Abstract
The circadian clock in cyanobacteria employs a posttranslational oscillator composed of a sequential phosphorylation–dephosphorylation cycle of KaiC protein, in which the dynamics of protein structural changes driven by temperature-compensated KaiC’s ATPase activity are critical for determining the period. On the other hand, circadian clocks in eukaryotes employ transcriptional feedback loops as a core mechanism. In this system, the dynamics of protein accumulation and degradation affect the circadian period. However, recent studies of eukaryotic circadian clocks reveal that the mechanism controlling the circadian period can be independent of the regulation of protein abundance. Instead, the circadian substrate is often phosphorylated at multiple sites at flexible protein regions to induce structural changes. The phosphorylation is catalyzed by kinases that induce sequential multisite phosphorylation such as casein kinase 1 (CK1) with temperature-compensated activity. We propose that the design principles of phosphorylation-dependent circadian-period determination in eukaryotes may share characteristics with the posttranslational oscillator in cyanobacteria.
The circadian clock produces near-24-h rhythms in physiological activities. From multicellular animals to plants, circadian clocks regulate cellular metabolic pathways and other physiological responses of the organism, such as sleep–wake cycles and immune responses (Levi and Schibler 2007; Bass and Takahashi 2010; Mohawk et al. 2012; Albrecht 2013; Greenham and McClung 2015; Lu et al. 2017; Sanchez and Kay 2017). The evolutionary advantage of a circadian clock was demonstrated by coculture experiments with prokaryotic cyanobacteria, in which strains with a circadian period matching the light–dark cycle of their environment became dominant in the population (Woelfle et al. 2004). This advantage should apply to many species; circadian rhythmicity of physiological activities is observed in evolutionarily distant species. Conserved features of the molecular mechanisms that drive circadian clocks in different phyla have been revealed through studies in a variety of organisms, including conventional models such as rodents in mammals, Drosophila melanogaster in insects, Arabidopsis thaliana in higher plants, Neurospora crassa in fungi, and Synechococcus elongatus in cyanobacteria. In all organisms, circadian rhythmicity is cell-autonomous. Even in mammals, various cell types show robust circadian rhythmicity in gene expression and physiological activity, even when cultured in vitro (Welsh et al. 1995; Yagita et al. 2001; Nagoshi et al. 2004; Yoo et al. 2004). Cell-autonomous circadian oscillation is produced by the system-level orchestration of transcription–translation feedback and/or a posttranslational interaction network (Gallego and Virshup 2007; Ukai and Ueda 2010; Millius and Ueda 2017), but the molecular components of the oscillators especially for transcriptional or translational regulators differ between species (Rosbash 2009; Brown et al. 2012; van Ooijen and Millar 2012; Takahashi 2017). However, the question arises whether a common design principle underlies the molecular components of circadian clocks in different organisms.
In most organisms, circadian molecular oscillators seem to be based on transcription–translation feedback loops (TTFLs). However, how circadian clocks maintain a robust 24-h oscillation is a mystery. The suprachiasmatic nucleus, a region of the brain central to circadian rhythm in mammals, can sustain oscillating gene expression for years when cultured in vitro (Yamazaki and Takahashi 2005). However, the periodic gene expression in artificial biological oscillators encoded as TTFLs is less stable when subjected to cell-intrinsic or cell-extrinsic changes (Elowitz and Leibler 2000; Danino et al. 2010). This observation suggests that an additional layer exists in the period-keeping mechanism of circadian cellular oscillators. Here, we review the circadian-clock features in various organisms from the perspective of TTFL-based design principles and posttranslational modifications of oscillator components. Through the comparison between a phosphorylation-based oscillator in cyanobacteria and multisite phosphorylation of clock components that can regulate the circadian period in eukaryotes, we propose that shared design principles can be found in the circadian period-determination mechanisms.
TRANSCRIPTION NEGATIVE FEEDBACK IN EUKARYOTIC CIRCADIAN OSCILLATORS
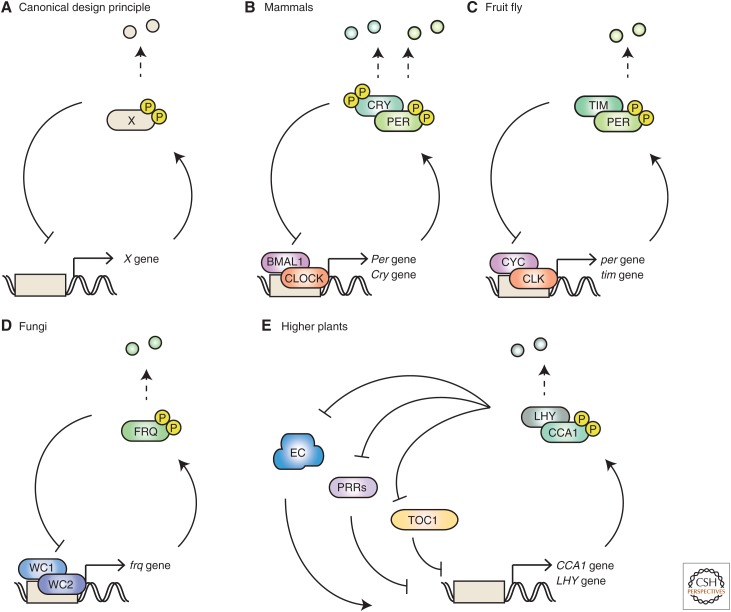
Figure 1 summarizes the major molecular components of circadian oscillators in different phyla. Eukaryote oscillators have conserved negative feedback loops in the transcription–translation network (Fig. 1A). The PERIOD protein (PER) is a key transcriptional repressor in mammals and Drosophila. PER binds and inhibits a basic helix–loop–helix transcription-activator complex consisting of CLOCK and BMAL1 in mammals (Fig. 1B). In Drosophila, PER inhibits a complex of CLOCK (CLK) and CYCLE (CYC), a BMAL1 ortholog (Fig. 1C). The CLOCK–BMAL1 (CLK-CYC in Drosophila) complex activates Per transcription; thus, the overall transcription network creates a negative feedback loop. PER binds other partners in various organisms to act on transcription activators. In Drosophila, PER represses its own transcription by binding TIMELESS (TIM), the vertebrate homolog of which maintains chromosomal integrity (Chou and Elledge 2006; Tanaka et al. 2009). In contrast, mammalian PER forms a complex with CRYPTOCHROME (CRY). The cryptochrome superfamily is widely conserved from bacteria to plants, but nonmammalian cryptochrome functions as a blue-light receptor rather than as a circadian transcription repressor (Lin and Todo 2005; Ozturk et al. 2007). Phylogenetically, the closest homolog of mammalian cryptochrome is 6–4 photolyase, which catalyzes the repair of damaged DNA residues using energy from blue light. These functional divergence of TIM and CRY in maintaining DNA integrity and circadian rhythmicity may be related to the evolutionarily close relationship of DNA-damage responses and circadian clocks in an environment with daily oscillations in sunlight.
Figure 1.
Shared structure of transcription–translation feedback loop (TTFL)-based eukaryotic circadian clocks. (A) Diagram showing the basic structure of a circadian negative-feedback transcription oscillator. The transcription repressor is regulated by multisite phosphorylation and proteasome-mediated proteolysis. (B–E) Major components of the circadian TTFL-based oscillator in each species. Note that the indicated components and pathways were selected based on the context of this review; the actual circadian systems are far more complex reaction networks involving numerous gene products. EC (evening complex) composed of ELF4, LUX, and ELF3.
The molecular components of circadian negative feedback loops differ between fungi and plants. White Collar complex (WCC) and FREQUENCY (FRQ) are the primary drivers of Neurospora circadian clocks (Fig. 1D). WCC activates FRQ transcription, which in turn inhibits WCC and creates the negative feedback loop. The canonical core feedback architecture of the circadian clock in plants is more complex (Fig. 1E). In Arabidopsis, the canonical model involves transcription feedback loops around two transcription repressors, CCA1 and LHY, which suppress the transcription of both activators and inhibitors of CCA1/LHY expression, creating an architecture combining positive and negative feedback.
Theoretical studies emphasize the importance of a negative feedback loop encoded as a TTFL in autonomous oscillation (Novak and Tyson 2008; Kim 2016). In a general sense, the period of TTFL-based oscillators is determined through the dynamics of protein quantity: the repression phase is initiated by the accumulation of transcription repressor(s) and ceases when the repressors have disappeared. Indeed, the circadian period is reported to be regulated through the delayed timing (phase) of the accumulation of mammalian transcriptional repressor Cry messenger RNA (mRNA) (Ukai-Tadenuma et al. 2011) or Per mRNA in mammals and Drosophila (Kadener et al. 2008; Fustin et al. 2013). These changes are regulated by the timing of transcription and the mRNA maturation process, independent of the biochemical characteristics of the CRY/PER proteins themselves. However, evidence suggests that the dynamics of mRNA abundance are less important in eliciting rhythmic transcription; noncircadian expression of the per gene in Drosophila (Ewer et al. 1988; Frisch et al. 1994; Vosshall and Young 1995; Yang and Sehgal 2001) and a constitutive supply of CRY protein in mammals can support circadian oscillation (Fan et al. 2007; Ukai-Tadenuma et al. 2011), indicating the importance of protein-level dynamics in the regulation of circadian clocks. Consistent with this view, rhythmic changes in mammalian PER levels were observed when PER was expressed under a constitutive promoter (Yamamoto et al. 2005; Fujimoto et al. 2006; Nishii et al. 2006).
In protein-level regulation of the circadian period, several theoretical models predict the importance of transcriptional repressor degradation rates (Gerard et al. 2009). In a simplified negative feedback TTFL model such as the Goodwin model, accelerating the degradation rate shortens the circadian period (Forger 2011), and this relationship is observed in model organisms. The degradation rate of core circadian components is fine-tuned by their phosphorylation and subsequent proteasome-dependent degradation. In mammals and Drosophila, the circadian period is shortened or lengthened by mutations that destabilize or stabilize PER, respectively (Vanselow et al. 2006; Chiu et al. 2008; Meng et al. 2008; Syed et al. 2011). Mammalian CRY also regulates the circadian period; genetic and pharmacological perturbations that stabilize CRY lengthen the period (Siepka et al. 2007; Hirota et al. 2012; Gao et al. 2013; Hirano et al. 2017), while destabilizing CRY shortens it (Hirano et al. 2013, 2016; Yoo et al. 2013). The causal relationship between CRY degradation and a shorter period was demonstrated by tuning the period through CRY-specific artificial proteolysis (Ode et al. 2017). In Neurospora, FRQ stability is also well correlated with the circadian period (Liu et al. 2000; Gorl et al. 2001; Baker et al. 2009; Tang et al. 2009).
Recent findings, however, indicate that period-determination processes can be independent of transcriptional-repressor clearance rates. Deleting a ubiquitin-ligase component leading to the proteolysis of FRQ stabilized FRQ but, surprisingly, had little effect on rhythmic FRQ expression (Larrondo et al. 2015). Several mutant alleles of mammalian CRY1 altered the circadian period but had little effect on CRY1’s stability (Ode et al. 2017). One chemical biology study designed a compound that stabilizes CRY1 and shortens the circadian period (Oshima et al. 2015). These studies suggest that the regulation of protein quantity (the rates of protein production and degradation) does not tell the entire story of circadian-period determination, and a question remains about the general applicability of the TTFL model in determining circadian periods (Gallego and Virshup 2007; Blau 2008).
POSTTRANSLATIONAL CIRCADIAN OSCILLATOR DESIGN PRINCIPLES IN CYANOBACTERIA
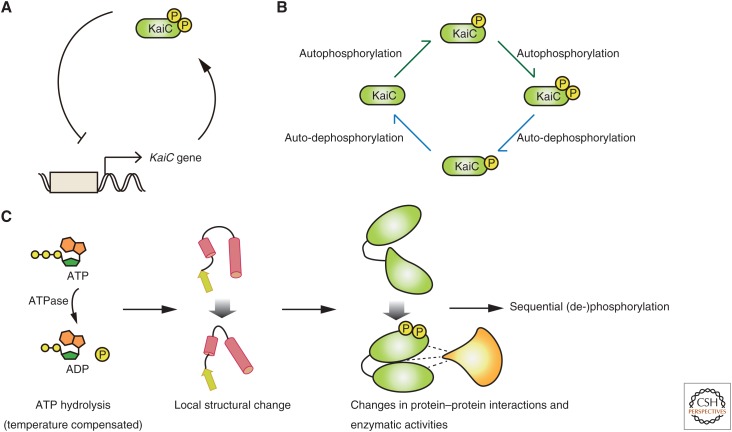
The prokaryotic cyanobacterial circadian clock is an example of circadian-period determination independent of protein-quantity dynamics. This clock is driven by the proteins KaiA, KaiB, and KaiC (Ishiura et al. 1998). KaiC represses transcription, and its targets include KaiC itself. Thus, it was proposed that KaiC forms a TTFL (Fig. 2A). However, a subsequent study showed that the circadian clock in cyanobacteria can operate in the absence of global transcription and translational activities (Tomita et al. 2005). Indeed, autonomous circadian rhythmicity in the phosphorylation status of purified KaiC can be reconstituted by incubating a mixture of KaiC, KaiA, and KaiB with ATP in vitro (Nakajima et al. 2005). Thus, KaiC provides a posttranslational oscillator (PTO) as well as a TTFL-based negative-feedback architecture. The coupling of PTO and TTFL-based oscillators appears to be important for cyanobacteria circadian clocks, depending on growth conditions (Kitayama et al. 2008; Teng et al. 2013). PTO rhythmicity requires fine-tuning the expression of KaiA, KaiB, and KaiC at specific ratios for oscillation in vitro (Nakajima et al. 2010). Their expression is also regulated by translational efficiency: nonoptimal codon usage in kaiBC genes tunes the amplitude of the circadian clock in response to growth conditions (Xu et al. 2013). However, detailed analysis of a reconstituted KaiC oscillator revealed a mechanism in which protein function itself creates the circadian oscillation and determines 24-h rhythmicity.
Figure 2.
The KaiA/B/C-dependent posttranslational circadian oscillator in cyanobacteria. (A) KaiC represses its own transcription, and thus forms a transcriptional negative-feedback loop. Although every reaction should be reversible, biased reaction rates between antagonizing kinase/phosphatase reactions are required to create a posttranslational oscillator (PTO). (B) A mixture of KaiA/B/C with ATP can reconstitute a PTO in vitro. (C) The period-determining step of the KaiC oscillator is temperature-insensitive ATP hydrolysis, which induces a structural change in KaiC.
KaiC is biochemically unique. It functions as a homohexamer and has autophosphorylation, autodephosphorylation, and ATPase activities. KaiC has two sites that are autophosphorylated and autodephosphorylated in a defined order, such that KaiC undergoes four distinct phosphorylation states over the course of the circadian cycle (Fig. 2B) (Nishiwaki et al. 2007; Rust et al. 2007). Each phosphorylation state has a distinct affinity to KaiA and KaiB. In turn, KaiA and KaiB regulate KaiC’s autophosphorylation and autodephosphorylation to autonomously switch between a “daytime” KaiC complex with autophosphorylation activity and a “nighttime” complex with autodephosphorylation activity. These switches include dynamic structural alterations in a flexible loop domain in KaiC (Chang et al. 2011; Tseng et al. 2014) and a fold switch in the KaiB structure (Chang et al. 2015).
The force that drives KaiC status switching is provided through KaiC-intrinsic ATP hydrolysis and ADP/ATP exchange. The ATP-hydrolysis rate is relatively slow (∼10 ATP molecules per day), is well correlated with the circadian period in vivo for several period-modulating mutants (Terauchi et al. 2007), and is unaffected by changes in incubation temperature (Terauchi et al. 2007; Murakami et al. 2008). Thus, in cyanobacteria, temperature-insensitive ATPase activity is proposed to support temperature compensation of the circadian period, a mechanism for maintaining a constant circadian period irrespective of environmental temperature. The crystal structure of KaiC reveals that the ATP-hydrolysis rate is determined by the position of ATP’s phosphorus atom and a water molecule (Abe et al. 2015). The unfavored position of the water molecule slows down ATP hydrolysis, which is coupled to an alternation of KaiC’s structure at ATP-binding surfaces, further slowing ATP hydrolysis (Abe et al. 2015) and driving a structural change in KaiC that induces it to interact with KaiA/B (Snijder et al. 2017; Tseng et al. 2017).
Based on the KaiC oscillator mechanism, we can deduce an abstractive design principle of the posttranslational circadian oscillator (Fig. 2C): energy from temperature-compensated ATP hydrolysis induces local structural alterations that induce biochemical changes in protein–protein interactions, enzymatic activities, and modification patterns. Transitions between protein states have a significant energy barrier that determines the length of the circadian period. In this scenario, the circadian period is not determined by the dynamics of protein quantity, but rather by the dynamics of status change or the quality of each protein molecule.
MULTISITE PHOSPHORYLATION DETERMINES THE PERIOD OF EUKARYOTIC CIRCADIAN CLOCKS
Although transcription-independent circadian oscillation marked by cellular redox states has been found in eukaryotic cells (O’Neill and Reddy 2011; O’Neill et al. 2011; Edgar et al. 2012; Reddy and Rey 2014), it is not known whether components in TTFL-based eukaryotic circadian oscillators can elicit posttranslational oscillation. If you recall, however, that the cyanobacteria circadian clock adopts a combination of PTO and TTFL-based oscillators, it may be useful to consider that some part of the design principles observed in the cyanobacteria circadian PTO might be also applicable to the regulation of eukaryotic circadian oscillators.
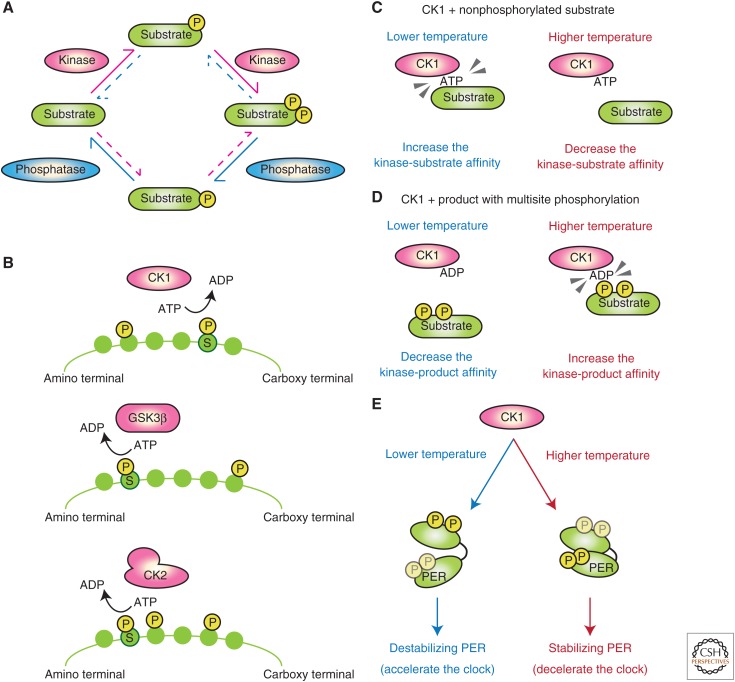
A PTO can theoretically be constructed using a generic kinase, phosphatase, and substrate (Jolley et al. 2012) with a similar oscillation mechanism to that proposed through the modeling of the KaiA/B/C oscillator (Fig. 3A) (Clodong et al. 2007; Rust et al. 2007; van Zon et al. 2007). A PTO constructed with a kinase, phosphatase, and substrate protein (assuming that processes are catalyzed in the Michaelis–Menten scheme) must use a substrate that is phosphorylated at multiple sites, because reversible phosphorylation at one site alone cannot produce autonomous oscillation in the substrate’s phosphorylation states (Angeli and Sontag 2008; Conradi and Shiu 2015). Interestingly, various lines of evidence show that several components of the eukaryotic circadian oscillators are also phosphorylated at multiple sites (Fig. 1), and that such multisite phosphorylation is critical for determining the oscillation period. Indeed, as discussed in the following sections, several features of design principles found in cyanobacterial PTO can be also found in the eukaryotic circadian substrates of the multisite phosphorylation and kinases catalyzing the phosphorylation.
Figure 3.
Multisite phosphorylation is the core mechanism of period determination in eukaryotic circadian clocks. (A) Multisite phosphorylation is a prerequisite for forming a posttranslational oscillator (PTO) from a generic kinase, a phosphatase, and a substrate. (B) The kinases responsible for period determination preferentially promote sequential phosphorylation. “S” denotes the target residues for phosphorylation. (C–D) Ueda and his colleagues found that raising the temperature decreases casein kinase 1 (CK1)’s affinity for the substrate (Shinohara et al. 2017). Raising the temperature also prompts CK1 to bind the phosphorylated product. These effects decrease the turnover number and thus decelerate the enzyme’s reaction rate. (E) Virshup, Forger, and their colleagues (2015) found that raising the temperature promotes PERIOD protein (PER) phosphorylation by CK1, which stabilizes PER and thereby lengthens the period (Zhou et al. 2015).
CK1, CK2, and Glycogen Synthase Kinase (GSK)3β
Casein kinase 1 (CK1) is a conserved component that markedly regulates the circadian-clock period in mammals and Drosophila. CK1ε was first characterized as a period-controlling kinase coded by the Drosophila gene doubletime, mutations of which shorten or lengthen the circadian period (Kloss et al. 1998; Price et al. 1998). CK1ε was subsequently identified as the molecule responsible for the shorter circadian period found in tau mutant hamsters (Lowrey et al. 2000). Another CK1 isoform, CK1δ, was identified as a mutation associated with a familial advanced sleep-phase syndrome (FASPS) pedigree (Xu et al. 2005). The genetic evidence for CK1’s contribution to period determination was further supported by high-throughput screening of small molecules or short interfering RNAs (siRNAs), which identified chemicals/siRNAs that lengthen the circadian period by inhibiting CK1δ/ε (Hirota et al. 2008, 2010; Isojima et al. 2009; Zhang et al. 2009; Chen et al. 2012). Screening revealed that GSK3β, casein kinase 2 (CK2), and CK1α also control the circadian period (Hirota et al. 2008, 2010; Maier et al. 2009; Zhang et al. 2009). Although other kinases such as AMP-activated protein kinase (AMPK), calmodulin-dependent protein kinase II (CaMKII), and mitogen-activated protein kinase (MAPK) regulate circadian clocks by incorporating various cellular and environmental signals (Gallego and Virshup 2007; Reischl and Kramer 2011), the importance of CK1, CK2, and GSK3β in period determination is highlighted both by screening studies and in the conservation of their roles across species. Among these kinases, CK1 may have one of the most prominent effects on the circadian period because the inhibition of CK1δ/ε can double the period up to 48 h (Isojima et al. 2009) and because CK1δ/ε-dependent phosphorylation in vitro and CK1δ/ε-dependent degradation in cellulo are both temperature-compensated (Isojima et al. 2009).
Kinases can phosphorylate multiple sites on a substrate in a random manner, phosphorylating each site with a random access, or in a sequential manner in which the kinase phosphorylates the target sites in a specific order (Yamada and Forger 2010). Intriguingly, CK1, GSK3β, and CK2 undergo sequential phosphorylation (Fig. 3B). CK1’s substrate consensus sequence is pS/T-X-X-S/T (pS/T denotes phosphorylated serine or threonine, X denotes any amino acids, and S/T denotes the target residue). The CK1 kinase preferentially phosphorylates residues downstream of prephosphorylated residues (Flotow et al. 1990). The consensus sequence of GSK3β is S/T-X-X-X-pS/T (Frame and Cohen 2001), and that of CK2 is S/T-X-X-D/E, where substituting pS into the +1 and +3 positions optimize the phosphorylation efficiency (St-Denis et al. 2015). Thus, all three of these important period-controlling kinases preferentially phosphorylate residues near prephosphorylated residues. When a kinase with this property acts on multiple and clustered target residues, phosphorylation events preferentially follow a defined order. For instance, CK1 preferentially phosphorylates sites sequentially, from the peptide amino terminus toward the carboxy terminus. This sequential phosphorylation also makes it easy to enrich local protein regions with phosphorylated residues.
PER
Transcription repressors in the eukaryotic circadian oscillator are regulated through multisite phosphorylation, catalyzed at least partly by the above-mentioned kinases. PER in fly and mammals is among the best-understood substrates of CK1δ/ε (DBT in Drosophila). PER is progressively phosphorylated as the circadian cycle progresses (Edery et al. 1994; Lee et al. 2001). The mammalian PER sequence contains a repeat of the CK1 motif. Once the amino-terminal side of the S-X-X-S repeat is phosphorylated (i.e., priming phosphorylation), CK1 can efficiently phosphorylate the downstream serine cluster (Vanselow et al. 2006). A mutation on the priming phosphorylation site, found in a FASPS pedigree (Toh et al. 2001), destabilizes the PER protein (Vanselow et al. 2006), indicating that phosphorylation at this site helps stabilize PER and regulate the circadian period. The kinase responsible for the priming site is unclear, but CK1δ/ε may be able to act on the nonphosphorylated PER-derived sequence (Isojima et al. 2009). Another PER phosphorylation domain is important for proteasome-dependent PER degradation (Eide et al. 2005; Shirogane et al. 2005; Shanware et al. 2011; Zhou et al. 2015). CK2 also controls the period of the mammalian circadian clock by phosphorylating PER (Tsuchiya et al. 2009). In Drosophila, DBT phosphorylates PER at multiple sites (Chiu et al. 2008). DBT-dependent multisite phosphorylation is primed by NEMO kinase (Chiu et al. 2011; Yu et al. 2011). In addition, CK2 (Lin et al. 2002; Akten et al. 2003; Nawathean and Rosbash 2004) and GSK3β (Martinek et al. 2001; Fang et al. 2007; Ko et al. 2010; Hara et al. 2011) regulate the action of Drosophila PER, although the effect is probably mediated partly through the phosphorylation of TIM bound with PER (Top et al. 2016).
CRY
Mammalian CRY also has multiple phosphorylation sites (Lamia et al. 2009; Gao et al. 2013; Ode et al. 2017) targeted by various kinases, including CK1δ/ε (Eide et al. 2002; Qin et al. 2015; Ode et al. 2017) and GSK3β (Kurabayashi et al. 2010). The multisite phosphorylation of CRY plays a critical role in period determination since combinatorial mutations in the phosphorylation sites results in the twofold change of the period length from 19 to 38 h (Ode et al. 2017). CRY phosphorylation sites appear to be concentrated in the P-loop and carboxy terminus, and mutations in several phosphorylation sites in these regions significantly affect the circadian period (Gao et al. 2013; Liu and Zhang 2016; Ode et al. 2017). Furthermore, a recent study identified a FASPS pedigree CRY mutation in which an alanine in the P-loop region is replaced by threonine (Hirano et al. 2016). This substitution might create a de novo phosphorylation site around the P-loop that modulates the circadian period. Indeed, the threonine-substitution phenotype can be recapitulated by aspartic-acid substitution, which mimics the phosphorylated residue (Hirano et al. 2016).
FRQ
Like PER, FRQ is progressively phosphorylated during the circadian cycle (Garceau et al. 1997) at multiple sites, many of which are presumably phosphorylated by CK1 and CK2 (Baker et al. 2009; Tang et al. 2009). CK1 stably binds FRQ and is responsible for its clustered phosphorylation (He et al. 2006; Baker et al. 2009; Querfurth et al. 2011). CK2 also directly phosphorylates FRQ (Yang et al. 2003; Mehra et al. 2009).
CCA1
In Arabidopsis, CCA1 is phosphorylated by CK2 to obtain the proper circadian period (Sugano et al. 1998, 1999; Daniel et al. 2004). Depleting CK2 or increasing its activity lengthens or shortens the circadian period, respectively (Sugano et al. 1999; Lu et al. 2011). Although the kinases responsible for phosphorylating CCA1 are not as well understood as those in other conventional model organisms, several circadian proteins exhibit circadian variations in their phosphorylation state (Fujiwara et al. 2008), suggesting that multisite phosphorylation is critical in regulating the circadian period in plants (Seo and Mas 2014). A study using the unicellular plant Ostreococcus tauri revealed that pharmacological inhibition of CK1 markedly lengthens the period over 8 h, suggesting that the central role of CK1 in determining the circadian period is conserved in the plant kingdom (van Ooijen et al. 2013).
Temperature Compensation
CK1 and CK2 and their substrates are important for the temperature compensation that preserves the circadian period. Mammalian CK1 is linked to temperature compensation (Tosini and Menaker 1998; Isojima et al. 2009; Zhou et al. 2015), and CK2 is at least partly responsible for the temperature compensation in Neurospora and higher plants (Mehra et al. 2009; Portoles and Mas 2010).
In general, increased temperature accelerates biochemical reactions such as phosphorylation and protein degradation, thus raising the temperature should accelerate clock speed. However, a previous study indicated that CK1δ/ε kinase activity markedly influences the speed of mammalian circadian clocks and is largely insensitive to temperature changes in vitro (Isojima et al. 2009). A subsequent study revealed that temperature-insensitive kinase activity is achieved through CK1δ/ε’s ability to bind substrate and phosphorylated products in a temperature-dependent manner (Shinohara et al. 2017). When CK1 acts on a substrate with a single phosphorylation site, which may be similar to the reaction step for catalyzing the priming phosphorylation site, raising the temperature decreases the affinity between the kinase and the substrate to be phosphorylated (Fig. 3C). In addition, when CK1 acts on a substrate with multiple phosphorylation sites, raising the temperature increases the affinity between the kinase and the phosphorylated product at multiple sites in the presence of ADP, reducing the turnover number of CK1 action (Fig. 3D). These temperature-sensitive affinity alterations reduce substrate binding and product release at higher temperatures. Therefore, the net kinase-reaction rate stays nearly constant despite temperature changes. Ueda and his colleagues successfully conferred temperature insensitivity on another temperature-sensitive kinase protein by introducing an evolutionarily conserved CK1-specific domain that seems to bind to substrates and phosphorylated product.
The presence of a temperature-insensitive kinase such as CK1 can provide another layer of regulation to achieve the circadian temperature compensation. Detailed analysis of the degradation kinetics of mammalian PER showed that raising the temperature stabilizes PER, which were proposed by the combination of temperature-insensitive and temperature-sensitive kinases (Zhou et al. 2015). At higher temperatures, CK1δ/ε preferentially phosphorylates PER on the PER-stabilizing domain (which contains the site of the mutation found in the FASPS pedigree), while at lower temperatures, the CK1δ/ε preferentially phosphorylates PER on the PER-destabilizing domain (Fig. 3E). Thus, increasing the temperature decelerates the PER degradation, slowing down the pace of the circadian clock. The similar temperature-dependent stability mechanism was also proposed in Neurospora FRQ. A study proposed that increasing the temperature accelerates FRQ stabilization and counteracts any temperature-dependent acceleration of the circadian clock (Mehra et al. 2009). This balance can be modulated by mutations of either CK2 or the CK2-phophorylation site on FRQ. These studies indicated that temperature-dependent kinase activity counteracts the canonical acceleration of clock speed in response to a rise in temperature by modulating the substrate degradation rate.
To summarize, the CK1δ/ε kinase reaction itself can be insensitive to temperature changes, providing the mechanism of temperature compensation at the systems’ component level. This temperature-insensitive kinase together with other temperature-sensitive kinases (or probably phosphatases) may provide the system-level tuning of FRQ/PER proteolysis, which further compensates the effect of temperature changes. These mechanisms are not mutually exclusive in circadian clocks (Hogenesch and Ueda 2011); however, the former model suggests that temperature-insensitive reactions are not exclusive to the circadian substrate. Rather, if the affinity of CK1δ/ε for phosphorylated products is important for temperature-insensitive kinase reactions, such temperature-compensated reactions may also function in other biological processes. In a study of temperature-compensated budding yeast metabolic oscillation, the oscillation period was sensitive to CK1 and GSK3β (Causton et al. 2015). Indeed, a budding yeast homologue of CK1 also preserves temperature-compensated kinase activity (Shinohara et al. 2017). Therefore, the CK1 enzyme might confer robust biochemical reaction rates on circadian clock and other systems to protect against environmental temperature fluctuations.
MULTISITE PHOSPHORYLATION AT FLEXIBLE PROTEIN REGIONS
The multisite phosphorylation process itself may contribute to period determination independent of the regulation of protein degradation. In a recent study in Neurospora, mutations in FRQ phosphorylation sites strongly affected the circadian period even in a genetic background in which FRQ’s stability was unchanged (Larrondo et al. 2015). In another study, a series of phosphorylation-site mutants of mammalian CRY severely affected the circadian period, even though CRY’s stability was only marginally affected (Ode et al. 2017). Furthermore, a deletion-mutant CRY found in delayed sleep-phase disorder extends the circadian period without a detectable change in the stability of the CRY protein (Patke et al. 2017). In plants, CK2 phosphorylation regulates CCA1 transcription without affecting the amount of CCA1 protein (Portoles and Mas 2010).
KaiC studies strongly suggest that circadian clock speed is encoded as the time necessary for changes in protein structure. On the other hand, a common feature of other circadian-period regulatory processes is that the period is largely determined by multisite phosphorylation by kinases that elicit clustered and sequential phosphorylation, although the events downstream of the phosphorylation may vary among circadian systems. Are there any links connecting the design principles of the KaiC oscillator and multisite phosphorylation-based oscillation? Serial enzymatic reactions such as multisite phosphorylation can produce cooperative responses and a sufficient delay to produce fully modified substrates, and can potentially determine period length (Salazar and Hofer 2009; Salazar et al. 2010; Ferrell and Ha 2014). Studies suggest that even apart from its time-related characteristics, multisite and clustered phosphorylation might be able to strongly modulate protein structure.
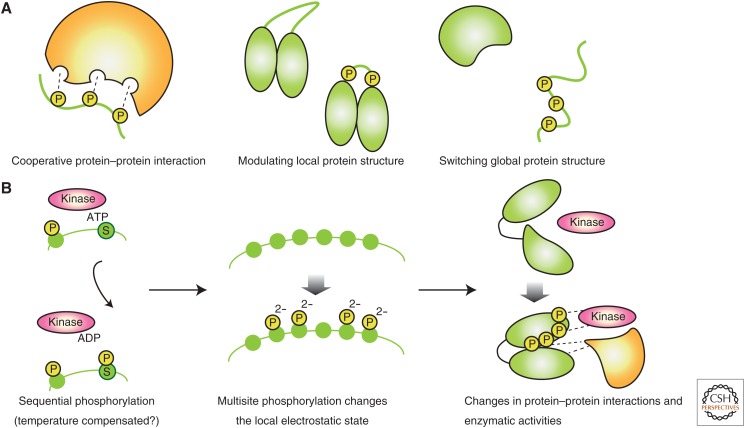
Multisite phosphorylation often affects protein structure and function by acting on intrinsically disordered regions (IDRs) of substrate proteins (Valk et al. 2014; Wright and Dyson 2015). In IDRs, the polypeptide structure is highly flexible, not rigid as in an α-helical conformation. IDRs tend to be located at the protein surface where they are accessible to kinases. Indeed, statistical analyses of proteomics data indicated that phosphorylation sites and that IDRs are located close together (Iakoucheva et al. 2004; Xie et al. 2007), and IDRs are often regulated by phosphorylation (Holt et al. 2009; Tyanova et al. 2013; Sharma et al. 2014). Multiple phosphorylations on IDRs dramatically alter the bulk electrostatic potential of the region and may induce dynamic structural changes (Fig. 4A) (Theillet et al. 2014). When the flexibility of an IDR strongly affects intramolecular interactions, multisite phosphorylation can change the overall structure of the proteins themselves. If the IDR is located at the interface of protein–protein interactions, such structural changes could affect binding affinity.
Figure 4.
Multisite phosphorylation affects protein structure and function. (A) Multisite phosphorylation, particularly around the flexible protein regions, affects protein–protein interactions and local/global protein structure. (B) The multisite phosphorylation process could be a period-determining step in eukaryotic circadian clocks. Reaction rates of the kinases responsible for these events can be temperature-insensitive. This abstractive principle may be compatible with the design principle of cyanobacterial circadian-period determination (see Fig. 2C).
Multisite phosphorylations on IDRs may have a critical role for circadian-period control. It has been suggested that FRQ is an intrinsically disordered protein (IDP) (Hurley et al. 2013). Multisite phosphorylation changes the structure of FRQ (Querfurth et al. 2011), and the nonphosphorylated FRQ is proposed to have a compact form through an interaction between the basic amino-terminal domain and the acidic carboxy-terminal domain. Multisite phosphorylation of the amino-terminal domain loosens the interaction, and the FRQ becomes more sensitive to limited protease digestion. The multisite phosphorylation on PER appears to be similar to FRQ. Although PER contains functionally important structured domains, such as Par-Arnt-Sim (PAS) domains and the CRY-binding domain (Hennig et al. 2009; Kucera et al. 2012; Nangle et al. 2014), most of the other regions are predicted to be disordered (Gustafson and Partch 2015). In both mammalian and Drosophila PER, phosphorylation states control the PER global structure, as indicated by an altered susceptibility to protease treatment (Chiu et al. 2011; Zhou et al. 2015). In contrast to FRQ and PER, the structure of the photolyase homology domain of cryptochrome superfamily proteins is well conserved, indicating that CRY is not an IDP. Nonetheless, clustered phosphorylation sites that are critical for controlling the circadian period are located near the flexible P-loop (Hirano et al. 2016; Liu and Zhang 2016; Ode et al. 2017). Multisite phosphorylation appears to additively regulate the circadian period, and the number of phosphorylation sites around this loop domain may serve as a cumulative timer for the circadian period (Ode et al. 2017). These lines of evidence suggest that structural changes in flexible regions, elicited by multisite phosphorylation, are a critical step in determining the circadian period.
Although the detailed mechanisms of period-determination remain unknown, the outcome of phosphorylation-dependent protein structural change could be coupled to TTFL-based architecture via proteolysis (Chiu et al. 2011; Zhou et al. 2015). Changes in the multiple phosphorylation state would also affect the protein–protein interaction. We further point out that the substrate with multiple phosphorylation sites can reciprocally regulate the action of kinases such as CK1. For example, PER protein alters the CK1’s substrate preference (Qin et al. 2015). Anionic small molecules and phosphorylated peptides can bind to the allosteric site of CK1 and thereby modulate the CK1’s enzymatic activity (Shinohara et al. 2017). These substrate–CK1 interactions may imply that phosphorylation events in a stable protein complex, including CK1, PER, and CRY, should not be interpreted as a simple mixture of kinases and substrates. Instead, the components in a kinase–substrate complex cooperatively regulate the process of multiple phosphorylation, providing the temperature-compensated and accurate period-determining information for the eukaryotic circadian clocks.
CONCLUDING REMARKS
The importance of multisite phosphorylation and structural changes in eukaryotic circadian-period control remind us of the unsolved questions: What is the source of the 24-h period? What are the rate-limiting steps? Why is the circadian period is so slow (a scale of minutes) compared with biochemical events in general (a scale of seconds), but still accurate (Akiyama 2012). Studies of the KaiC oscillator revealed a structural basis for slow kinetics (Abe et al. 2015). In contrast, a structural basis for its accuracy remains unknown. Although transcription–translation dynamics are at the core of noncyanobacterial circadian clocks, we may well find phosphorylation-dependent structural changes at the core of timekeeping mechanisms in these organisms. In addition, a recent study proposed that the slow cis/trans isomerization of specific peptide bonding in BMAL1 is a timekeeping mechanism in mammalian circadian clocks (Gustafson et al. 2017). Such rate-limiting events, encoded as protein structures, could be induced directly (via control of the local electrostatic state) or indirectly (via protein–protein interactions) through multisite phosphorylation. Furthermore, the temperature-compensation property of reaction speed could be encoded via multisite phosphorylation by CK1 (Isojima et al. 2009; Shinohara et al. 2017).
From this perspective, the circadian timekeeping mechanism in the KaiC protein oscillator and eukaryotic circadian oscillator may converge as the progressive regulation of protein structure powered by temperature-compensated enzymatic activities—KaiC’s ATPase activity and CK1’s phosphorylation activity (Fig. 4B), thus encoding circadian-period determination as a quality rather than quantity of protein dynamics (Larrondo et al. 2015; Ode et al. 2017). The rate-limiting step should be encoded as a protein structural property to keep the period length constant regardless of fluctuations in expression (Dibner et al. 2009), especially for low-abundance circadian-transcription factors (Forger and Peskin 2005; Narumi et al. 2016). To evaluate this perspective, we must execute a structure-based analysis of the clock proteins, and reconstruct and design circadian timekeeping steps in eukaryotic circadian systems.
ACKNOWLEDGMENTS
Studies from our laboratory introduced in this manuscript were supported in part by Japan Agency for Medical Research and Development (AMED)-Core Research for Evolutional Science and Technology (CREST), CREST, Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS), the Basic Science and Platform Technology Program for Innovative Biological Medicine, Japan Society for the Promotion of Science (JSPS) (25221004, 23115006, 25830146, 16K14653), Rikagaku Kenkyusho (Institute of Physical and Chemical Research, Japan—RIKEN), and the Takeda Science Foundation.
Footnotes
Editors: Paolo Sassone-Corsi, Michael W. Young, and Akhilesh B. Reddy
Additional Perspectives on Circadian Rhythms available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abe J, Hiyama TB, Mukaiyama A, Son S, Mori T, Saito S, Osako M, Wolanin J, Yamashita E, Kondo T, et al. 2015. Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349: 312–316. [DOI] [PubMed] [Google Scholar]
- Akiyama S. 2012. Structural and dynamic aspects of protein clocks: How can they be so slow and stable? Cell Mol Life Sci 69: 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. 2003. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6: 251–257. [DOI] [PubMed] [Google Scholar]
- Albrecht U. 2013. Circadian clocks and mood-related behaviors. In Handbook of experimental pharmacology (ed. Kramer A, Merrow M), pp. 227–239. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Angeli D, Sontag ED. 2008. Translation-invariant monotone systems, and a global convergence result for enzymatic futile cycles. Nonlinear Anal Real World Appl 9: 128–140. [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. 2009. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell 34: 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. 2010. Circadian integration of metabolism and energetics. Science 330: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J. 2008. PERspective on PER phosphorylation. Genes Dev 22: 1737–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Kowalska E, Dallmann R. 2012. (Re)inventing the circadian feedback loop. Dev Cell 22: 477–487. [DOI] [PubMed] [Google Scholar]
- Causton HC, Feeney KA, Ziegler CA, O’Neill JS. 2015. Metabolic cycles in yeast share features conserved among circadian rhythms. Curr Biol 25: 1056–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YG, Kuo NW, Tseng R, LiWang A. 2011. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci 108: 14431–14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YG, Cohen SE, Phong C, Myers WK, Kim YI, Tseng R, Lin J, Zhang L, Boyd JS, Lee Y, et al. 2015. Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349: 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, Ye ZY, Pan HL, Takahashi JS. 2012. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci 109: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. 2008. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 22: 1758–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I. 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ. 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci 103: 18143–18147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clodong S, Duhring U, Kronk L, Wilde A, Axmann I, Herzel H, Kollmann M. 2007. Functioning and robustness of a bacterial circadian clock. Mol Syst Biol 3: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradi C, Shiu A. 2015. A global convergence result for processive multisite phosphorylation systems. Bull Math Biol 77: 126–155. [DOI] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM. 2004. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci 101: 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. 2010. A synchronized quorum of genetic clocks. Nature 463: 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Sage D, Unser M, Bauer C, d’Eysmond T, Naef F, Schibler U. 2009. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. 1994. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci 91: 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. 2002. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iε. J Biol Chem 277: 17248–17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. 2005. Control of mammalian circadian rhythm by CKIε-regulated proteasome-mediated PER2 degradation. Mol Cell Biol 25: 2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338. [DOI] [PubMed] [Google Scholar]
- Ewer J, Rosbash M, Hall JC. 1988. An inducible promoter fused to the period gene in Drosophila conditionally rescues adult per-mutant arrhythmicity. Nature 333: 82–84. [DOI] [PubMed] [Google Scholar]
- Fan Y, Hida A, Anderson DA, Izumo M, Johnson CH. 2007. Cycling of CRYPTOCHROME proteins is not necessary for circadian-clock function in mammalian fibroblasts. Curr Biol 17: 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. 2007. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1). Genes Dev 21: 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE Jr, Ha SH. 2014. Ultrasensitivity part II: Multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends Biochem Sci 39: 556–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. 1990. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem 265: 14264–14269. [PubMed] [Google Scholar]
- Forger DB. 2011. Signal processing in cellular clocks. Proc Natl Acad Sci 108: 4281–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. 2005. Stochastic simulation of the mammalian circadian clock. Proc Natl Acad Sci 102: 321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame S, Cohen P. 2001. GSK3 takes centre stage more than 20 years after its discovery. Biochem J 359: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. 1994. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12: 555–570. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Yagita K, Okamura H. 2006. Does mPER2 protein oscillate without its coding mRNA cycling?: Post-transcriptional regulation by cell clock. Genes Cells 11: 525–530. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE. 2008. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155: 793–806. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148. [DOI] [PubMed] [Google Scholar]
- Gao P, Yoo SH, Lee KJ, Rosensweig C, Takahashi JS, Chen BP, Green CB. 2013. Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J Biol Chem 288: 35277–35286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, Dunlap JC. 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469–476. [DOI] [PubMed] [Google Scholar]
- Gerard C, Gonze D, Goldbeter A. 2009. Dependence of the period on the rate of protein degradation in minimal models for circadian oscillations. Philos Trans A Math Phys Eng Sci 367: 4665–4683. [DOI] [PubMed] [Google Scholar]
- Gorl M, Merrow M, Huttner B, Johnson J, Roenneberg T, Brunner M. 2001. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J 20: 7074–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, McClung CR. 2015. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16: 598–610. [DOI] [PubMed] [Google Scholar]
- Gustafson CL, Partch CL. 2015. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry 54: 134–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson CL, Parsley NC, Asimgil H, Lee HW, Ahlbach C, Michael AK, Xu H, Williams OL, Davis TL, Liu AC, et al. 2017. A slow conformational switch in the BMAL1 transactivation domain modulates circadian rhythms. Mol Cell 66: 447–457 e447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Koh K, Combs DJ, Sehgal A. 2011. Post-translational regulation and nuclear entry of TIMELESS and PERIOD are affected in new timeless mutant. J Neurosci 31: 9982–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee HC, Yang Y, Liu Y. 2006. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20: 2552–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S, Strauss HM, Vanselow K, Yildiz O, Schulze S, Arens J, Kramer A, Wolf E. 2009. Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol 7: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152: 1106–1118. [DOI] [PubMed] [Google Scholar]
- Hirano A, Shi G, Jones CR, Lipzen A, Pennacchio LA, Xu Y, Hallows WC, McMahon T, Yamazaki M, Ptacek LJ, et al. 2016. A Cryptochrome 2 mutation yields advanced sleep phase in humans. eLife 5: 16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A, Braas D, Fu YH, Ptacek LJ. 2017. FAD regulates CRYPTOCHROME protein stability and circadian clock in mice. Cell Rep 19: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D, et al. 2010. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol 8: e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. 2008. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc Natl Acad Sci 105: 20746–20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, et al. 2012. Identification of small molecule activators of cryptochrome. Science 337: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Ueda HR. 2011. Understanding systems-level properties: Timely stories from the study of clocks. Nat Rev Genet 12: 407–416. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Larrondo LF, Loros JJ, Dunlap JC. 2013. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ. Mol Cell 52: 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281: 1519–1523. [DOI] [PubMed] [Google Scholar]
- Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, et al. 2009. CKIε/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci 106: 15744–15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley CC, Ode KL, Ueda HR. 2012. A design principle for a posttranslational biochemical oscillator. Cell Rep 2: 938–950. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M. 2008. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol 6: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK. 2016. Protein sequestration versus Hill-type repression in circadian clock models. IET Syst Biol 10: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. 2008. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev 22: 1513–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. 2010. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3β/SGG in circadian clocks. J Neurosci 30: 12664–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera N, Schmalen I, Hennig S, Ollinger R, Strauss HM, Grudziecki A, Wieczorek C, Kramer A, Wolf E. 2012. Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc Natl Acad Sci 109: 3311–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y. 2010. DYRK1A and glycogen synthase kinase 3β, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol 30: 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo LF, Olivares-Yanez C, Baker CL, Loros JJ, Dunlap JC. 2015. Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination. Science 347: 1257277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855–867. [DOI] [PubMed] [Google Scholar]
- Levi F, Schibler U. 2007. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 47: 593–628. [DOI] [PubMed] [Google Scholar]
- Lin C, Todo T. 2005. The cryptochromes. Genome Biol 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. 2002. A role for casein kinase 2α in the Drosophila circadian clock. Nature 420: 816–820. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhang EE. 2016. Phosphorylation regulating the ratio of intracellular CRY1 protein determines the circadian period. Front Neurol 7: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Loros J, Dunlap JC. 2000. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci 97: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Liu H, Knowles SM, Li J, Ma L, Tobin EM, Lin C. 2011. A role for protein kinase casein kinase2 α-subunits in the Arabidopsis circadian clock. Plant Physiol 157: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, McClung CR, Zhang C. 2017. Tick tock: Circadian regulation of plant innate immunity. Annu Rev Phytopathol 55: 287–311. [DOI] [PubMed] [Google Scholar]
- Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A. 2009. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev 23: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769–779. [DOI] [PubMed] [Google Scholar]
- Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, Dunlap JC. 2009. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, et al. 2008. Setting clock speed in mammals: The CK1 ε tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millius A, Ueda HR. 2017. Systems biology-derived discoveries of intrinsic clocks. Front Neurol 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. 2012. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M. 2008. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells 13: 387–395. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. 2004. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Ito H, Kondo T. 2010. In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett 584: 898–902. [DOI] [PubMed] [Google Scholar]
- Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, Zheng N. 2014. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife 3: e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi R, Shimizu Y, Ukai-Tadenuma M, Ode KL, Kanda GN, Shinohara Y, Sato A, Matsumoto K, Ueda HR. 2016. Mass spectrometry-based absolute quantification reveals rhythmic variation of mouse circadian clock proteins. Proc Natl Acad Sci 113: E3461–E3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M. 2004. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell 13: 213–223. [DOI] [PubMed] [Google Scholar]
- Nishii K, Yamanaka I, Yasuda M, Kiyohara YB, Kitayama Y, Kondo T, Yagita K. 2006. Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: A real-time analysis. Neurosci Lett 401: 44–48. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. 2007. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J 26: 4029–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak B, Tyson JJ. 2008. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 9: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ode KL, Ukai H, Susaki EA, Narumi R, Matsumoto K, Hara J, Koide N, Abe T, Kanemaki MT, Kiyonari H, et al. 2017. Knockout-rescue embryonic stem cell-derived mouse reveals circadian-period control by quality and quantity of CRY1. Mol Cell 65: 176–190. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. 2011. Circadian clocks in human red blood cells. Nature 469: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. 2011. Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Yamanaka I, Kumar A, Yamaguchi J, Nishiwaki-Ohkawa T, Muto K, Kawamura R, Hirota T, Yagita K, Irle S, et al. 2015. C-H activation generates period-shortening molecules that target cryptochrome in the mammalian circadian clock. Angew Chem Int Ed Engl 54: 7193–7197. [DOI] [PubMed] [Google Scholar]
- Ozturk N, Song SH, Ozgur S, Selby CP, Morrison L, Partch C, Zhong D, Sancar A. 2007. Structure and function of animal cryptochromes. Cold Spring Harb Symp Quant Biol 72: 119–131. [DOI] [PubMed] [Google Scholar]
- Patke A, Murphy PJ, Onat OE, Krieger AC, Ozcelik T, Campbell SS, Young MW. 2017. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169: 203–215 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portoles S, Mas P. 2010. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Qin X, Mori T, Zhang Y, Johnson CH. 2015. PER2 differentially regulates clock phosphorylation versus transcription by reciprocal switching of CK1ε activity. J Biol Rhythms 30: 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth C, Diernfellner AC, Gin E, Malzahn E, Hofer T, Brunner M. 2011. Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol Cell 43: 713–722. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Rey G. 2014. Metabolic and nontranscriptional circadian clocks: Eukaryotes. Annu Rev Biochem 83: 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Kramer A. 2011. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett 585: 1393–1399. [DOI] [PubMed] [Google Scholar]
- Rosbash M. 2009. The implications of multiple circadian clock origins. PLoS Biol 7: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. 2007. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318: 809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar C, Hofer T. 2009. Multisite protein phosphorylation—From molecular mechanisms to kinetic models. FEBS J 276: 3177–3198. [DOI] [PubMed] [Google Scholar]
- Salazar C, Brummer A, Alberghina L, Hofer T. 2010. Timing control in regulatory networks by multisite protein modifications. Trends Cell Biol 20: 634–641. [DOI] [PubMed] [Google Scholar]
- Sanchez SE, Kay SA. 2017. The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Mas P. 2014. Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanware NP, Hutchinson JA, Kim SH, Zhan L, Bowler MJ, Tibbetts RS. 2011. Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J Biol Chem 286: 12766–12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, D’Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, Mann M. 2014. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep 8: 1583–1594. [DOI] [PubMed] [Google Scholar]
- Shinohara Y, Koyama YM, Ukai-Tadenuma M, Hirokawa T, Kikuchi M, Yamada RG, Ukai H, Fujishima H, Umehara T, Tainaka K, Ueda HR. 2017. Temperature-sensitive substrate and product binding underlie temperature-compensated phosphorylation in the clock. Mol Cell 67: 783–798. [DOI] [PubMed] [Google Scholar]
- Shirogane T, Jin J, Ang XL, Harper JW. 2005. SCFβ-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem 280: 26863–26872. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. 2007. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Schuller JM, Wiegard A, Lossl P, Schmelling N, Axmann IM, Plitzko JM, Forster F, Heck AJ. 2017. Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science 355: 1181–1184. [DOI] [PubMed] [Google Scholar]
- St-Denis N, Gabriel M, Turowec JP, Gloor GB, Li SS, Gingras AC, Litchfield DW. 2015. Systematic investigation of hierarchical phosphorylation by protein kinase CK2. J Proteomics 118: 49–62. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM. 1998. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci 95: 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. 1999. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci 96: 12362–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed S, Saez L, Young MW. 2011. Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. J Biol Chem 286: 27654–27662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18: 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Kubota Y, Tsujimura T, Kumano M, Masai H, Takisawa H. 2009. Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes Cells 14: 949–963. [DOI] [PubMed] [Google Scholar]
- Tang CT, Li S, Long C, Cha J, Huang G, Li L, Chen S, Liu Y. 2009. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc Natl Acad Sci 106: 10722–10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SW, Mukherji S, Moffitt JR, de Buyl S, O’Shea EK. 2013. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. Science 340: 737–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. 2007. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci 104: 16377–16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet FX, Binolfi A, Frembgen-Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley PB, Gierasch L, Pielak GJ, et al. 2014. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem Rev 114: 6661–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040–1043. [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. 2005. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251–254. [DOI] [PubMed] [Google Scholar]
- Top D, Harms E, Syed S, Adams EL, Saez L. 2016. GSK-3 and CK2 kinases converge on timeless to regulate the master clock. Cell Rep 16: 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosini G, Menaker M. 1998. The tau mutation affects temperature compensation of hamster retinal circadian oscillators. Neuroreport 9: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Tseng R, Chang YG, Bravo I, Latham R, Chaudhary A, Kuo NW, Liwang A. 2014. Cooperative KaiA-KaiB-KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J Mol Biol 426: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng R, Goularte NF, Chavan A, Luu J, Cohen SE, Chang YG, Heisler J, Li S, Michael AK, Tripathi S, et al. 2017. Structural basis of the day–night transition in a bacterial circadian clock. Science 355: 1174–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Akashi M, Matsuda M, Goto K, Miyata Y, Node K, Nishida E. 2009. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal 2: ra26. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Cox J, Olsen J, Mann M, Frishman D. 2013. Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS Comput Biol 9: e1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai H, Ueda HR. 2010. Systems biology of mammalian circadian clocks. Annu Rev Physiol 72: 579–603. [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. 2011. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144: 268–281. [DOI] [PubMed] [Google Scholar]
- Valk E, Venta R, Ord M, Faustova I, Koivomagi M, Loog M. 2014. Multistep phosphorylation systems: Tunable components of biological signaling circuits. Mol Biol Cell 25: 3456–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, Millar AJ. 2012. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem Sci 37: 484–492. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Hindle M, Martin SF, Barrios-Llerena M, Sanchez F, Bouget FY, O’Neill JS, Le Bihan T, Millar AJ. 2013. Functional analysis of casein kinase 1 in a minimal circadian system. PLoS ONE 8: e70021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. 2006. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev 20: 2660–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zon JS, Lubensky DK, Altena PR, ten Wolde PR. 2007. An allosteric model of circadian KaiC phosphorylation. Proc Natl Acad Sci 104: 7420–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Young MW. 1995. Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron 15: 345–360. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. 1995. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697–706. [DOI] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. 2004. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr Biol 14: 1481–1486. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. 2007. Functional anthology of intrinsic disorder. 3: Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res 6: 1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. 2005. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434: 640–644. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH. 2013. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, van Der Horst GT, Okamura H. 2001. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292: 278–281. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Forger D. 2010. Multiscale complexity in the mammalian circadian clock. Curr Opin Genet Dev 20: 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Yagita K, Okamura H. 2005. Role of cyclic mPer2 expression in the mammalian cellular clock. Mol Cell Biol 25: 1912–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Takahashi JS. 2005. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol 393: 288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. 2001. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453–467. [DOI] [PubMed] [Google Scholar]
- Yang Y, Cheng P, He Q, Wang L, Liu Y. 2003. Phosphorylation of FREQUENCY protein by casein kinase II is necessary for the function of the Neurospora circadian clock. Mol Cell Biol 23: 6221–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Houl JH, Hardin PE. 2011. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol 21: 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW III, Janes J, et al. 2009. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Kim JK, Eng GW, Forger DB, Virshup DM. 2015. A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol Cell 60: 77–88. [DOI] [PubMed] [Google Scholar]