SUMMARY
To understand the assembly and functional outcomes of protein–RNA regulation, it is crucial to precisely identify the positions of such interactions. Cross-linking and immunoprecipitation (CLIP) serves this purpose by exploiting covalent protein–RNA cross-linking and RNA fragmentation, along with a series of stringent purification and quality control steps to prepare complementary DNA (cDNA) libraries for sequencing. Here we describe the core steps of CLIP, its primary variations, and the approaches to data analysis. We present the application of CLIP to studies of specific cell types in genetically engineered mice and discuss the mechanistic and physiologic insights that have already been gained from studies using CLIP. We conclude by discussing the future opportunities for CLIP, including studies of human postmortem tissues from disease patients and controls, RNA epigenetic modifications, and RNA structure. These and other applications of CLIP will continue to unravel fundamental gene regulatory mechanisms while providing important biologic and clinically relevant insights.
1. INTRODUCTION
A crucial step in understanding RNA-related gene regulation and its relationship to disease is identifying how RNAs are bound and hence regulated by RNA-binding proteins (RBPs) in specific cell types and subcellular compartments. Driving our interest in this topic has been the growing list of human neurologic diseases that have RNA dysregulation at their core (Conlon and Manley 2017). The first method developed for this purpose used antibodies against the spliceosomal Sm proteins (lupus autoimmune sera) to identify the small nuclear RNAs (snRNAs), which interact with Sm proteins within the abundant small nuclear ribonucleoprotein complexes (snRNPs) (Lerner and Steitz 1979). This method, later referred to as RIP (RNP/RNA immunoprecipitation), relies on immunoprecipitation (IP) of an RBP under conditions that preserve RNPs (Niranjanakumari et al. 2002). However, RIP can suffer from low specificity, partly because it preserves protein–protein interactions and can therefore purify multiple RBPs in complex with their bound RNAs, and partly because RNA–protein complexes can reassociate in vitro (Mili and Steitz 2004).
To understand the assembly of RNPs, a method to identify direct protein–RNA interactions with high specificity was required. Moreover, it was important to also identify the positions on RNAs that are recognized by a specific RBP to identify the RNA sequences and structures where regulation takes place. We have developed cross-linking and immunoprecipitation (CLIP) for this purpose by exploiting zero-length covalent protein–RNA cross-linking and RNA fragmentation, along with a series of stringent purification and quality control steps, followed by the preparation of a complementary DNA (cDNA) library for sequencing (Ule et al. 2003; Darnell 2010). With CLIP, only RNA fragments that are cross-linked to the specific RBP are isolated under well-optimized conditions that avoid copurification of additional RBPs. CLIP data can be validated with the use of appropriate negative controls, such as the use of cells or tissue that lack the RBP of interest or have not been cross-linked (Licatalosi and Darnell 2010). CLIP is now generally accepted as a highly reliable method for identifying the positions of endogenous RNA–protein interactions (Conlon and Manley 2017).
Initially, CLIP relied on Sanger sequencing to identify 340 sequences corresponding to RNA interactions of splicing factors Nova1 and Nova2 in the mouse brain (Ule et al. 2003). Despite the limited number of these initial sequences, many were located next to alternative exons that turned out to be regulated by Nova proteins, thus demonstrating the capacity of CLIP to identify functionally important binding sites. Subsequent development of HITS-CLIP (high-throughput sequencing of RNA isolated by CLIP) was then applied to further studies of the functional interactions of Nova and several other RBPs with neuronal transcripts in the mouse brain (Licatalosi et al. 2008; Darnell 2013).
This work established a general strategy to link functional studies with a transcriptome-wide map of protein–RNA interactions. Such maps validated regulatory rules that had been bioinformatically predicted, in which the position of binding of RBPs within primary transcripts largely determines the outcome of RNA regulation (Ule et al. 2006; Licatalosi et al. 2008; Licatalosi and Darnell 2010). These rules are now recognized as a general feature of pre-mRNA processing, applicable to dozens of different RBPs (Witten and Ule 2011). Following these approaches, CLIP has been applied to a large number of different RBPs in a variety of cell types, organs, and species.
2. THE CLIP METHOD
2.1. The Fundamentals
Many variant protocols of CLIP have been derived during the last decade. Their detailed variations have been reviewed elsewhere (Darnell 2010; Lee and Ule 2018), and therefore we will focus here only on the primary steps that are common to most protocols (Fig. 1). A feature common to all protocols is that RBPs are cross-linked by ultraviolet (UV) light to their bound RNAs within the intact endogenous environment. Acutely harvested cells, triturated or pulverized tissues, or even whole organisms such as Caenorhabditis elegans are immediately exposed to UV light, which cross-links RBPs to their bound RNAs in their native (intracellular) state. We have adopted use of the term “CLIP” as a general way to refer to all protocols that use UV cross-linking and protein purification of directly bound native RNA–protein complexes.
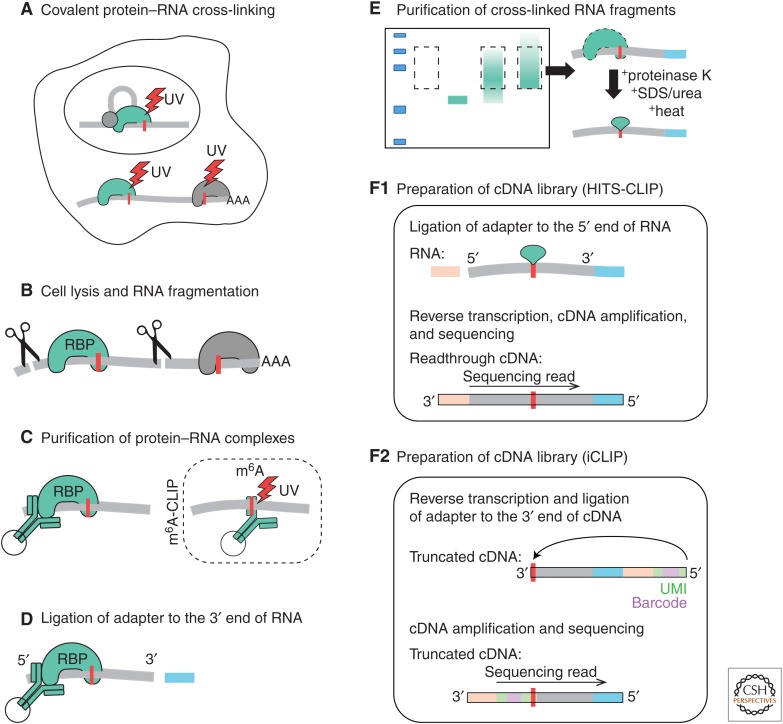
Figure 1.
The core steps of cross-linking and immunoprecipitation (CLIP). The steps A–E are common across most variants of CLIP. Methods that identify the sites of RNA methylation cross-link purified RNA to an antibody specific to a methylated base, as in m6A-CLIP, and then proceed from step C onward. The first option to prepare the complementary DNA (cDNA) library is as in high-throughput sequencing (HITS)-CLIP (F1), where the cDNA library is prepared by ligating an adapter to the 5′ end of RNA to allow amplification and sequencing of readthrough cDNAs, but not truncated cDNAs. The second option is as in individual nucleotide resolution CLIP (iCLIP) (F2), in which an adapter is ligated to the 3′ end of cDNAs, which allows amplification of both readthrough and truncated cDNAs (only a truncated cDNA is shown here, because this type most often represents >90% of the iCLIP cDNA library) (Sugimoto et al. 2012; Haberman et al. 2017). The unique molecular identifiers (UMIs) and experimental barcodes, which are introduced in iCLIP during reverse transcription (RT), are color coded in green and violet. The RNA fragments and their corresponding cDNA inserts are in gray. (Adapted based on previous publication by Lee and Ule 2018.)
Most CLIP variants exploit the capacity of UV light to promote formation of covalent bonds between RBPs and their direct RNA-binding sites. Unlike formaldehyde cross-linking that is used in chromatin immunoprecipitation (ChIP) and some variants of RIP, UV does not cross-link proteins to each other. UV cross-linking requires direct (Angstrom) contact between an amino acid and a nucleobase, and therefore ensures that only direct protein–RNA interactions are preserved. The high strength of the covalent bond allows a stringent buffer containing ionic detergents to be used for cell lysis, which disrupts most non-cross-linked protein–protein and protein–RNA interactions. The stringent buffer also increases the accessibility of RNA for RNase fragmentation and minimizes the chance of copurifying multiple associated RBPs during later IP, thus increasing data specificity. RNA fragmentation is crucial to avoid copurifying multiple RBPs that cross-link to the same RNAs, and to provide insight into the position of RNA-binding sites, because the RNA fragment contains the cross-link site. A well-optimized RNase concentration is important to minimize the constraints at the sites of RNase cleavage, which can lead to biased binding site assignment (Haberman et al. 2017).
Purification of endogenous (i.e., untagged) RBPs normally requires that antibodies are available for efficient IP. The need for stringent purification and quality control varies to some extent on the type and expression of the RBP being studied, but in general this improves signal to noise of detection of directly bound RNAs. Some RBPs participate in stable RNPs that may not efficiently dissociate under standard CLIP IP conditions, thus increasing their risk of copurifying multiple RBPs, and careful protocols have articulated how to optimize the stringency of CLIP IPs (see Darnell 2012). Even a minor copurification of an abundant RBP that cross-links with high efficiency can lead to a major loss of specificity if the RBP of interest is of low abundance or cross-links poorly. In such cases, RBPs can be epitope-tagged to allow purification under denaturing conditions (Granneman et al. 2009; Huppertz et al. 2014). With epitope tagging, it is important to avoid overexpression, which can give artificial binding to unnatural RNA partners, and to test whether the function, stability, and localization of the tagged RBP remain unperturbed.
To prepare cDNA libraries from the CLIP RNA fragments, an adapter is ligated to the 3′ end of the RNA. The sequence of the adapter is complementary to the primers that are later used in reverse transcription (RT) and polymerase chain reaction (PCR). This is followed by purification of the protein–RNA complexes with the use of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and membrane transfer. Both steps reduce the contamination of non-cross-linked RNAs and excess adapters, which normally run at a lower molecular weight (MW) in the gel than fragments cross-linked to the RBP; moreover, free (contaminating) RNA does not bind well to the membrane. The labeled protein–RNA complex can be visualized on the membrane, serving to optimize RNA fragmentation and to control for the specificity of purified complexes. Labeling can be achieved radioactively with 32P on the 5′ end of the RNA (Ule et al. 2003) or the adapter (Chi et al. 2009), or by covalently coupling an infrared dye to the adapter, which allows visualization with infrared imaging (irCLIP) (Zarnegar et al. 2016). Under well-optimized RNase conditions, the protein–RNA complexes migrate on the gel diffusely above the apparent MW of the immunoprecipitated RBP, because the diverse sizes of RNA fragments variably affect the migration of the RBP–RNA complex. These complexes are then excised from the appropriate region of the membrane according to recommended guidelines (Ule et al. 2005; Huppertz et al. 2014), and the RBP is digested with proteinase K, which leaves only a short peptide at the cross-link site and releases the RNA fragments into solution. To prepare a cDNA library from these RNA fragments, the original CLIP protocol ligated a second adapter to the 5′ end of RNA fragments. This approach has been used when CLIP was first combined with HITS-CLIP (Fig. 1). With the HITS-CLIP protocol, it is important that the cDNAs read through the cross-link site during the RT to ensure that they contain both adapters, which is required for the amplification and sequencing of cDNAs.
2.2. Primary Variations in Clip Protocols
The first aspect that has been varied in the CLIP methods is the cross-linking of protein–RNA complexes. Most variants of CLIP expose cells or tissues to the UVC wavelength (254 nm), a procedure that usually takes less than a minute with cells placed on ice (Ule et al. 2003) or frozen in liquid nitrogen. Photoactivatable ribonucleoside-enhanced (PAR)-CLIP introduces a variation in the cross-linking strategy (Hafner et al. 2010), in which live cells are preincubated for hours with PARs4-thiouridine (4SU) or 6-thioguanosine (6SG), which label the RNA in vivo and enable protein–RNA cross-linking to be performed with UVA wavelength (365 nm). A mass spectrometry study has compared this protocol with UVC cross-linking to conclude that the efficiency of the two approaches is quite similar across all RBPs (Castello et al. 2012). The advantage of UVA cross-linking is its decreased capacity to induce DNA damage as compared with UVC, although in most CLIP experiments samples are chilled or frozen during ∼30-sec-long UV cross-linking and then immediately solubilized, so that biologic consequences of DNA damage are minimal. The disadvantages of PAR-CLIP are its limitation to biological systems in which the photoactivatable nucleosides can be efficiently incorporated, and its recommended use of >100 million cells (Danan et al. 2016), whereas one million or less are used by standard variants of CLIP. Moreover, prolonged preincubation with 6SG or 4SU can cause cellular toxicity, including stress responses and inhibition of ribosomal RNA (rRNA) synthesis (Burger et al. 2013; Huppertz et al. 2014), and therefore care needs to be taken to monitor the cellular response to these ribonucleosides (Burger et al. 2013; Huppertz et al. 2014).
For methods aimed at studying RNA methylation, it is also possible to use a mutant RNA methylase enzyme, which forms a covalent bond with RNA spontaneously without the need for UV-induced cross-linking, as has been performed by 5-methylcytosine m6A individual-nucleotide-resolution CLIP (m5C-miCLIP) for transcriptome-wide mapping of m5C modification sites (Sugimoto et al. 2012; Hussain et al. 2013). Alternatively, proteins can be cross-linked to purified RNA with UV light in vitro. This has been exploited by protocols that cross-link an antibody that binds to a methylated base to study RNA methylation, such as m6A-CLIP (Fig. 1) (Chen et al. 2015; Ke et al. 2015; Linder et al. 2015). The antibody–RNA complex is then captured on protein A/G magnetic beads, and the sample continues to the rest of the CLIP protocol.
The second aspect of CLIP methods in which major variations have been explored involves ligating adapters to the fragmented RNA. By modifying the ligation conditions, it is possible to enable proximity ligation of the two strands of RNA duplexes that are bound by double-strand RNA-binding proteins (dsRBPs). When the two strands get ligated, the resulting sequencing reads are referred to as “hybrid reads,” because they need to be split into two sequences that map to separate loci in the genome. This approach has been exploited to study intermolecular duplexes, such as small nucleolar RNA (snoRNA)–rRNA, messenger RNA–microRNA (mRNA–miRNA) and long noncoding RNA (lncRNA)–mRNA, as well as intramolecular RNA–RNA duplexes bound by dsRBPs (Chi et al. 2009; Kudla et al. 2011; Imig et al. 2014; Moore et al. 2015; Sugimoto et al. 2015). Further technical details of these protocols, their data analysis, and relationship to other methods to study RNA secondary structure are reviewed in more depth elsewhere (Sugimoto et al. 2017).
A major advance in analysis of RNA structure was made with the development of ligation conditions to allow hybrid CLIP (hiCLIP), which ligates the two RNA strands with an additional adapter to gain control over the intermolecular ligation. This is achieved with the use of two types of RNA adapters, which are ligated to RNA duplexes: RNA adapter A with an irreversible 3′ block (dideoxynucleotide), and RNA adapter B with an enzymatically removable 3′ block (phosphate). Whereas the RNA duplexes remain bound to dsRBPs during IP, the two adapters are first ligated in equimolar concentration; thus, ∼50% of the duplexes are expected to contain a different adapter at each strand. The 3′ block of adapter B is then removed, and the next ligation reaction is performed to ligate the two arms of RNA duplexes via the adapter B. This results in hybrid reads that contain the two strands of RNA duplexes separated by the adapter B, which allows unambiguous identification of the two arms of the hybrid reads. The method has been used to identify RNA duplexes bound by Staufen 1 in human embryonic kidney 293 (HEK293) cells, which identified intermolecular mRNA–mRNA and mRNA–lncRNA duplexes in addition to RNA duplexes that connect complementary parts of the same RNA. In the coding sequence, these complementary parts were generally proximally located, although in untranslated regions (UTRs) they were more often located >100 nucleotides (nt) apart. Initial studies with a reporter assay indicated a role for such “long-range” duplexes in the regulation of mRNA stability (Sugimoto et al. 2015), and their broader functional importance remains to be examined.
The final major variations in CLIP involve the preparation of the cDNA library. Individual nucleotide resolution CLIP (iCLIP) ligates the second adapter to the 3′ end of the cDNA via circularization (König et al. 2010). This enables amplification of cDNAs that prematurely truncate at the cross-linked nucleotide, in addition to the readthrough cDNAs, thus increasing the sensitivity of the method. Seventeen other published protocols similarly amplify truncated cDNAs, including bromodeoxyuridine (BrdU)-CLIP, enhanced CLIP (eCLIP), and irCLIP (Weyn-Vanhentenryck et al. 2014; Van Nostrand et al. 2016; Zarnegar et al. 2016). In iCLIP and most other CLIP methods, additional sequences are part of the adapter oligonucleotide, including an experimental barcode that enables multiplexing of different experiments, and a unique molecular identifier (UMI) that enables quantification of unique cDNAs to remove the artifacts of variable PCR amplification (Fig. 1) (König et al. 2010). Following adapter ligation, cDNAs are then purified and amplified by PCR, which is achieved in multiple ways by variant protocols, as described in detail elsewhere (Lee and Ule 2018).
2.3. Clip Data Analysis
The primary advantage of CLIP is that it allows investigators to narrow down the very large number of computationally possible binding sites for a given RBP to a smaller subset of sites with evidence of direct RBP contact in the relevant cell type. This was clearly illustrated in the ability of Argonaute (Ago)-miRNA-mRNA CLIP to identify true miRNA-binding sites from among a larger set of computationally predicted sites, improving accuracy (Chi et al. 2009) and leading to unexpected discoveries (Chi et al. 2012; Luna et al. 2015; Moore et al. 2015).
The first steps in analyzing CLIP sequencing data involve using the experimental barcodes to demultiplex the cDNA libraries, mapping the data to the genome, and identifying unique cDNAs with the use of UMIs, if available. Moreover, multiple methods can be used to normalize the binding patterns relative to RNA abundance (Lee and Ule 2018). Depending on whether the protocol amplified truncated cDNAs, the peaks of RNA-binding sites can be assigned in two primary ways. For HITS-CLIP and other protocols that only amplify readthrough cDNAs, the full position of mapped reads is examined. The most likely position of the cross-link site can be identified within a proportion of the reads by identifying cross-linking-induced mutation sites, such as C to T transitions when 4SU is used in PAR-CLIP (Hafner et al. 2010), or deletions and other types of mutations in HITS-CLIP (Zhang and Darnell 2011). In iCLIP and 17 other protocols that amplify truncated cDNAs, cross-link sites are located at the start of these cDNAs. The truncated cDNAs strongly dominate the iCLIP libraries, and thus analysis of cDNA starts enables the nucleotide-resolution mapping of the cross-link sites, especially when combined with high-resolution peak-calling algorithms (Sugimoto et al. 2012; Haberman et al. 2017). The simplest peak-calling approach is to identify clusters of cross-linking events with cDNA counts that are significantly higher compared with randomized data in the same genomic regions (König et al. 2010).
Although peak analysis of RBP–RNA-binding sites has been of great value in understanding binding specificity and demarcating discrete points of regulation (Chakrabarti et al. 2017), not all regulatory factors have been shown to bind to individual discrete peaks. For example, heterogeneous nuclear ribonucleoprotein C (hnRNPC) binds widely across the transcriptome, with a periodicity suggesting the possibility of nucleosomal-like organization on RNA transcripts, which also reflects its binding to the dual U-rich sequences in antisense Arthrobacter luteus restriction endonuclease (Alu) elements (König et al. 2010; Zarnack et al. 2013). Fragile X mental retardation protein (FMRP), the protein lost in the commonest inherited form of intellectual disability, the fragile X syndrome, was found by CLIP to bind to a subset of transcripts across their entire coding length (Darnell et al. 2011). This was then shown to result from dual interactions of FMRP with the ribosome and the mRNA to cause ribosomal elongation arrest and translational pausing on its target transcripts. In such instances, more complex computational modeling may be needed in addition to peak analysis to understand how the RBPs recognize specific target RNAs.
Paralleling the genomic mapping and peak assignment, an essential concomitant is to examine the quality of data. Analysis of negative controls is particularly important for interpreting the data specificity. For example, purification can be performed from cells in which the RBP is absent, such as knockout (KO) cells, or when using tag-based purification, cells that do not express a tagged protein. As an alternative, nonspecific serum or immunoglobulin G (IgG) can be used for IP, or the RBP can be purified from non-cross-linked cells. Ideally, these controls should not produce any clearly detectable signal during SDS–PAGE analysis, and the number of unique cDNAs in their resulting libraries should be at least 100-fold fewer than in the specific experiments (König et al. 2010).
The sensitivity and specificity of data can be further examined by assessing the total number of unique cDNAs in the sequencing library and the peaks, respectively. To monitor the specificity of CLIP, a suitable peak-calling program needs to be chosen according to the CLIP protocol used to produce the data (Chakrabarti et al. 2017). A low number of peaks indicates either that the cDNAs are dispersed along the transcripts or that they are concentrated in a small number of abundant RNAs (such as rRNA)—this could either reflect low preference of the RBPs for specific sites on longer RNAs or low quality of data. Thus, some understanding of the expected binding properties of the RBP can help when interpreting CLIP data.
Finally, an extremely informative way to examine CLIP data sensitivity and specificity is to assess binding peaks with orthogonal data in the format of RNA maps (Licatalosi et al. 2008; Licatalosi and Darnell 2010), which allow the investigator to examine whether binding is enriched at exons or mRNAs that are differentially regulated by the RBP. Analysis of cross-link peaks in the format of RNA maps is particularly valuable when comparing multiple datasets for the same RBP. For example, data produced by iCLIP of polypyrimidine tract-binding protein 1 (PTBP1) led to a larger number of peaks around regulated exons than data produced by other protocols, although the number of unique cDNAs in iCLIP is equal or smaller (Chakrabarti et al. 2017).
3. CONDITIONALLY TAGGED (cTAG)-CLIP
A major forefront in biological research is to understand the functional roles of individual cell types in a multicellular organism. Cell type–specific regulation of RNA expression and processing emerges as a common phenomenon from recent RNA profiling studies in selected cell populations (Bhate et al. 2015; Yap et al. 2016; Zhang et al. 2016). Importantly, it is clear from these studies that many cell type–specific RNA regulatory events are not identifiable in whole tissue analysis. Therefore, developing new technologies with cell type resolutions is critical to advance our understanding of cell type–specific RNA regulation.
To meet the challenge of applying CLIP to study cell type–specific biology, we developed the conditionally tagged (cTag) CLIP technology, which uses a genetically engineered mouse that carries a latent green fluorescent protein (GFP)-tagged allele of the RBP of interest. The latent GFP-tagged allele is “off” by default, leaving the native allele unaffected, and only turns “on” in the presence of Cre recombinase (Fig. 2A) (Hwang et al. 2017). Importantly, the GFP-tagged allele is expressed by homologous recombination into the endogenous locus, such that the quantitative, qualitative (alternate isoforms), spatial, and temporal expression of GFP-tagged RBP is minimally disruptive to the biology of the system and can be precisely controlled through Cre expression to achieve cell type specificity. Cell type–specific CLIP analysis for the RBP of interest can then be performed using a mix of high-affinity GFP antibodies to immunoprecipitate the GFP-tagged RBPs under stringent conditions (Fig. 2B) only from cells expressing Cre recombinase.
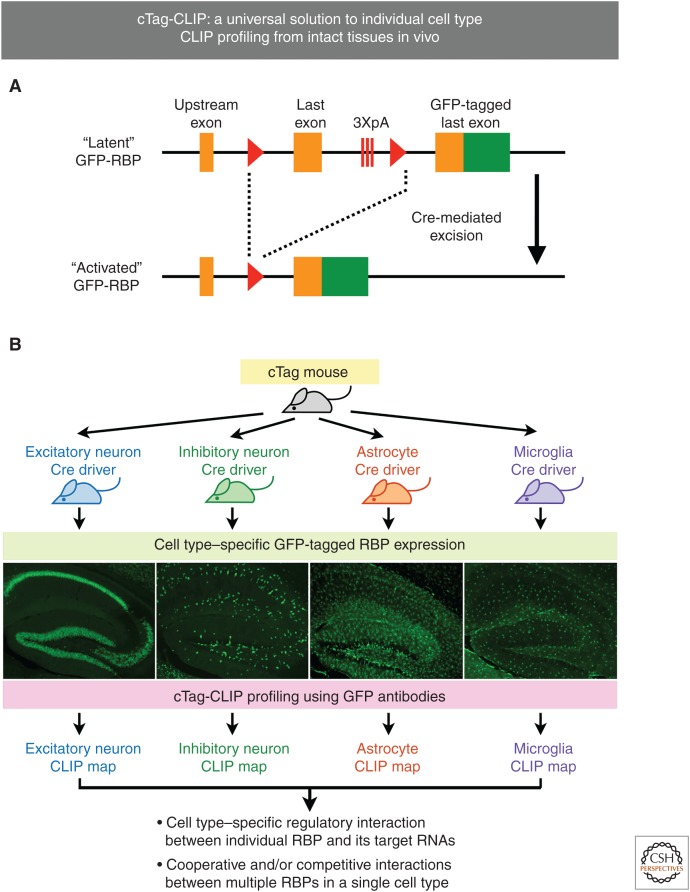
Figure 2.
Conditionally tagged (cTag)-cross-linking and immunoprecipitation (CLIP). (A) The basic design of a cTag mouse. An extra copy of the last coding exon fused with green fluorescent protein (GFP) sequence (green) is inserted downstream from sequences encoding the RNA-binding protein (RBP) of interest. To prevent readthrough, triple poly(A) sites (3XpA) are inserted before the GFP-tagged exon. Cre-mediated excision of the endogenous last exon results in expression of GFP-tagged RBP from its native locus. Arrowheads indicate the locus of X-over P1 (loxP) sites. (B) Illustrative examples of cTag-CLIP with results from the cTag-poly(A)-binding protein (PABP) mouse (Hwang et al. 2017).
The cTag-CLIP technology has several important features to ensure a proper representation of endogenous RNA–protein interactions but at the same time to maintain a broad applicability. First, because cTag mice are created using a “knock-in” approach, the proper cellular expression of RBPs and the RNA-RBP stoichiometry is likely to be maintained, both of which are difficult to achieve with either traditional or even bacterial artificial chromosome (BAC) transgenes (Fig. 2A) (He et al. 2012; Gerfen et al. 2013). Second, the utilization of a mix of high-affinity GFP antibodies (Heiman et al. 2014) allows cTag-CLIP analysis to be performed with a standardized and optimized high-stringency protocol for different RBPs (Hwang et al. 2017). The standardization eliminates the need for RBP-specific antibodies and the subsequent experimental optimization to achieve high-stringency IP for individual RBPs. Furthermore, it also provides an equal footing for cross-comparison of cTag-CLIP data from different RBPs to facilitate data integration. Third, its compatibility with the Cre/Lox system allows direct application of existing or newly developed Cre mouse lines, which makes it a versatile tool for wide-ranging biological investigations.
We have since successfully established a series of cTag mice to perform cTag-CLIP analysis for different RBPs in multiple cell types. We validated this approach using cTag poly(A)-binding protein (PABP; gene symbol: Pabpc1) to generate the cTag-PABP mouse and to establish the cTag-PAPERCLIP technique (Hwang et al. 2017). cTag-PAPERCLIP uses PABP as a biological filter to specifically clone distal mRNA 3′ ends to generate comprehensive mRNA alternative polyadenylation (APA) maps. With cTag-PAPERCLIP, we performed an in-depth APA analysis of individual cell types in vivo in the adult mouse brain to show the dynamics of APA regulation and discovered a previously unknown role of APA in diversifying protein expression in the brain.
Several observations from this cTag-PAPERCLIP study exemplify the technical advances and, more importantly, biological implications of the cTag-CLIP technology, which are detailed below.
cTag-CLIP achieves desired cell type specificity to provide a powerful means to dissect cell type–specific biology: The ubiquitous expression of PABPC1 presents an ideal test case for the capabilities of cTag-CLIP technology in achieving high cell type specificity. To evaluate the cell type specificity at the anatomical level, we examined the expression patterns of PABP-GFP in brains from four mouse models generated by crossing the cTag-PABP mouse to four Cre drivers with different cell type specificity (excitatory neurons, inhibitory neurons, astrocytes and microglia [Fig. 2B]). The analyses indeed confirmed the distinct expression patterns of PABP-GFP from the four mouse models and their consistency with the corresponding cell type markers. Next, we evaluated the cell type specificity at the molecular level by analyzing the four resulting RNA profiles from each mouse model generated by cTag-PAPERCLIP. These cTag-PAPERCLIP profiles correlated with independently generated cell type–specific RNA sequencing (RNA-seq) profiles and showed expected segregation of known cell type–specific marker genes by the Cre activities. Overall, these data show that cTag-CLIP indeed accomplishes its goals: (a) specific expression of the tagged RBP in the desired cell populations, and (b) generation of cell type–specific CLIP maps. Importantly, cTag-CLIP data obtained from cTag mouse models are very consistent with data generated in wild-type mouse models through a variety of methods in multiple occasions (Hwang et al. 2017; Jereb et al. 2018; Saito et al. 2018), showing that the cTag-CLIP approach in general does not interfere with normal RBP biology and hence provides a powerful means for dissecting cell type–specific biology.
cTag-CLIP has excellent power to “zoom in” on less abundant cell types to discover cell type–specific regulatory events: Our in-depth cTag-PAPERCLIP analysis of APA patterns across four major brain cell types and between different cellular states in microglia (quiescent vs. activated) revealed novel cell type–specific regulatory events. For example, we showed that cell type–specific RBPs directly shape the distinct APA patterns of Itsn1, Map4, and Cdc42 between neurons and glia/nonneural tissues. Moreover, in activated microglia, we identified that Araf switches from a truncated APA isoform (which generates a kinase-deficient protein) to a full-length APA isoform (which generates a kinase-intact protein). Importantly, these findings nicely illustrate how gaining resolution at the cell type level allows cTag-CLIP to generate new biological insights, as these four genes are expressed in multiple cell types in the brain, and it would be difficult for whole tissue analysis to pick up these cell type–specific regulatory events.
cTag-CLIP well preserves native transcriptome states and has an advantage in profiling sensitive cell types: One distinctive feature of cTag-CLIP is its “built-in” cell type specificity imparted through genetic expression of cell type–restricted Cre in cTag mice (Fig. 2B). As a result, cTag-CLIP can use the whole tissue as the input material without going through the process of tissue dissociation and cell type purification, which is necessary for common RNA profiling methods such as RNA-seq to achieve cell type specificity. Tissue dissociation and cell type purification disrupts the tissue microenvironment, causes cellular stress, and may alter gene expression (Cardona et al. 2006; Okaty 2011). Most importantly, cTag-CLIP kept the expression of microglia activation markers and immediate early genes (which respond to cellular stress) low compared with RNA-seq profiling from ex vivo purified microglia. These results show that cTag-CLIP better preserves native transcriptome states of individual cell types and therefore is suitable for accurate in vivo RNA profiling.
The success of our initial cTag-CLIP studies starts to reveal a diversity of RNA regulatory events in individual cell types. Generation and integration of cTag-CLIP maps for different RBPs in multiple cell types will provide essential information to refine the existing rules of RNA–protein interaction for individual cell types. Importantly, with the cTag-CLIP maps, it is now feasible to deduce the cooperative and/or competitive interactions among different RBPs for the same RNA-binding sites in a single cell type to construct high-order RNA–protein interaction maps. Recent improvement in generating knock-in mouse models from advances in synthetic biology and gene-targeting technology will speed up the process of expanding the cTag mice repertoire (Shalem et al. 2015; Wright et al. 2016). Moreover, continued identification of cell type–specific markers and technical advancement enhancing the sensitivity of CLIP technique will broaden the application of cTag-CLIP to new and rare cell types (Tasic et al. 2016; Lee and Ule 2018).
4. INSIGHTS FROM CLIP AND FUTURE DIRECTIONS
The development of CLIP has been driven largely by the need to understand the regulation of pre-mRNA processing, although it can be applied to all forms of posttranscriptional RNA regulation, including translation as shown with CLIP studies of FMRP. For studies of pre-mRNA, it is crucial to map the binding of nuclear RBPs to introns, which are generally of low abundance and of transient nature. To allow studies of RBP interactions on introns, specificity of the method had to be maximized, which led to the inclusion of covalent cross-linking followed by several stringent purification and quality control steps. Although the success of this approach was shown by the very first studies of Nova, CLIP also led to an unexpected finding that nuclear RBPs have many thousands of high-affinity binding sites, most of which are far from exons (Ule et al. 2003; Licatalosi et al. 2008). In recent years, it has become clear that many abundant nuclear RBPs, such as hnRNPC, often bind to specific transposable elements or other repetitive sequences that are present in introns, where they repress processing of cryptic exons and polyadenylation sites (Zarnack et al. 2013; Sibley et al. 2016). While this repressive binding safeguards the transcriptome from newly emerging intronic sites of RNA processing, it may also create an opportunity for a gradual evolutionary emergence of new alternative exons on mutations that weaken the repressive binding (Attig et al. 2016). Thus, although the large number of intronic-binding sites of nuclear RBPs initially came as a surprise, it is becoming clear that this plays an important role in the control of rarely included cryptic exons, including those mediating nonsense-mediated decay (Eom et al. 2013), and in the evolution of gene expression (Sibley et al. 2016).
4.1. Molecular and Physiologic Insights from CLIP
Following identification of in vivo binding sites, further integration with orthogonal functional data can then allow interpretation of regulatory mechanisms that operate in cells and organs. For example, the integrative approach to examine binding profiles around regulated exons and polyadenylation sites has been visualized in the form of RNA maps (Ule et al. 2006; Licatalosi et al. 2008; Witten and Ule 2011). This union of biochemical CLIP data with functional evidence via the bioinformatic generation of global “RNA maps” led to the recognition that the position of protein-binding sites in transcripts provided information relevant to their mechanisms and regulatory outcomes (Ule et al. 2003; Licatalosi et al. 2008). For example, it revealed that nuclear RBPs often regulate splicing and 3′ processing via similar positional principles, with repressive binding located close to the sites of RNA processing, and enhancing binding located further downstream (Licatalosi et al. 2008; Rot et al. 2017). The insights from such analysis can complement predictive programs such as TargetScan, which predicts possible binding sites for miRNAs by incorporating information on evolutionary conservation and general binding rules; for example, CLIP could identify important miRNA-binding sites that are unique to human (Chi et al. 2009) or are located at noncanonical sequences that do not conform to general rules (Chi et al. 2012; Moore et al. 2015).
Many studies have shown that RNP assembly on the nascent and mature mRNA, and the resulting regulatory mechanisms are coordinated at many levels (Singh et al. 2015), and CLIP has solidified these observations. Many RBPs shuttle between nucleus and cytoplasm, which allows messenger ribonucleoprotein (mRNP) assembly to start in the nucleus on pre-mRNAs and then continue to influence cytoplasmic regulation. For example, although CLIP identified roles for Nova proteins in nuclear pre-mRNA processing (Ule et al. 2003; Licatalosi et al. 2008), it also revealed 3′ UTR-binding sites in the vicinity of factors involved in alternative polyadenylation; again using genetics, such binding was found to be functional in determining the use of APA sites (Ule et al. 2003; Licatalosi et al. 2008). Moreover, CLIP, together with immunoelectron microscopy (EM) localization of Nova in the dendrite, identified a new link between nuclear regulation and synaptic RNA localization within neurons (Racca et al. 2010). This showed that an RNA regulatory factor was governed by its binding affinity to motifs that can be located at various positions in the transcriptome, and that a mammalian splicing factor could also bind and regulate mature mRNA biology.
Biologically, some of the more powerful conclusions from CLIP biochemistry have come from pairing it with genetic manipulations. CLIP studies have been most definitive in vivo in mice in which individual (Darnell 2013) and, more recently, multiple (Saito et al. 2018) RBPs have been genetically deleted. Similarly, CLIP studies have been successful in tissue culture cells in which RBPs have been knocked down. Such genetic studies have been complemented by genetic rescue experiments. This was illustrated in early studies demonstrating the requirement of Nova for splicing of the Z+ isoform of the agrin gene in motor neurons (Ule et al. 2005), and as a result the Nova1 KO mice have abnormal neuromuscular junctions and are paralyzed (Ruggiu et al. 2009). Moreover, Nova proteins regulate an alternative exon in Dab1 (Ule et al. 2006), an important regulator of reelin signaling in newly born migrating neurons (Yano et al. 2010). Importantly, specific phenotypes of Nova KO mice can be rescued by expressing the appropriately spliced isoform; interestingly, when this was performed by mating Nova1 KO mice to transgenic mice constitutively expressing Z+ agrin, the normal neuromuscular junction was reestablished, but the mice remained paralyzed, illustrating the multilayered aspect of RBP-RNA regulation in vivo (Ruggiu et al. 2009). Beyond studies of Nova, CLIP has been successfully applied to characterize the function and assembly of many additional disease-relevant RBPs in the brain and other tissues of model organisms (Darnell 2013; Modic et al. 2013).
4.2. CLIP: Future Directions
One of the most recent applications of CLIP has been to undertake studies of epigenetic modifications of RNA. These were pioneered with studies of m6A-RNA modifications, using a variant of CLIP in which the modified nucleotide itself was directly cross-linked, in vitro, to m6A-specific antibodies (Ke et al. 2015; Linder et al. 2015; Patil et al. 2018). This provides a potentially general strategy for analyzing what is an expanding number of posttranscriptional RNA modifications important in cell function and human disease (Jonkhout et al. 2017).
The same general principles of CLIP experiments apply in analyses of RNA modifications as in traditional CLIP: unbiased, genome-wide genetic studies allow demarcation of specific sites, here using direct modification-specific antibodies to cross-link the modified RNA nucleotide. For example, m6A-CLIP was used to identify terminal exons as the most common positions for m6A modification on RNA transcripts, rather than exons harboring stop codons (Ke et al. 2015); to show primary functions of m6A in RNA turnover rather than regulation of alternative splicing (Ke et al. 2017); and to clarify our current understanding of the biologic role of m6A-RNA modifications (Ke et al. 2017; Darnell et al. 2018).
Importantly, CLIP has been shown to be of value also for studies of human postmortem tissues (Tollervey et al. 2011). This is important as such tissues are often frozen in liquid nitrogen. RNA–protein complexes appear to survive such conditions well, and allow CLIP to be applied to tissues relevant for a range of human disease (e.g., cancer, immunologic, or neurologic disease) using a range of human tissue (tumor [Vanharanta et al. 2014], blood [Loeb et al. 2012], and brain [Tollervey et al. 2011; Scheckel et al. 2016], respectively). The value of CLIP has also been shown to unravel the dynamics of RNPs under disease conditions by comparing tissue from disease patients and controls (Tollervey et al. 2011), or monitoring the impact of viral infection on host transcripts in biology and human disease (Flynn et al. 2015; Luna et al. 2015, 2017; Moore et al. 2018).
Demarcation of RNA structure using CLIP was originally pioneered with hiCLIP to analyze Staufen–RNA interactions (Sugimoto et al. 2015) and subsequently developed with a variety of complementary approaches such as PARIS (psoralen analysis of RNA interactions and structures; Lu et al. 2016, 2018) as discussed above. Such analyses are important in their own right and in their potential to allow more general understanding of structures accessible to RBP regulation, as determined from CLIP analyses. The combined understanding of protein-binding sites, RNA structure, and evolutionary constraints promises to expand our ability to predict genetic variants using Bayesian (Zhang et al. 2010) and machine-learning tools (Barash et al. 2010) and ultimately apply such understanding to interpretation of those variants that have an impact on disease phenotypes.
Since the very initial development of CLIP, a common theme of many CLIP studies has been that they combine discovery of fundamental mechanisms of gene regulation along with biologic and clinically relevant insights. CLIP holds great promise for studies aimed at defining the functionally important RNA sites across the transcriptome, unraveling the mechanisms of RNP assembly and function, and uncovering the role of these mechanisms in diseases.
Acknowledgments
The authors thank their many colleagues for fruitful discussions and contributions over the years—too many to cite here despite our best efforts. In particular, J.U. thanks Flora Lee for her help and feedback on the cross-linking and immunoprecipitation (CLIP) methods section. This work was supported by the National Institutes of Health (NIH) grant 5RC2NS069473-02 (R.B.D.), the European Research Council (617837-Translate), and the Wellcome Trust (103760/Z/14/Z) (J.U.). R.B.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Attig J, Ruiz de Los Mozos I, Haberman N, Wang Z, Emmett W, Zarnack K, König J, Ule J. 2016. Splicing repression allows the gradual emergence of new Alu-exons in primate evolution. eLife 5: e19545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. 2010. Deciphering the splicing code. Nature 465: 53–59. [DOI] [PubMed] [Google Scholar]
- Bhate A, Parker DJ, Bebee TW, Ahn J, Arif W, Rashan EH, Chorghade S, et al. 2015. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat Commun 6: 8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Mühl B, Kellner M, Rohrmoser M, Gruber-Eber A, Windhager L, Friedel CC, Dölken L, Eick D. 2013. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol 10: 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AE, Huang D, Sasse ME, Ransohoff RM. 2006. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc 1: 1947–1951. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Chakrabarti AM, Haberman N, Praznik A, Luscombe NM, Ule J. 2017. Data science issues in understanding protein-RNA interactions. bioRxiv 10.1101/208124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, Han D, et al. 2015. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew Chem 54: 1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB. 2012. An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EG, Manley JL. 2017. RNA-binding proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev 31: 1509–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan C, Manickavel S, Hafner M. 2016. PAR-CLIP: A method for transcriptome-wide identification of RNA binding protein interaction sites. Methods Mol Biol 1358: 153–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. 2010. HITS-CLIP: Panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R. 2012. CLIP (cross-linking and immunoprecipitation) identification of RNAs bound by a specific protein. Cold Spring Harb Protoc 2012: 1146–1160. [DOI] [PubMed] [Google Scholar]
- Darnell RB. 2013. RNA protein interaction in neurons. Ann Rev Neurosci 36: 243–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB, Ke S, Darnell JE Jr. 2018. Pre-mRNA processing includes N6 methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics.” RNA 24: 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, Zhang C, Wang H, Lay K, Fak J, Noebels JL, Darnell RB. 2013. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife 2: e00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Martin L, Spitale RC, Do BT, Sagan SM, Zarnegar B, Qu K, et al. 2015. Dissecting noncoding and pathogen RNA–protein interactomes. RNA 21: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. 2013. GENSAT BAC Cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80: 1368–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D. 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman N, Huppertz I, Attig J, König J, Wang Z, Hauer C, Hentze MW, et al. 2017. Insights into the design and interpretation of iCLIP experiments. Gen Biol 18: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. 2012. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. 2014. Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP). Nat Protoc 9: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: Protein–RNA interactions at nucleotide resolution. Methods 65: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, et al. 2013. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Saito Y, Park CY, Blachère NE, Tajima Y, Fak JJ, Zucker-Scharff I, Darnell RB. 2017. cTag-PAPERCLIP reveals alternative polyadenylation promotes cell-type specific protein diversity and shifts Araf isoforms with microglia activation. Neuron 95: 1334–1349.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imig J, Brunschweiger A, Brümmer A, Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M, Hall J. 2014. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19–miR-106a interaction. Nat Chem Biol 11: 107–114. [DOI] [PubMed] [Google Scholar]
- Jereb S, Hwang HW, Van Otterloo E, Govek EE, Fak JJ, Yuan Y, Hatten ME, Darnell RB. 2018. Differential 3′ processing of specific transcripts expands regulatory and protein diversity across neuronal cell types. bioRxiv 10.1101/245886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. 2017. The RNA modification landscape in human disease. RNA 23: 1754–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, et al. 2015. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29: 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. 2011. Cross-linking, ligation, and sequencing of hybrids reveals RNA–RNA interactions in yeast. Proc Natl Acad Sci 108: 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FCY, Ule J. 2018. Advances in CLIP technologies for studies of protein-RNA interactions. Mol Cell 69: 354–369. [DOI] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. 1979. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci 76: 5495–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. 2010. RNA processing and its regulation: Global insights into biological networks. Nat Rev Gen 11: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 48: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Gong J, Zhang QC. 2018. PARIS: Psoralen analysis of RNA interactions and structures with high throughput and resolution. Methods Mol Biol 1649: 59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna JM, Scheel TKH, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, et al. 2015. Hepatitis C virus RNA functionally sequesters miR-122. Cell 160: 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna JM, Barajas JM, Teng KY, Sun HL, Moore MJ, Rice CM, Darnell RB, Ghoshal K. 2017. Argonaute CLIP defines a deregulated miR-122-bound transcriptome that correlates with patient survival in human liver cancer. Mol Cell 67: 400–410.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10: 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modic M, Ule J, Sibley CR. 2013. CLIPing the brain: Studies of protein–RNA interactions important for neurodegenerative disorders. Mol Cell Neurosci 56: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Scheel TKH, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. 2015. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Comm 6: 8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Blachère NE, Fak JJ, Park CY, Sawicka K, Parveen S, Zucker-Scharff I, Moltedo B, Rudensky A, Darnell R. 2018. ZFP36 RNA-binding proteins restrain T-cell activation and anti-viral immunity. bioRxiv 10.1101/247668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods 26: 182–190. [DOI] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. 2011. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS One 6: e16493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Pickering BF, Jaffrey SR. 2018. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol 28: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. 2010. The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front Neural Circuits 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rot G, Wang Z, Huppertz I, Modic M, Lenče T, Hallegger M, Haberman N, Curk T, von Mering C, Ule J. 2017. High-resolution RNA maps suggest common principles of splicing and polyadenylation regulation by TDP-43. Cell Rep 19: 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, Herbst R, Kim N, Jevsek M, Fak JJ, Mann MA, Fischbach G, Burden SJ, Darnell RB. 2009. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci 106: 3513–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yuan Y, Zucker-Scharff I, Fak JJ, Tajima Y, Licatalosi DD, Darnell RB. 2018. Neuron-specific cTag-CLIP reveals cell-specific diversity of functional RNA regulation in the brain. bioRxiv 10.1101/244905. [DOI] [Google Scholar]
- Scheckel C, Drapeau E, Frias MA, Park CY, Fak J, Zucker-Scharff I, Kou Y, et al. 2016. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. eLife 5: e10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. 2015. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Gen 16: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CR, Blazquez L, Ule J. 2016. Lessons from non-canonical splicing. Nat Rev Gen 17: 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: Past and present trends in mRNP fashion. Ann Rev Biochem 84: 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, König J, Hussain S, Zupan B, Curk T, Frye M, Ule J. 2012. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Gen Biol 13: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, Luscombe NM, Ule J. 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Chakrabarti AM, Luscombe NM, Ule J. 2017. Using hiCLIP to identify RNA duplexes that interact with a specific RNA-binding protein. Nat Protoc 12: 611–637. [DOI] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, et al. 2016. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, et al. 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215. [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, et al. 2005. Nova regulates brain-specific splicing to shape the synapse. Nat Gen 37: 844–852. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586. [DOI] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massagué J. 2014. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife 3: e02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, et al. 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6: 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J. 2011. Understanding splicing regulation through RNA splicing maps. Trends Genet 27: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA. 2016. Biology and applications of CRISPR systems: Harnessing nature's toolbox for genome engineering. Cell 164: 29–44. [DOI] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. 2010. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66: 848–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Xiao Y, Friedman BA, Je HS, Makeyev EV. 2016. Polarizing the neuron through sustained co-expression of alternatively spliced isoforms. Cell Rep 15: 1316–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. 2016. irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods 13: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotech 29: 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, et al. 2016. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166: 1147–1162.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]