two Pyrus species UBC genes showed that the numbers of introns in the 83 PbrUBC and 84 PcpUBC genes varied from 0 (PbrUBC44, PcpUBC34, PbrUBC33, PcpUBC14, PbrUBC32, PbrUBC72, PcpUBC02, PcpUBC34, PcpUBC59, and PcpUBC63) to 16 (PcpUBC29) (Figure S1). Additionally, we found that most of the Pyrus UBC genes clustered in the same subfamily contained highly similar gene structure maps, including intron numbers and exon length. For instance, PbrUBC53 and PcpUBC44 in the subfamily C contained four introns, and PbrUBC10 and PbrUBC37 in the subfamily B had five introns. We also scanned the conserved motifs in these UBC genes, and found that motif 2, −3, and −15 encoded the UBC domain (Figure S2). To sum up, the gene structures and conserved motifs of UBC genes were basically consistent with the above evolutionary relationship.
In general, the different protein isoforms produced by alternative splicing may affect the diversity of transcriptomics and proteomics, ultimately affecting gene expression regulation and protein function. In our study, the occurrence of alternative splicing events was revealed in the UBC family during evolution, such as PcpUBC06, PcpUBC13, PcpUBC19, PcpUBC31, PcpUBC32, and PcpUBC50 (Table 1). The mRNAs of PcpUBC13.1/PcpUBC13.2, PcpUBC19.1/PcpUBC19.2, PcpUBC32.1/PcpUBC32.2, and PcpUBC50.1/PcpUBC50.2, which are produced by variable splicing, are different in the 3′-end. However, the mRNAs of PcpUBC06.1/PcpUBC06.2 and PcpUBC31.1/PcpUBC31.2, which are produced by variable splicing, are different in the 5′-end (Figure S3). These results suggested that changes in the transcript sequence of the UBC gene caused by alternative splicing events may have an effect on the interaction ability and function of the encoded proteins.
3.3. Chromosomal Distribution and Gene Duplication of UBC Genes in Two Pyrus Species
In the present study, 167 genes were identified as members of the UBC gene family, with 83 PbrUBC genes in P. bretschneideri and 84 PcpUBC genes in P. communis (Table 1). Then, we determined the chromosomal distribution of each UBC gene. As shown in Figure 3 and Table 1, the distribution of 167 UBC genes on the chromosome is random, and some of them were located on scaffolds. The genome maps of the UBC genes suggested that PbrUBC genes were dispersed across all chromosomes; however, PcpUBC genes were mainly found on 16 out of 17 chromosomes, except for chromosome 1. In the P. bretschneideri genome, chromosome 15 had the maximum number of PbrUBC genes (13), while chromosome 7 contained only one gene (PbrUBC15) gene. In the P. communis genome, both chromosome 3 and 14 contained the most PcpUBC genes, followed by chromosome 7 (6) and 15 (6) (Figure 3).
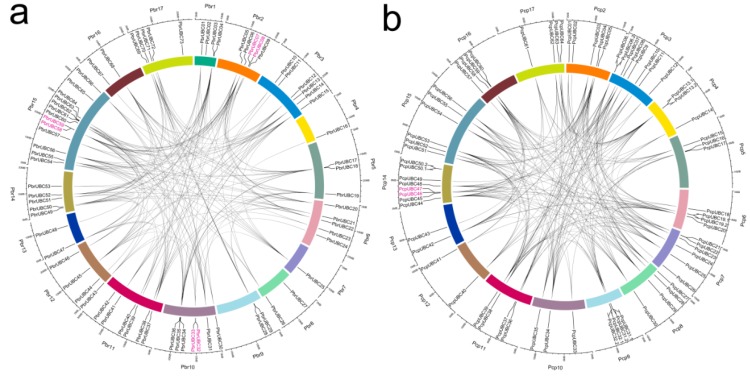
Figure 3.
Localization and duplication of UBC genes in the P. bretschneideri (a) and P. communis (b) genome, respectively. The localizations of PbrUBCs and PcpUBCs mapped on the P. bretschneideri and P. communis genome, respectively, were obtained from Circos software [40]. The P. communis and P. bretschneideri chromosomes are labeled Pcp and Pbr, and are represented by different color boxes, respectively. Red regions indicate tandem duplication, and grey lines represent segment duplication. The outermost scale represents the megabases (Mb).
Gene duplication contributes to the expansion of gene family members and diversification of protein functions. In general, if two genes are collinear, they are considered to have evolved from a duplication event. In order to further investigate the expansion mechanism of UBC gene family members, the occurrence of segmental duplication and tandem duplication events were analyzed during the evolution of this gene family. Finally, 198 and 215 duplication events (Figure 3) of the P. bretschneideri and P. communis UBC genes were identified, respectively. Among these duplication gene pairs, four and one gene pairs were identified to have evolved from tandem duplications in P. bretschneideri and P. communis, respectively, and the remaining gene pairs were involved in segmental duplications. Additionally, a series of several-for-one duplication events in P. bretschneideri and P. communis UBC genes were observed, such as PbrUBC07/PbrUBC26, PbrUBC07/PbrUBC20, PcpUBC12/PcpUBC34, and PcpUBC12/PcpUBC48, and it is envisaged that these genes may contribute to the expansion of UBC gene family members during evolution. The pear genome shared two whole-genome duplication (WGD) events, the ancient WGD occurred in ~140 MYA (Millions of years ago) (Ks ~ 1.5–1.8) and the recent WGD occurred in 30–45 MYA (Ks ~ 0.15–0.3). Subsequently, 15 and 14 duplication gene pairs (Table S2) were identified as being derived from ancient WGDs and recent WGDs in the P. communis genome, respectively. In the P. bretschneideri genome, 17 duplication gene pairs were evolved from the recent WGDs, and 13 from the ancient WGDs. These results suggested that two WGDs contribute to the expansion of UBC gene family members in the Pyrus genome.
3.4. Evolutionary Patterns in Two Pyrus Species
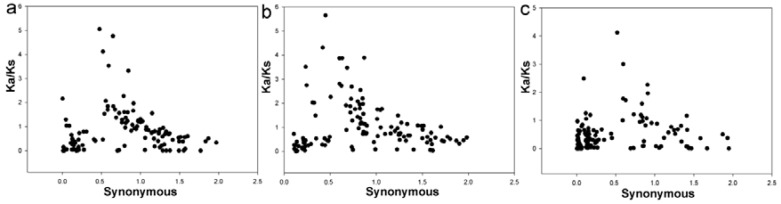
To investigate the evolutionary divergence and patterns of the UBC genes in P. bretschneideri and P. communis, the selection pressures of 198 paralogous gene pairs in P. bretschneideri, 215 paralogous gene pairs in P. communis, and 129 orthologous gene pairs in P. bretschneideri and P. communis were analyzed. All gene pairs, including paralogous and orthologous, are listed in Table S1. To avoid the risk of saturation [41], we removed any Ks values >2.0 in our study. In P. bretschneideri, 99 paralogous pairs contained Ka/Ks ratios below one, while the remaining gene pairs had ratios greater than one (Table S2). In P. communis, 94 paralogous pairs had Ka/Ks ratios below one, while the remaining gene pairs had ratios greater than one. The maximum Ka/Ks value was 5.055 in P. bretschneideri (PbrUBC04-PbrUBC70) and 5.65 in P. communis (PcpUBC51-PcpUBC58) (Table S1 and Figure 4). Among these orthologous pairs, we found that the most of the gene pairs had Ka/Ks ratios that were below one, indicating that these genes (which evolved from a common ancestor) have undergone purify selection with slow evolution at the protein level. Remarkably, these genes might also evolve through positive selection (Ks = 0; Ka ≠ 0, such as PbrUBC32-PcpUBC32), negative selection (i.e., Ka = 0; Ks ≠ 0, such as PcpUBC42-PcpUBC58), and strongly negative selection (i.e., Ka = Ks = 0, such as PbrUBC09-PcpUBC03) due to these gene pairs being subject to strong constraints (Table S2).
Figure 4.
The distribution of Ka (nonsynonymous), Ks (synonymous), and Ka/Ks values of paralogous and orthologous gene pairs. (a–c) represent Pbr-Pbr, Pcp-Pcp, and Pbr-Pcp gene pairs, respectively. The X- and Y-axes denote the synonymous distance and Ka/Ks ratio for each pair, respectively.
3.5. Expression Profiles of UBC Genes in Pyrus Fruit Development
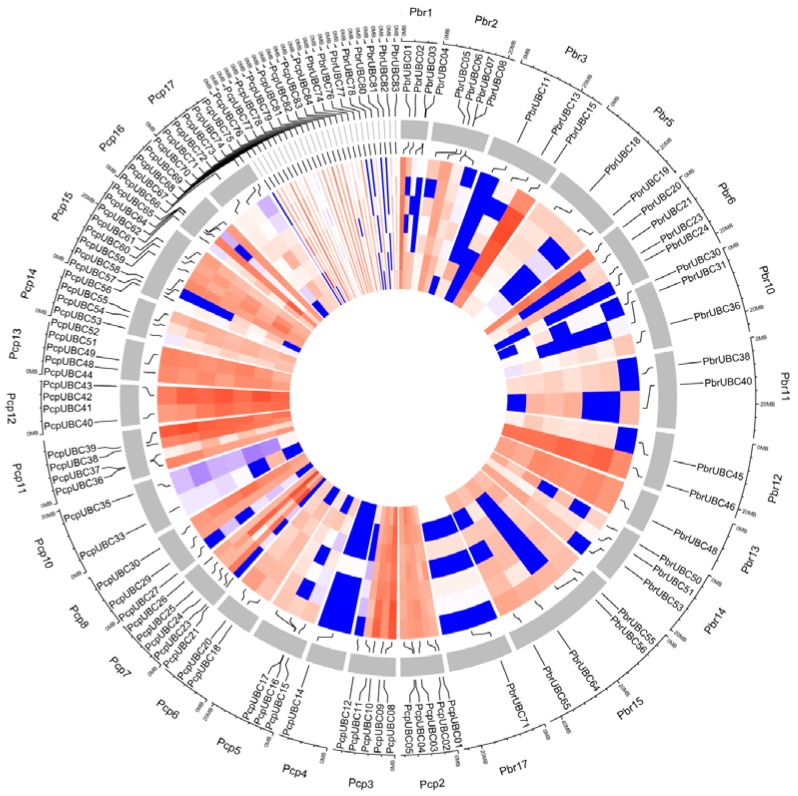
The genome sequences of both P. bretschneideri and P. communis provided an excellent opportunity to further study gene expression. Previous studies have shown that UBC genes may play an important role during fruit development [11,12]. To further understand the potential roles of PbrUBC and PcpUBC genes during pear fruit development, we obtained the transcriptome data of these UBC genes and built a heat map. From the transcriptome data results, it was apparent that 50.6% (42/83) PbrUBCs and 25% (21/84) PcpUBCs were not detected in each fruit developmental stage, suggesting their activity in other organs, such as the flower, root, or leaf. In P. bretschneideri, 41 PbrUBC genes were expressed in one or more developmental stages (Figure 5 and Table S2). Among them, 17 PbrUBC genes were expressed in all P. bretschneideri fruit development stages, indicating that these genes might be very important for the development and maturation of fruit. In P. communis, 47 PcpUBC genes were expressed in all P. communis fruit development stages, implying that these have functional activity in all fruit development stages. Remarkably, we found that the different isoforms produced by alternative splicing were not expressed in the period of pear fruit development, suggesting that the alternative splicing events might not play a role during pear fruit development. Additionally, we found that some UBC genes continuously increased or reduced at one or several stages, such as PbrUBC24 and PbrUBC80, which were highly expressed in Fruit_stage3 (55 days after full blooming), and PcpUBC14, which was highly expressed at Fruit_stage5 (115 days after full blooming), implying that these genes might be very important for fruit-specific developmental stages.
Figure 5.
Expression of UBC genes from P. bretschneideri and P. communis during fruit development and ripening, including Fruit_stage1 (15 days after full blooming (DAB)), Fruit_stage2 (30 DAB), Fruit_stage3 (55 DAB), Fruit_stage4 (85 DAB), Fruit_stage5 (115 DAB), Fruit_stage6 (mature stage), and Fruit_stage7 (fruit senescence stage). The color scale represents normalized log 2-transformed, where grey indicates a medium level, blue indicates a low level, and red indicates a high level. Circos software was used to visualize the heat map. The FPKM values of PbrUBCs and PcpUBCs are presented in Table S3. The outermost ring represents Fruit_stage7, followed by Fruit_stage6, Fruit_stage5, Fruit_stage4, Fruit_stage3, and Fruit_stage2, and the innermost ring represents Fruit_stage1.
3.6. Comparison of the Expression Patterns of UBC Genes in Two Pyrus Species
Pear is one of the leading cultivated fruit trees of temperate regions, and the fruit is the focus of this study due to its economic value. Homologous genes may have gene functional redundancy or divergence during evolution [42]. In the present study, to gain insight into the degree of expression diversity of UBC gene family members between P. bretschneideri and P. communis, their expression correlations were estimated using Pearson’s correlation coefficient (r). Remarkably, we only considered the homologous genes which were expressed in at least one pear fruit development stage (Table S4). Twenty-two orthologous gene pairs (such as PbrUBC36-PcpUBC17, PbrUBC20-PcpUBC42, and PbrUBC06-PcpUBC54) were found to be non-divergent, five orthologous gene pairs (such as PbrUBC18-PcpUBC17, PbrUBC01-PcpUBC69, PbrUBC31-PcpUBC35, PbrUBC07-PcpUBC55, and PbrUBC80-PcpUBC21) were ongoing divergent, and the remaining orthologous gene pairs (such as PbrUBC50-PcpUBC53, PbrUBC19-PcpUBC35, PbrUBC04-PcpUBC21, and PbrUBC20-PcpUBC58) were divergent (Table S3). These results suggested that most of the UBC orthologous gene pairs have undergone functional divergence.
3.7. Expression Profiles of PbrUBC Genes Respond to Drought Stress
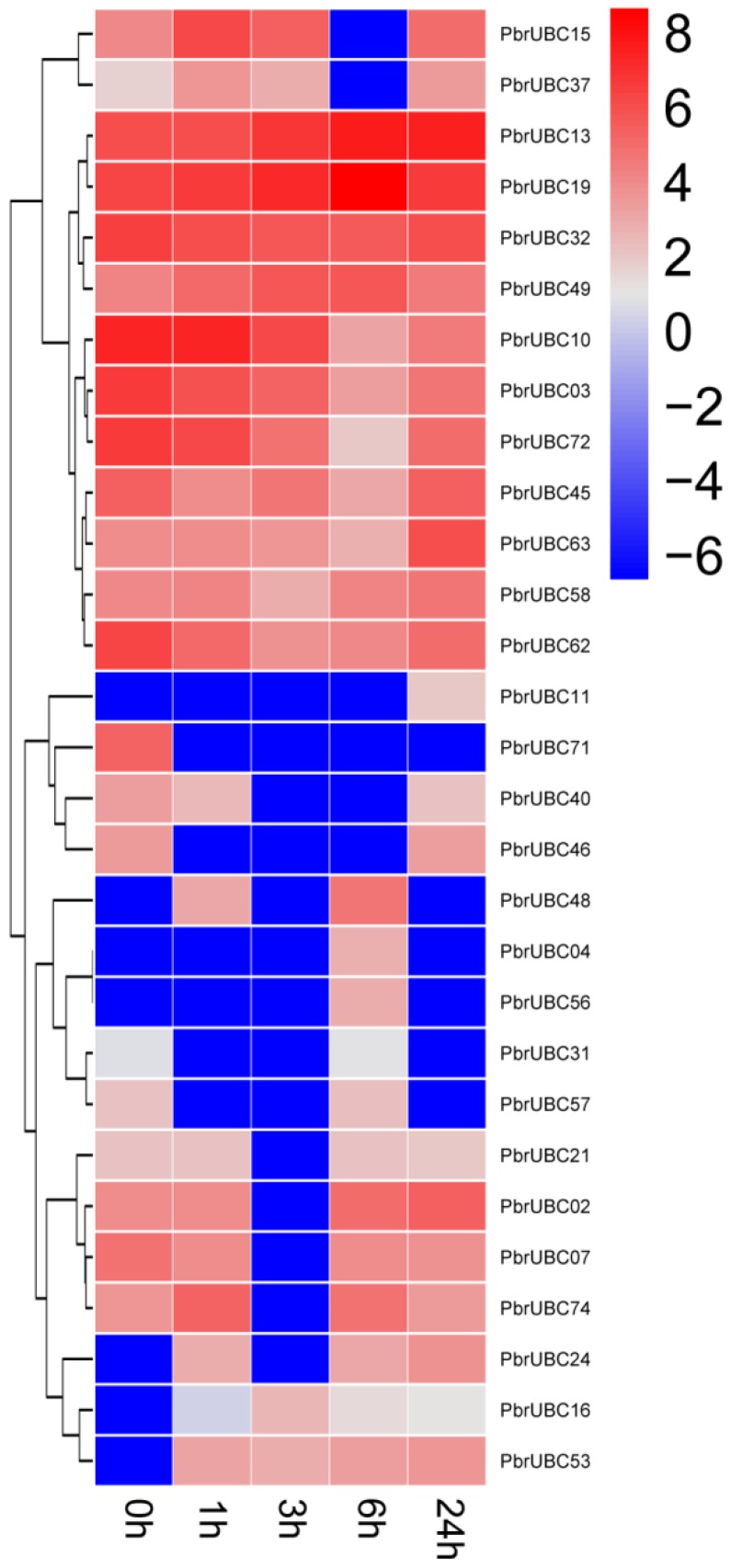
As a major abiotic stress, drought can affect plant productivity, growth, and development. Previous studies have shown that plants can enhance their drought tolerance by regulating gene transcription [9,15,16]. To identify UBC genes with a potential role in the drought stress response of P. bretschneideri, we carried out the expression analysis for 83 PbrUBC genes under drought stress. From the transcriptome data results, we found that only 34.9% (28/83) PbrUBCs were expressed under drought stress (Figure 6). Under drought stress treatment, five PbrUBC genes (PbrUBC02, PbrUBC10, PbrUBC15, PbrUBC37, and PbrUBC74) were up-regulated at early time points; however, they were down-regulated after a long period of stress treatment, indicating the existence of a possible feedback regulatory mechanism. Two (PbrUBC49 and PbrUBC63) and five PbrUBC (PbrUBC03, PbrUBC07, PbrUBC13, PbrUBC32, and PbrUBC62) genes under drought stress treatment were up- and down-regulated, respectively (Figure 6). Our data indicated that these genes might be important for drought stress responses and will help to select candidate genes for functional analysis under drought stress.
Figure 6.
Expression analysis of PbrUBC genes under drought stress treatment. The color scale represents normalized log 2-transformed, where grey indicates a medium level, blue indicates a low level, and red indicates a high level. The treatments were indicated at the bottom of each column, and the genes are located on the right.
4. Discussion
As a part of the ubiquitin proteasome system, ubiquitin-conjugating enzymes have been proved to play an important role in plant growth and development [1,11,21]. Although members of the UBC gene family have potential functional significance, they are relatively few in higher plants. Pear is widely cultivated in temperate regions due to its high nutritional and economic value. For pears, the fruit is the focus of this study. Previous studies have shown that the UBC gene family plays a very important role in fruit development and ripening [11,12], but is still excluded in two Pyrus species (P. communis and P. bretschneideri).
In our study, 83 PbrUBCs and 84 PcpUBCs genes were identified from the P. bretschneideri and P. communis genome, respectively. The number of PbrUBCs and PcpUBCs is much larger than 75, 74, 52, 48, 48, and 34 UBC genes previously reported from Zea mays, Musa nana, Solanum lycopersicum, Oryza sativa, Arabidopsis thaliana, and Carica papaya, respectively [1,8,9,10,11,12]. The genome sizes of Zea mays, Musa nana, Solanum lycopersicum, Oryza sativa, Arabidopsis thaliana, and Carica papaya are ~2300, ~523, ~900, ~466, ~125, and ~372 Mb, respectively. Then, we found that the genome sizes of S. lycopersicum and O. sativa are 7.2 and 3.7 times larger than that of A. thaliana, respectively; however, the genomes of these species have a similar number of UBCs, including 52, 48, and 48, respectively. In addition, the genome size of Z. mays is 4.67 times larger than that of Pyrus species (i.e., P. bretschneideri and P. communis), but the genomes of both P. bretschneideri (83) and P. communis (84) have a larger number of UBCs compared to Z. mays (75) and other studied species. Therefore, we speculate that the difference in the number of UBC genes is not related to the size of the genome.
Alternatively, gene duplication events, including segmental and tandem duplication, play a significant role in the expansion of gene family members in the genome. Two WGD events, including recent WGD [23] and ancient WGD [43], were shared by both the P. bretschneideri and P. communis genome during evolution. In order to understand the contribution of gene duplication events to the expansion of UBC family members in two Pyrus species, we analyzed the expansion mechanism of both the PbrUBC and PcpUBC gene family. In the P. bretschneideri genome, 192 PbrUBC gene pairs were determined to be involved in segmental duplication events and four gene pairs were identified that were involved in tandem duplication events. Similarly, 211 and one UBC gene pairs were involved in segmental duplication and tandem duplication events in the P. communis genome, respectively. These data indicate that the common expansion mechanism of the UBC gene family is mainly segmental duplication events, which is shared by both PbrUBCs and PcpUBCs. Therefore, we can infer that the expansion of UBC gene family members may not completely depend on independent duplications of individual sequences, and it may also be the result of rearrangement events and segmental chromosome duplication. A growing number of studies have shown that segmental duplications play a major role in the expansion of the pear gene family, such as the VQ, MYB, PRX, PHD, and WOX gene families [22,42,44,45,46].
UBC genes have been demonstrated to play an important role in plant growth and development, and physiological processes. For instance, OsUBC1 from O. sativa involves cellular responses to abiotic and biotic stresses [47], and the expression of five A. thaliana UBC genes (AtUBC13, AtUBC17, AtUBC20, AtUBC26, and AtUBC31) and three O. sativa UBC genes (OsUBC2, OsUBC5, and OsUBC18) is significantly down-regulated under drought and salt stress treatments; however, three OsUBC genes (OsUBC13, OsUBC15, and OsUBC45) are significantly up-regulated [21]. In the present study, we found that 34.9% (28/83) of PbrUBCs can respond to drought stress treatment at the transcriptional level, implying these genes play essential roles in responsive to drought stress in P. bretschneideri, such as PbrUBC02, PbrUBC10, PbrUBC15, PbrUBC37, and PbrUBC74, which were up-regulated at early time points. In the O. sativa and Z. mays, similar expression changes among UBC genes were also observed, including the expression of 34 ZmUBC genes that changed significantly and were up-regulated during early time points. These data indicated that these UBC genes might have important roles under drought stress treatment during P. bretschneideri development. The function of UBC genes in plant development and response stress has been well studied, but little is known about the role of protein ubiquitination in fruit development and ripening, except for M. nana, S. lycopersicum, and C. papaya. In M. nana, five UBC genes (MaUBC1, MaUBC9, MaUBC70, MaUBC68, and MaUBC71) presented about 10-fold to 40-fold higher expression levels at the fifth stage than at other stages of fruit ripening; however, seven other UBC genes (MaUBC8, MaUBC16, MaUBC17, MaUBC33, MaUBC34, MaUBC56, and MaUBC61) presented continuously increasing expression during all the fruit development stages [9]. In S. lycopersicum, six UBC genes (SlUBC6, SlUBC8, SlUBC24, SlUBC32, SlUBC41, and SlUBC42) were directly regulated by RIN, a fruit-ripening regulator [11]. In C. papaya, 13 (CpUBC4, CpUBC6, CpUBC7, CpUBC8, CpUBC9, CpUBC11, CpUBC12, CpUBC14, CpUBC16, CpUBC19, CpUBC20, CpUBC28, and CpUBC34) and two (CpUBC2 and CpUBC10) were up-regulated and down-regulated during C. papaya fruit ripening stages, respectively [12]. Our results suggested that PbrUBC82, PcpUBC10, and PcpUBC62, orthologs of MaUBC3 and MaUBC8 and AtUBC36, respectively (Figure S4), contained high expression levels during the fruit developmental period. S. lycopersicum SlUBC6 orthologs in two Pyrus species PbrUBC18 and PcpUBC17 were directly regulated by RIN (a fruit-ripening regulator), as reported in S. lycopersicum, and in two Pyrus species, were also highly expressed in fruits, suggesting a major role in fruit development. Additionally, we found that orthologs from different species exhibit different expression profiles. SlUBC32, MaUBC72, MaUBC47, PcpUBC44, PbrUBC53, PcpUBC31, and CpUBC5, which belong to the same subfamily (Figure S4), contained different expression profiles. For example, MaUBC72 was expressed during all M. nana fruit development, while its P. communis and S. lycopersicum orthologous genes, SlUBC32 and PcpUBC44/-31, were not expressed during fruit development. The contrary expression patterns suggested that these genes have different regulatory mechanisms in the development of plant fruits. Taken together, the present study indicated that some PbrUBCs and PcpUBCs might contribute to the regulation of fruit development and ripening processes. For UBC genes, the expression of most UBC orthologous gene pairs from P. bretschneideri and P. communis has undergone functional divergence, indicating functional redundancy evolved from a common ancestry for some orthologous gene pairs, and from neo-functionalization or sub-functionalization for others.
Acknowledgments
We extend our thanks to the reviewers and editors for their careful reading and helpful comments on this manuscript. This study was supported by The National Natural Science Foundation of China (grant 31640068). The funding bodies were not involved in the design of the study; in collection, analysis, and interpretation of data; and in writing the manuscript.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4409/7/7/77/s1, Figure S1: Structure of both PbrUBC and PcpUBC genes. UTR, exons, and introns are indicated by green, yellow, and grey, respectively. The length of PbrUBCs and PcpUBCs can be estimated using the scale at the bottom. Figure S2: Conserved protein motifs of both PbrUBC and PcpUBC proteins. All motifs were scanned by MEME software using the complete amino acid sequences of all P. bretschneideri and P. communis UBC proteins documented in Figure S2: Using the scale at the bottom, the length of UBC protein and motif can be estimated. Figure S3: Schematic depictions of the alternatively spliced UBC genes. The introns and exons are represented by thin lines and green rectangles, respectively. The length of the alternatively spliced UBC genes can be estimated using the scale at the bottom. Figure S4: Phylogenetic tree of the UBC proteins from P. bretschneideri, P. communis, A. thaliana, M. nana, C. papaya, and S. lycopersicum. Complete alignments of all UBC proteins from P. bretschneideri, P. communis, M. acuminate, C. papaya, S. lycopersicum, and A. thaliana were carried out using the MUSCLE program. The tree was constructed using the Maximum Likelihood (ML) method with IQ-TREE, and HsUfc1 from Homo sapiens was used as the out-group. Table S1. Homologous analyses of UBC genes in P. bretschneideri and P. communis. The homologous analyses of UBC genes in P. bretschneideri and P. communis were carried out using MCscanX software. Table S2: Ka/Ks analysis of between UBC gene pairs in Pyrus species. The “add_ka_and_ks_to_collinearity.pl” script of MCscanX software was used to detect the non-synonymous (Ka)/synonymous (Ks) substitution ratio. Table S3: The FPKM (fragments per kilobaseof exon per million fragments mapped) values of PbrUBC and PcpUBC genes during fruit development and ripening. Table S4: Divergence analysis of orthologous UBC gene pairs among P. bretschneideri and P. communis. The Pearson’s correlation coefficient (r) was used to estimate the similarity between the expression patterns of the orthologous gene pair. In general, r > 0.5, 0.3 < r < 0.5, and r < 0.3 suggest non-divergence, ongoing divergent, and divergent, respectively.
Author Contributions
Y.C. (Yunpeng Cao) and Y.C. (Yongping Cai) designed and performed the experiments; Y.C. (Yunpeng Cao) and D.M. analyzed the data; D.M., Q.J., Y.L., Y.C. (Yunpeng Cao), M.A., Y.C. (Yu Chen), and Y.C. (Yongping Cai) contributed reagents/materials/analysis tools; Y.C. (Yongping Cai) wrote the paper. All authors reviewed and approved the final submission.
Funding
This research was funded by National Natural Science Foundation of China (31640068).
Conflicts of Interest
The authors declare that they have no competing interests.
Ethics Approval and Consent to Participate
The experiments did not involve endangered or protected species. No specific permits were required for these locations/activities because the pears used in this study were obtained from a horticultural field in Dangshan, which are demonstration orchards at Auhui Agricultural University.
References
- 1.Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.-S., Deng X.-W., Callis J. Genome analysis and functional characterization of the e2 and ring-type e3 ligase ubiquitination enzymes of arabidopsis. Plant Physiol. 2005;139:1597–1611. doi: 10.1104/pp.105.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callis J., Vierstra R.D. Protein degradation in signaling. Curr. Opin. Plant Boil. 2000;3:381–386. doi: 10.1016/S1369-5266(00)00100-X. [DOI] [PubMed] [Google Scholar]
- 3.Peters J.-M., Harris J.R., Finley D. Ubiquitin and the Biology of the Cell. Springer Science & Business Media; Berlin, Germany: 2013. [Google Scholar]
- 4.Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 5.Van Wijk S.J., Timmers H.M. The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 6.Michelle C., Vourc’h P., Mignon L., Andres C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 2009;68:616–628. doi: 10.1007/s00239-009-9225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones D., Crowe E., Stevens T.A., Candido E.P.M. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: Ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Boil. 2001;3:research0002.1–0002.15. doi: 10.1186/gb-2001-3-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae H., Kim W.T. Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochem. Biophys. Res. Commun. 2014;444:575–580. doi: 10.1016/j.bbrc.2014.01.098. [DOI] [PubMed] [Google Scholar]
- 9.Dong C., Hu H., Jue D., Zhao Q., Chen H., Xie J., Jia L. The banana e2 gene family: Genomic identification, characterization, expression profiling analysis. Plant Sci. 2016;245:11–24. doi: 10.1016/j.plantsci.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Jue D., Sang X., Lu S., Dong C., Zhao Q., Chen H., Jia L. Genome-wide identification, phylogenetic and expression analyses of the ubiquitin-conjugating enzyme gene family in maize. PLoS ONE. 2015;10:e0143488. doi: 10.1371/journal.pone.0143488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Wang W., Cai J., Zhang Y., Qin G., Tian S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Boil. 2014;15:548. doi: 10.1186/s13059-014-0548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jue D., Sang X., Shu B., Liu L., Wang Y., Jia Z., Zou Y., Shi S. Characterization and expression analysis of genes encoding ubiquitin conjugating domain-containing enzymes in Carica papaya. PLoS ONE. 2017;12:e0171357. doi: 10.1371/journal.pone.0171357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L., Ménard R., Berr A., Fuchs J., Cognat V., Meyer D., Shen W.H. The E2 ubiquitin-conjugating enzymes, Atubc1 and Atubc2, play redundant roles and are involved in activation of Flc expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009;57:279–288. doi: 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- 14.Cui F., Liu L., Zhao Q., Zhang Z., Li Q., Lin B., Wu Y., Tang S., Xie Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell. 2012;24:233–244. doi: 10.1105/tpc.111.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G.-A., Chang R.-Z., Qiu L.-J. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol. Boil. 2010;72:357–367. doi: 10.1007/s11103-009-9575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan X., Mo A., Liu S., Yang L., Li L. Constitutive expression of a peanut ubiquitin-conjugating enzyme gene in Arabidopsis confers improved water-stress tolerance through regulation of stress-responsive gene expression. J. Biosci. Bioeng. 2011;111:478–484. doi: 10.1016/j.jbiosc.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Chung E., Cho C.-W., So H.-A., Kang J.-S., Chung Y.S., Lee J.-H. Overexpression of VrUBC1, a mung bean e2 ubiquitin-conjugating enzyme, enhances osmotic stress tolerance in Arabidopsis. PLoS ONE. 2013;8:e66056. doi: 10.1371/journal.pone.0066056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Schmidt W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 2010;62:330–343. doi: 10.1111/j.1365-313X.2010.04150.x. [DOI] [PubMed] [Google Scholar]
- 19.Wen R., Newton L., Li G., Wang H., Xiao W. Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Boil. 2006;61:241–253. doi: 10.1007/s11103-006-0007-x. [DOI] [PubMed] [Google Scholar]
- 20.Zolman B.K., Monroe-Augustus M., Silva I.D., Bartel B. Identification and functional characterization of arabidopsis peroxin4 and the interacting protein peroxin22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.E Z., Zhang Y., Li T., Wang L., Zhao H. Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS ONE. 2015;10:e0122621. doi: 10.1371/journal.pone.0122621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Y., Han Y., Li D., Lin Y., Cai Y. Myb transcription factors in Chinese pear (Pyrus bretschneideri rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016;7:577. doi: 10.3389/fpls.2016.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J., Wang Z., Shi Z., Zhang S., Ming R., Zhu S., Khan M.A., Tao S., Korban S.S., Wang H. The genome of the pear (Pyrus bretschneideri rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mistry J., Finn R.D., Eddy S.R., Bateman A., Punta M. Challenges in homology search: Hmmer3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2011;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chagné D., Crowhurst R.N., Pindo M., Thrimawithana A., Deng C., Ireland H., Fiers M., Dzierzon H., Cestaro A., Fontana P., et al. The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’) PLoS ONE. 2014;9:e92644. doi: 10.1371/journal.pone.0092644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I., Doerks T., Bork P. Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zdobnov E.M., Apweiler R. Interproscan—An integration platform for the signature-recognition methods in interpro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 29.Jung S., Ficklin S.P., Lee T., Cheng C.H., Blenda A., Zheng P., Yu J., Bombarely A., Cho I., Ru S. The genome database for rosaceae (GDR): Year 10 update. Nucleic Acids Res. 2014;42:1237–1244. doi: 10.1093/nar/gkt1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Boil. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.-H., Jin H., Marler B., Guo H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C., Xia R., Chen H., He Y. Tbtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018 doi: 10.1101/289660. [DOI] [Google Scholar]
- 34.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The meme suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon A., Hannon G. Fastx-Toolkit. Fastq/a Short-Reads Pre-Processing Tools. 2003. Unpublished work.
- 36.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Boil. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with Tophat and Cufflinks. Nat. Protoc. 2012;7:562. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanc G., Wolfe K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yim W.C., Lee B.-M., Jang C.S. Expression diversity and evolutionary dynamics of rice duplicate genes. Mol. Genet. Genom. 2009;281:483–493. doi: 10.1007/s00438-009-0425-y. [DOI] [PubMed] [Google Scholar]
- 40.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maher C., Stein L., Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16:510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y., Meng D., Abdullah M., Jin Q., Lin Y., Cai Y. Genome wide identification, evolutionary, and expression analysis of VQ genes from two Pyrus species. Genes. 2018;9:224. doi: 10.3390/genes9040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fawcett J.A., Maere S., Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Y., Han Y., Meng D., Li D., Jin Q., Lin Y., Cai Y. Structural, evolutionary, and functional analysis of the class iii peroxidase gene family in Chinese Pear (Pyrus bretschneideri) Front. Plant Sci. 2016;7:1874. doi: 10.3389/fpls.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y., Han Y., Meng D., Li G., Li D., Abdullah M., Jin Q., Lin Y., Cai Y. Genome-wide analysis suggests the relaxed purifying selection affect the evolution of WOX genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 2017;8:78. doi: 10.3389/fgene.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Y., Han Y., Meng D., Abdullah M., Li D., Jin Q., Lin Y., Cai Y. Systematic analysis and comparison of the PHD-Finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct. Integr. Genom. 2018:1–13. doi: 10.1007/s10142-018-0609-9. [DOI] [PubMed] [Google Scholar]
- 47.Jeon E.H., Pak J.H., Kim M.J., Kim H.J., Shin S.H., Lee J.H., Kim D.H., Oh J.S., Oh B.-J., Jung H.W. Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2012;427:309–314. doi: 10.1016/j.bbrc.2012.09.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
P. bretschneideri fruit developmental stage 1 (Fruit_stage1: 15DAB), Accession: SRX1595645; P. bretschneideri Fruit_stage2 (30DAB), Accession: SRX1595646; P. bretschneideri Fruit_stage3 (55DAB), Accession: SRX1595647; P. bretschneideri Fruit_stage4 (85 DAB), Accession: SRX1595648; P. bretschneideri Fruit_stage5 (115 DAB), Accession: SRX1595650; P. bretschneideri Fruit_stage6 (mature stage), Accession: SRX1595651; P. bretschneideri Fruit_stage7 (fruit senescence stage), Accession: SRX1595652; P. communis Fruit_stage1 (15DAB), Accession: SRX1595636; P. communis Fruit_stage2 (30DAB), Accession: SRX1595637; P. communis Fruit_stage3 (55DAB), Accession: SRX1595638; P. communis Fruit_stage4 (85 DAB), Accession: SRX1595639; P. communis Fruit_stage5 (115 DAB), Accession: SRX1595640; P. communis Fruit_stage6 (mature stage), Accession: SRX1595641; P. communis Fruit_stage7 (fruit senescence stage), Accession: SRX1595644. P. bretschneideri drought-tolerant for 0 h, Accession: SRX4110141; P. bretschneideri drought-tolerant for 1 h, Accession: SRX4110140; P. bretschneideri drought-tolerant for 3 h, Accession: SRX4110143; P. bretschneideri drought-tolerant for 6 h, Accession: SRX4110142; P. bretschneideri recovery for 24 h, Accession: SRX4110139.