Abstract
Malignant gliomas are heterogeneous diseases in genetic basis. The development of sequencing techniques has identified many gene rearrangements encoding novel oncogenic fusions in malignant glioma to date. Understanding the gene fusions and how they regulate cellular processes in different subtypes of glioma will shed light on genomic diagnostic approaches for personalized treatment. By now, studies of gene fusions in glioma remain limited, and no medication has been approved for treating the malignancy harboring gene fusions. This review will discuss the current characterization of gene fusions occurring in both adult and pediatric malignant gliomas, their roles in oncogenesis, and the potential clinical implication as therapeutic targets.
Introduction
Gene fusions are hybrid of two coding or regulatory DNA sequences between genes. They are pathognomonic mutations resulting from genomic rearrangements such as translocations, deletions, duplications, or inversions. This phenomenon was first described in chronic myeloid leukemia (CML) when researchers used quinacrine fluorescence and Giemsa staining, finding a cellular oncogene ABL1 on chromosome 9 fused to BCR on chromosome 22, forming BCR-ABL1 fusion gene on the newly formed Philadelphia chromosome. Later studies found that this gene fusion had changed this previously deadly type of cancer into the nearly cured disease using tyrosine kinase inhibitors [1], [2], [3], [4], [5]. Since then, with the development of biomedical technologies, more and more gene fusions were detected with further understanding of their mechanisms and biological functions. At present, more than 10,000 fusion genes in cancer have been found and indexed in the Mitelman Database of Chromosome Aberrations and Gene Fusions (2017) (http://cgap.nci.nih.gov/Chromosomes/Mitelman).
Despite most of fusions appearing to be passenger mutations without oncogenic functions, some of the fusions do constitute strong driver alterations in the initial steps of tumor development, while other fusions may play important roles in tumor progression [6], [7], [8]. For example, EML4-ALK fusion causes transformation of non–small-cell lung cancer, and COL1A1-PDGFRB fusion contributed directly to growth and progression of dermatofibrosarcoma protuberans [9], [10], [11], [12].
The deep insight into oncogenic gene fusions will not only help extend our understanding of tumor biology but also provide robust therapeutic targets for selected tumor types [13]. The tyrosine kinase inhibitor (TKI) imatinib, designed for treating CML with BCR-ABL1 fusion, was approved by US Food and Drug Administration (FDA) in 2001 and achieved excellent efficacy with improved lifespan and quality of life of patients [14]. The successful clinical practice has spurred interest in developing drugs targeting the products of gene fusions in multiple tumors. Subsequently, several new kinase inhibitors were approved by the FDA, such as crizotinib and ceritinib for the treatment of non–small cell lung cancer with ALK fusions, and ponatinib used to treat both CML and acute lymphoblastic leukemia patients with BCR-ABL1 fusion [7].
Gliomas are the most common primary brain tumors with high recurrence and mortality rates [15]. Over the past decade, molecular targeted therapy has underscored the promise of managing the malignant gliomas. One of the challenges in understanding and treating gliomas has been the clinical and molecular heterogeneity associated with this disease [16]. Currently, genomic and transcriptomic analyses are defining the molecular architecture of gliomas and have uncovered a subset of patients carrying gene fusions [8]. Despite relatively low-frequency rate in gliomas, gene fusion has a disproportionate importance in investigating the mechanism of tumorigenesis and provides potential targeted therapies for selected patients with specific fusions [8]. In this review, we summarize the current characterization of gene fusions occurring in both adult and pediatric malignant gliomas and discuss the potential clinical implication of oncogenic fusions as therapeutic targets.
Types of Gene Fusions
Gene fusions are two or more genes that form chimeric genes involving parts of each gene. The formation of fusion genes in different cells can occur through different mechanisms [17]. Previous studies have demonstrated that fusion transcripts can be not only produced by chromosomal rearrangement at the DNA level but also formed through trans-splicing and cis-splicing between neighboring genes.
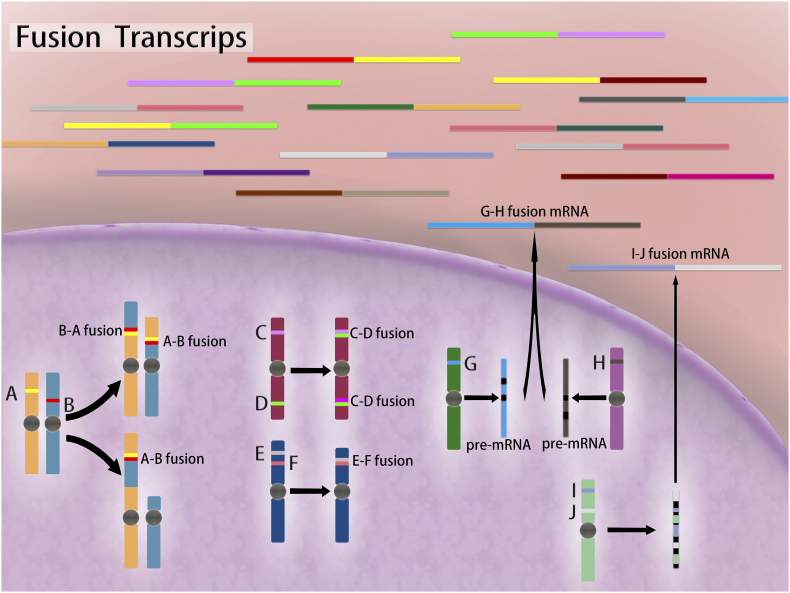
There are four basic types of chromosomal rearrangements that could lead to gene fusions (Figure 1): (1) a genomic region between two genes located on the same strand is deleted, and a fusion gene arises through deletion; (2) a genomic region is duplicated one or more times on chromosomal is the tandem duplication; (3) a chromosomal translocation involves the translocation of genomic regions on different chromosomes; and (4) two genes located on opposite strands of a chromosome form an infusion gene, and inversion event is suspected to happen [17].
Figure 1.
Gene fusions leading to chimeric transcripts through a variety of mechanisms. Fusion genes can form at chromosomal level, where translocation, insertion, inversion, and deletion occur and produce B-A and A-B fusion mRNAs, A-B fusion mRNA, C-D fusion mRNA, and E-F fusion mRNA, respectively. Fusion transcripts can also be caused by trans-splicing (G-H fusion mRNA) or cis-splicing (I-J fusion mRNA) events at posttranscriptional level. Black blocks represent introns. Cyan block in the pre-mRNA represents intergenic region.
Detection of Gene Fusions
Advances in sequencing technologies have refined the “driver” mutation in tumors and facilitated a move toward “precision medicine.” These technologies were discussed below with their own advantages and disadvantages summarized in Table 1.
Table 1.
Different Detection Methods of Gene Fusions
| Detection Methods | Advantages | Disadvantages |
|---|---|---|
| FISH | Gold standard; rapid; capable of detecting rearrangements without knowing fusion partners | Expensive; not uniformly available in all laboratories; low multiplexing ability |
| IHC | Low cost; easy to perform; small sample requirement, retained tissue morphology; widespread availability | Time consuming; possible false-negative findings because of low sensitivity; low multiplexing ability |
| RT-PCR | Rapid; sensitive; low detection limit; identification of specific fusion partners | Unable to detect unknown fusion partners; require specialized equipment and lab settings; rely on the quality of RNA; low multiplexing ability |
| Gene array | Genome-wide profile and high-resolution analysis without prior cell culturing | Need confirmation using RT-PCR or FISH to avoid false-positive findings |
| WGS | Comprehensive and unbiased characterization of genomic alterations, capable of detecting novel fusions. | Short read length; technical artifacts; limited coverage; high false-positive signal; expensive |
| RNA-Seq | Low cost; quick turnaround time; detect multiple alternative splice variants caused by a fusion event | Unable to monitor nontranscribed regions; complicated data analysis |
Immunohistochemistry (IHC)
IHC is the most commonly used method in most pathology laboratories [18]. It is inexpensive, is easy to perform and requires only a small biopsy sample, and that tissue morphology is retained during analysis. However, it cannot detect unknown fusions, while the technique is time consuming with relatively low efficacy [19]. Also, not all fusion genes have primary antibodies with high affinity and specificity. Therefore, IHC is not applicable to genes without well-performing antibodies.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
RT-PCR is a fast and sensitive method that is routinely used in detecting low levels of gene fusions. The rapid turnaround time, low amount of tissue requirement, and identification of specific fusion partners are advantages of RT-PCR. However, the requirement of fusion-specific primers results in inability to detect rearrangements involving unknown fusion partners [20]. Another limitation of RT-PCR is that when applied in formalin-fixed, paraffin-embedded tissues, false-negative results might be due to highly degraded RNA available [18].
Fluorescence In Situ Hybridization (FISH)
FISH is a rapid, expensive, gold standard method that can detect gene fusions on both DNA and RNA levels [21]. The advantage of FISH is that it can be performed on formalin-fixed, paraffin-embedded tissues and nondividing cells (interphase nuclei), and it is also capable of detecting rearrangements without prior knowledge of 5′ fusion partners compared with RT-PCR [7], [18], [22]. However, this technique is not uniformly available in all laboratories [18]; thus, it is used as a confirmatory tool rather than a screening tool [23], [24], [25]. A lot of gene fusions were identified by using FISH probes, e.g., ETV6/AML1 fusion in acute lymphoblastic leukemia [26], JAZF1-JJAZ1 fusion in endometrial stromal tumors [27], and EML4-ALK in lung cancer [28].
Currently, as researches in oncogenic fusion continue, the need to detect multiple targets at the same time is becoming more and more urgent, calling for high-throughput methods such as gene array and next-generation sequencing (NGS). The low multiplexing ability makes IHC, PT-PCR, and FISH only as orthogonal confirmatory methods for detection of gene fusions.
Gene Array
The usage of gene array-based technologies provided new options for detection of gene fusions from 1990s [29]. This technique could enable a genome-wide profile and offer high-resolution analysis on the expression level of DNA or exons without the requirement of prior cell culturing [30]. Numerous novel gene fusions were detected in different cancer types using high-throughput array-based technologies, like PAX3-NCOA1 in alveolar rhabdomyosarcoma tumors [31] and HEY1-NCOA2 in mesenchymal chondrosarcoma [32]. However, this method cannot be used alone because of the sheer number of breakpoints found typically in malignancies and by extensive constitutional copy number variation; thus, it always needs confirmation by FISH or RT-PCR to avoid false-positive findings [33]. Also, not all gene fusions will lead to abnormalities in expression of involved parts of genes; these fusions are hard to be detected by gene array. With the development of deep sequencing, gene array-based technology is not used as widely as before.
Next-Generation Sequencing
NGS has provided a radically new means to identify novel or known fusions either in genomic DNA or transcriptome sequencing datasets. This technique contains several subtypes involving whole genome sequencing (WGS), whole exome sequencing, and whole transcriptome sequencing (RNA-Seq). WGS and RNA-Seq are the most commonly used NGS in this field and have achieved important discoveries [34], [35], [36], [37], [38].
WGS offers the large comprehensive and unbiased characterization of genomic alterations in genomes, especially cancer genomes [39]. Despite being the most powerful sequencing technology, WGS falls short because of to the short read length, technical artifacts, limited coverage, and high false-positive signal resulting from sequencing errors [40], [41], [42]. Besides, despite significantly decreased cost of NGS in the last few years, the cost of WGS is still more expensive than RNA-Seq [43].
RNA-Seq is the most commonly selected technique for transcript detection of fusion gene owing to its low cost and quick turnaround time. More importantly, it can detect multiple alternative splice variants caused by a fusion event [39] and focuses on the expressed regions of the genome, which makes the detected fusions more biologically relevant in general. However, RNA-Seq technology only sequences about 2% of the entire genome compared with WGS [44], and it cannot monitor fusion events involving nontranscribed regions [45], which is a double-edged sword that detecting fusion events do not occur at the DNA level [41].
In general, it is reasonable to believe that development and progression of fusion gene detection technologies will contribute to the future studies on diagnosis and personalized treatment of human cancers.
The Landscape of Gene Fusions in Subtypes of Malignant Gliomas
Glioblastoma (GBM)
GBM accounts for 82% of malignant gliomas [46]. Patients with GBMs have a uniformly poor prognosis, with a median survival of approximately 14 to 16 months after multimodal treatments [47]. Considering the poor prognosis and therapeutic effect, it is of significance to develop novel and effective treatment strategies. Gene fusions are frequent genomic abnormalities in GBM; previous studies have shown that two major specific genomic hotspots where fusion frequently occurs are on chromosomes 7p and 12q, while other regions with a higher frequency of fusions are on chromosomes 1, 4, 6, and 19 [48]. Although fusions occur in approximately 30% to 50% of GBM patient samples, only a few of them are found in more than one patient with oncogenic biological function (Table 2). Potentially druggable gene fusions in all GBMs are discussed below, some of which have shown good preliminary antitumor effects in preclinical and clinical trials.
Table 2.
Recurrent gene fusions with verified biological functions in malignant gliomas
| Tumor Subtype | Fusion gene symbol | Incidence Rate | Activated Signaling Pathway | Prognostic or Diagnostic role | Target Medicine |
|---|---|---|---|---|---|
| Glioblastoma | FGFR1-TACC1 | 1.1% | MAPK, ERK, PI3K, and STAT3 |
Ponatinib BGJ398 Erdafitinib AZD4547 |
|
| FGFR3-TACC3 | 1.2%-8.3% | ||||
| EGFR-SEPT14 | 4% | STAT3 | Lapatinib Erlotinib |
||
| EGFR-PSPH | 2.2% | ||||
| NTRK fusions | 1.2-1.7 % | NGF/TrkA- | Entrectinib | ||
| Larotrectinib | |||||
| PTPRZ1-MET | 3% | Unfavorable prognosis | |||
| pHGG | NTRK fusions | 4% in DIPGs, 10% in NBS-pHGGs 40% in NBS-pHGGs within 3 years old |
PI3K, MAPK | Entrectinib | |
| MET fusions | 3-7% in pGBM, 10% in pHGG | MAPK | Foretinib Crizotinib |
||
| Ependymoma | RELA fusion | 70% in pediatric supra-tentorial ependymoma patients | NF-κB | Unfavorable prognosis |
Fibroblast Growth Factor Receptor (FGFR) Fusions
The tyrosine kinase coding domains of FGFR genes (FGFR1 to FGFR3) were found to be fused to the transforming acidic coiled-coil (TACC) coding domains of TACC1 or TACC3, respectively, forming the FGFR-TACC fusions [49]. TACC proteins are characterized by a coiled-coil domain at the C-terminus, known as the TACC domain, which regulates localization to the centrosome and mitotic spindle [50], [51]. It is hypothesized that TACC proteins are oncogenic in several human cancers including GBM [52], [53]. The FGFR family plays an important role in cell growth, differentiation, and angiogenesis, and its activation usually contributes to carcinogenic events [54]. The oncogenic activities of the FGFR-TACC fusion protein are greater than the sum of its parts when introduced into astrocytes or stereotactically transduced in the mouse brain [49].
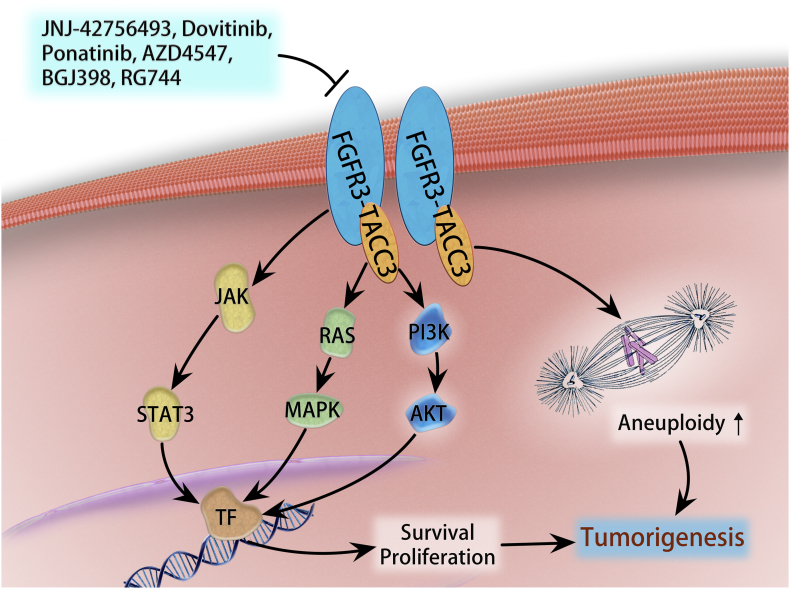
FGFR-TACC1 fusion was found in 1 out of 88 primary GBMs [49]. The incidence of FGFR3-TACC3 is considerably higher than the frequency of FGFR1-TACC1. The FGFR3-TACC3 fusion gene is a consequence of a 70-kb tandem duplication on chromosome 4q16.3 [49] and 1.2% to 8.3% of GBM patients harboring FGFR3-TACC [54], [55], [56], [57]. FGFR3-TACC3 fusion protein displays oncogenic activity that promotes tumorigenesis and enhances tumor progression [49], [56]. The abnormal expression of FGFR3-TACC3 in certain cell type was reported to activate mitogen-activated protein kinase (MAPK), extracellular signal–regulated kinase (ERK), phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), and signal transducer and activator of transcription 3 (STAT3) pathways (Figure 2) [56], [58], [59]. FGFR-TACC fusions may identify a subset of GBM patients who would benefit from targeted FGFR kinase inhibition.
Figure 2.
FGFR3-TACC3 fusion-mediated oncogenic pathways in GBM. FGFR3-TACC3 protein can constitutively and automatically activate tyrosine kinase domain, which exerts oncogenic function weakly through PI3K/ATK, RAS/MAPK, and STAT3 signaling and may localize to mitotic spindle poles to trigger aneuploidy.
Drugs targeting FGFR-TACC fusion genes have shown promising therapeutic potential in several types of cancer models, while clinical trials are still ongoing. To date, preclinical studies have shown a reduction of tumor growth and an increase of apoptosis upon treatment with the pan-FGFR inhibitor ponatinib in the U87MG cell line and the mouse xenograft model, suggesting a potential application of ponatinib as a chemotherapeutic option against GBM cells [60]. A phase II clinical trial studying ponatinib on recurrent GBM is now ongoing (NCT02478164); however, it is not specifically designed for FGFR fusion–positive patients.
Erdafitinib (JNJ-42756493), an oral ATP-competitive pan-FGFR selective inhibitor, inhibits tyrosine phosphorylation of activated FGFR at nanomolar concentrations [61]. It was applied to treat two GBM patients with FGFR3-TACC3 in a phase I trial and showed a tolerable toxicity and clear antitumor activity [61]. BGJ398, an FGFR1-3 kinase inhibitor, is evaluated in a phase II study in recurrent GBM with FGFR-TACC fusions and/or FGFR mutation (NCT01975701). Another clinical trial involving AZD4547 (a selective inhibitor of FGFR tyrosine kinase family) in recurrent glioma patients with FGFR fusion is under recruiting (NCT02824133). Given the encouraging outcome of FGFR inhibition in preclinical studies and earlier trials, further studies are warranted to identify the most effective FGFR inhibitors targeting FGFR-TACC fusion so as to translate them into clinical trials.
Epidermal Growth Factor Receptor (EGFR) Fusions
EGFR is a transmembrane protein which serves as a receptor for the epidermal growth factor family [62]. Overexpression of EGFR is frequently found in GBM, and EGFR-targeted therapy represents a promising anti-GBM therapy [63]. Besides the FGFR3-TACC3 fusions, EGFR emerged as the most frequent recurrent in-frame fusion in GBM [64]. EGFR is commonly fused to intron 9 of SEPT14 or to the gene PSPH, and most of them presented a carboxyl-terminal truncation [65].
EGFR-SEPT14 fusion gene is an in-frame fusion with C-terminal deletion of EGFR [48] and was found in about 4% of GBMs [64]. This fusion gene has been shown to activate STAT3 signaling, thus increasing tumor proliferation, migration, and stemness [64]. EGFR-PSPH fusion was also found in GBM, and the frequency of which was 2.2% (3/135), whereas signaling of EGFR-PSPH fusions remains unclear [64]. Interestingly, most of the EGFR-SEPT14 or -PSPH fusions lack EGFRvIII (exon 2-7 deletion) expression, which may be the reason that the C-terminal deletion of EGFR via gene fusion could serve as an alternative mechanism of EGFR activation [65].
Several modes of targeted therapies have been used to target EGFR including TKIs, antibody-based therapy, immunotherapy, and preclinical trials of RNA therapies. TKIs are small-molecule inhibitors, which bind to the ligand-binding site on the extracellular domain, and are the most clinically advanced EGFR-targeting therapy to date [66]. Preclinical studies suggest a beneficial role when EGFR-SEPT14 was targeted by TKIs like lapatinib and erlotinib; tumors with EGFR genomic alterations show delay on growth. However, clinical trials of EGFR inhibitors failed to show a definitive survival superiority in GBM, probably because of nonspecific patient selection; only a small portion of study populations were harboring EGFR fusions [67], [68], [69], [70]. Future studies may focus on targeting patients purely with EGFR-fusions based on individualized genomic examination.
Neurotrophic Tyrosine Kinase Receptor Type (NTRK) Fusions
NTRK encodes the tropomyosin receptor kinase (Trk) receptor family. TrkA, B, and C receptors are encoded by NTRK1, NTRK2, and NTRK3 genes, respectively. The rearrangements of the Trk receptor family play important roles in the oncogenesis in many types of tumor including glioma as well as NSCLC, colon cancer, and papillary thyroid cancer. Several drugs targeting cancers harboring these gene rearrangements are within clinical trials, yet none has been approved by the FDA so far [71].
NTRK1 is a well-known oncogene that is found commonly in many human cancers but largely lacked in GBMs. In the RNA-Seq data of The Cancer Genome Atlas (TCGA), 2 out of 162 GBM (1.2%) patients were found to have NTRK1 fused with two genes, neurofascin (NFASC) and brevican (BCAN), both of which are expressed in neuronal tissues. The fusion gene showed elevated expression of NTRK1, as well as neuro growth factor (NGF)–triggered activation of the NGF/TrkA- downstream pathway. Transduction of the NFASC-NTRK1 fusion gene in cell model increased proliferation both in vitro and in vivo [72]. What's more, both patients with NTRK fusion showed EGFR amplification (less than two-folds), indicating some potential linkage between these two genes. In another study involving 115 GBMs, Zongli et al. found two novel NTRK1 fusions in two separate GBM patients, CHTOP-NTRK1and ARHGEF2-NTRK1, both of which are in-frame fusions. Interestingly, the CHTOP-NTRK1 case harbored IDH1 R132 mutation, while the ARHGEF2-NTRK1 showed EGFR amplification [73]; the biological functions of these two novel fusions are not clear yet.
Although more data need to be accumulated to support the efficiency of Trk inhibitors in clinical trials, it is very promising that targeting the Trk family of receptor tyrosine kinases will help glioma patients with NTRK fusions. Entrectinib (RXDX-101) is a pan-Trk inhibitor with good blood-brain barrier penetration. In an open-label, multicenter, global, Phase II basket study (STARTRK-2) examining the use of entrectinib in patients having different types of tumors with the Trk gene rearrangements, the only pontine astrocytoma patient harboring BCAN-NTRK1 fusion showed 45% tumor volume reduction after dosing entrectinib [74]. Larotrectinib (LOXO-101) is a selective pan-TRK inhibitor that has no significant activity outside of the Trk family. Preliminary results from NAVIGATE Phase 2 larotrectinib trial showed its potential role in treating NTRK fusion-positive recurrent GBM. A 35-year-old woman with recurrent multifocal GBM harboring in-frame EML4-NTRK3 fusion was treated with larotrectinib [75]. The periventricular tumor shrank significantly (from 67 × 52 mm to 8 × 4 mm), while tumor at frontal and occipital lobes had no shrinkage. The trial is continuing to further understand this biological effect.
Other Fusions
There are several other gene fusions detected; however, the functional outcomes of these fusions are yet to be studied. The FIG-ROS1 fusion was the first identified fusion gene in GBM cell lines; an intrachromosomal homozygous deletion on 6q21 caused the FIG-ROS locus, this fusion encodes an in-frame fusion protein that has an active kinase activity, suggesting an oncogenic role [76]. Nameeta et al. explored genomic data of 185 GBM patient samples from both TCGA and Ivy center and found in total 27 genes that were fusion partners in more than one patient sample, including some novel finding of fusion involving noncoding genes. For example, noncoding RNA RP11-745C15.2 was found fusing with LANCL2 gene in two patients, one in TCGA database and the other in Ivy database. The same noncoding RNA was fused with EGFR in two other TCGA patients. Both fusions lead to C-terminal truncation of the fused gene [48]. In a study carried out based on Chinese Glioma Genome Atlas database, a novel PTPRZ1-MET fusion was found in 3% (3/99) of GBM patients. This fusion can induce elevation expression and phosphorylation of MET oncoprotein, and was associated with poor prognosis [77]. Currently, there have been no studies featuring targeting these gene fusions in adult GBM patients.
Pediatric High-Grade Glioma (pHGG)
pHGGs account for approximately 8% to 12% of all childhood central nervous system tumors, with a distinct biological entity compared with adult HGGs [78]. Most of the pHGGs are histologically anaplastic astrocytoma or GBM; the general prognosis is poor, especially for tumors that are not amenable to resection like diffuse intrinsic pontine glioma (DIPG) [79]. Gene fusions were commonly seen in pHGG including DIPG, although most of them were not recurrent, with uncertain influence on tumor initiating and progressing (Table 2).
NTRK Fusions
In a study done by Wu et al. analyzing transcriptome and WGS data from 127 pHGG patients, the NTRK fusions, which were detected also in adult GBM with a low frequency as described above, were found in 4% of DIPGs, 10% of non–brain stem pHGGs, and, most notably, 40% of non–brain stem pHGGs in infants within 3 years old [80].
A total of five NTRK fusion genes were found: TPM3-NTRK1, BTBD1-NTRK3, ETV6-NTRK3, VCL-NTRK2, and AGBL4-NTRK2. The first two fusion proteins even showed the ability to induce high-grade astrocytoma in Tp53-null mice models with PI3K and MAPK pathway activation. Taken together, these results indicated that NTRK fusion genes may act as important tumorigenic drivers in infant pHGGs. Based on these findings, a phase I/Ib study is conducted in pediatric population to evaluate entrectinib in primary central nervous system tumors (NCT02650401).
MET Fusions
MET gene encodes the protein c-Met, which possesses tyrosine kinase activity and is involved in different types of cancers. MET amplification has been described in about 3% to 7% of pediatric GBMs [81]. A recently published study from the International Cancer Genome Consortium PedBrain Tumor Project found an up to 10% chance of MET fusions with activated MAPK signaling.
The whole-genome sequencing of tumor and blood DNA was done in 53 pediatric GBM samples, as well as 5 pediatric GBM cell lines; researchers found 5 partners that fused with MET. They were TFG-MET, CLIP2-MET, PTPRZ1(exon1)-MET, PTPRZ1(exon1-2)-MET, and PTPRZ1(exon1-8)-MET. What's more, all these samples with MET fusion also harbor impaired cell cycle regulation resulting from Tp53 mutation or CKDN2A and CDKN2B deletion.
The TGF-MET fusion showed strong oncogenicity when induced to neonatal mice with impaired cell cycle (Ckdn2a-dificient or p53-null) in very short periods; histopathology confirmed that the tumor was high-grade glioma expressing fusion protein as well as phosphorylated MET and ERK. Notably, foretinib, an experimental inhibitor targeting MET and vascular endothelial growth factor receptor 2, decelerated MET fusion–driven tumor growth in vivo. The therapeutic effect of foretinib was further confirmed in another CLIP2-MET fusion SCID mice model.
Based on these findings, crizotinib, an FDA-approved inhibitor targeting ALK as well as MET and ROS1, was used in an 8-year-old GBM patient with PTPRZ1-MET fusion; this therapy led to substantial tumor shrinkage at 2 months, but treatment-resistant lesions were also detected with rapid progression that finally caused death of this patient, indicating that pediatric GBM remains a heterogenetic lesion that needs combined therapies on multitargets to help maintain a long-term response [82].
DHX57-TMEM178-MAP4K3 Fusion
Carvalho et al. found and validated a novel fusion complex including three genes in a DNA copy number profiling study of 100 pHGG cases. The fusion gene DHX57-TMEM178-MAP4K3 was found on chromosome 2p22.1 of an anaplastic astrocytoma case; it comprised exons 1-12 of DHX57, exons 2-4 of TMEM178, and exons 13-34 of MAP4K3. The resulting fusion gene contains key regulatory domains from all three proteins. Although the biological function is yet to be studied, the oncogenic capacity of all three components draws the hypothesis that this DHX57-TMEM178-MAP4K3 gene may be associated with tumor cell growth and proliferation [83].
Ependymomas
Ependymomas are glial tumors that arise from the cells lining the ventricles and central canal within the spinal cord. They can arise on various age groups and make up 3.6% of malignant primary central nervous system tumors [46]. Ependymomas have a variable prognosis, probably because of their different genomic landscape including transcriptional profiles, DNA copy number alterations, et al. The molecular classification of ependymoma involving nine subgroups helps make therapeutic strategies and prognosis prediction than classical histopathology [84]. Gene fusions were used to characterize two specific subgroups which were discussed below.
V-rel Avian Reticuloendotheliosis Viral Oncogene Homolog A (RELA) Fusion
By whole-genome sequencing in 41 tumors with matched normal blood and RNA sequencing in 77 tumors, Parker et al. found that a novel fusion of two genes in chromosome 11q13.1, RELA and a poorly characterized gene, C11orf95. This fusion resulted from chromothripsis, leading to constitutive aberrant activation of the nuclear factor-kB (NF-κB) signaling pathway (Table 2). After inducing the fusion gene to the neural stem cell, it rapidly generated tumor in xenograft models [85].
Another study from an independent cohort including 500 ependymoma data confirmed that C11orf95-RELA fusion is mostly seen in pediatric patients with a median age of 8 years old, but only in supratentorial ependymomas and not in posterior fossa or spinal ependymomas [84]. The incidence is more than 70% [84], [85], [86], and most tumors harboring fusion are World Health Organization (WHO) grade II to III tumors. About 15% patients with C11orf95-RELA fusion also have homozygous deletions of CDKN2A, which indicates poor prognosis for ependymomas [84], [87]. In the newly released 2016 WHO Classification of Tumors of Central Nervous System, ependymomas with C11orf95-RELA fusion were put into a specific diagnostic entity [88], indicating high-risk biological behavior, with a 10-year overall survival of around 50% and a 10-year progression-free survival of around 20% [84].
YAP1 Fusions
YAP1 fusions were found in the same studies with RELA fusion; it is regarded as the character of another subgroup of supratentorial ependymoma, which is distinct from RELA fusion–positive ones [84]. There are two partners found to be fused with YAP1 gene in chromosome 11. The most common fusion partner is MAMLD1, identified in 86% (6/7), followed by FAM118B, identified in 14% (1/7) of the cases. The exact function of these fusions is still not clear and remains to be investigated. Unlike RELA fusion, tumors harboring YAP1 fusion showed no evidence of chromothripsis and had relatively stable genomes. YAP1 fusion was found mostly in ependymoma patients with a median age at diagnosis of 1.4 years, with a relatively favorable prognosis and with a 5-year progression-free survival of 66% and overall survival of 100% [84]. The detailed mechanism why the fusion is related with good prognosis is not clear yet.
Conclusion and Perspectives
Malignant gliomas are characterized by extensive genomic instability [89], while most of them are associated with a corresponding increase in fusion transcript frequency [8], [90]. The advent of high-throughput technologies, especially NGS, has revolutionized the detection of gene fusions, with an overwhelming majority identified in the past 5 years [7]. Discovery of gene fusions in gliomas benefits from the new wave of technology, as fusions are identified in approximately 30% to 50% of GBMs and 47% of DIPGs and non–brain stem HGGs [48], [80]. To capture exactly what proportion oncogenic drivers account for among all gene fusions, the significant organization is urgently required with the rapid increase of gene fusion data. Excitedly, concerted efforts have been made to develop computational algorithms and tools involving Oncofuse and Pegasus for separating driver fusions from passenger mutations with sequencing data [91], [92]. Furthermore, bioinformatics approaches also have elucidated many aspects of oncogenic gene fusions, from the genetic changes and a causative importance of fusion events to the structural properties and regulatory functions of fusion proteins [90]. Thus, the computational study will catalyze advancements in oncogenic gene fusion identification and guide experimental validation studies in glioma.
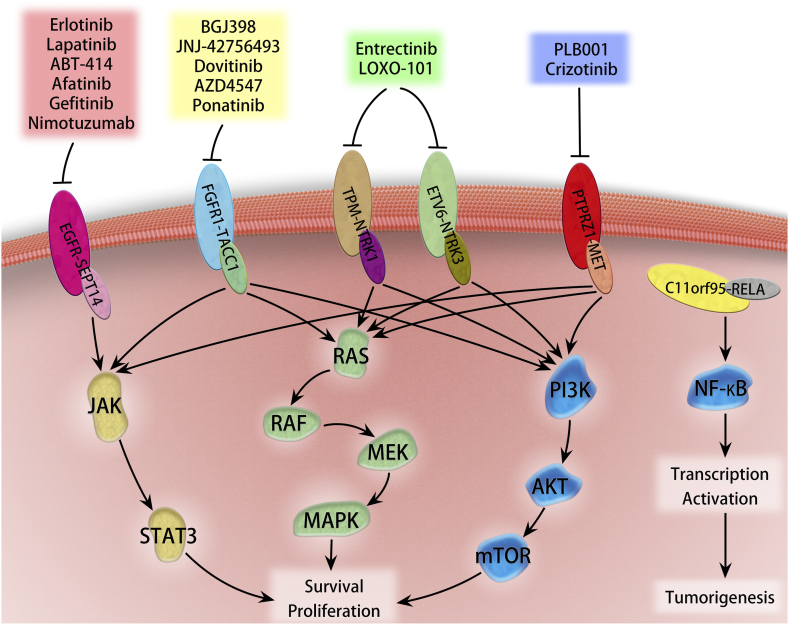
Fusions with well-characterized kinases make up a significant part of oncogenic drivers, which is attributed to their susceptibility to kinase inhibitors [57], [93]. Recently, a deep-sequencing data study reported 4.4% of GBMs and 1.5% low-grade gliomas harboring potentially druggable kinase fusions [8]. Consistently, a more recent analysis of RNA-seq confirmed a similar prevalence (9.7%) of potentially targetable fusions in gliomas, of which 11% (24/226) of GBMs, 12% (5/42) of anaplastic astrocytomas, and 8% (2/25) of grade II astrocytomas carried targetable fusions [94]. As demonstrated, the portion of kinase gene involved in the fusion encoded the intact domain which is conserved across all fusions and essential for activity [95]. It is reasonable that drugs targeting primary oncoproteins have great promise to be applied directly to clinical samples which are addicted to the oncogenic fusion kinases in malignant gliomas (Figure 3). Several drugs have been granted approval by FDA as standard therapy for tumor patients harboring specific gene fusions [96], and in malignant glioma patients, clinical trials are still ongoing, with some exciting preliminary findings (Table 3). The successful targeting of oncogenic fusion kinases is now driving a major paradigm shift in oncology, whereby somatic genetic alterations rather than the histological subtype can provide the guidance for selecting therapeutic strategies. Although the frequency of recurrent fusion transcripts is generally substantially low [7], the subset of different types of glioma stratified by the druggable oncogenic fusions will benefit from the precise treatment approaches.
Figure 3.
Overview of downstream signaling linked to selected driver fusions and potential intervention of specific targeting inhibitors. Glioma-associated oncogenic fusion proteins commonly activate canonical pathways, such as JAK/STAT3, ERK/MAKP, PI3K/ATK, and NF-κB, involved in tumor survival, proliferation, and tumorigenesis. Some of the proteins have been explored as therapeutic targets to inhibit glioma.
Table 3.
Gene Fusions as Therapeutic Targets in Malignant Gliomas
| Fusion Gene | Medicine | Clinical Trial Status |
|---|---|---|
| FGFR fusion | BGJ398 | Phase II study in recurrent GBM patients with FGFR-TACC fusions and/or FGFR mutation. (ongoing, NCT01975701) |
| Erdafitinib (JNJ-42756493) | Phase I study in adult participants with advanced or refractory solid tumors or lymphoma. (ongoing, NCT01703481) - Two GBM patients with FGFR3-TACC3 showed clear anti-tumor activity. |
|
| AZD4547 | Phase I/II study in relapsed/refractory glioma patients positive for an FGFR fusion (recruiting, NCT02824133) | |
| NTRK fusion | Entrectinib (RXDX-101) | Phase II basket study in patients having different types of tumors with the TRK gene rearrangements (recruiting, NCT02568267) - The only pontine astrocytoma patient harboring BCAN-NTRK1 fusion showed 45% tumor volume reduction after dosing entrectinib; Phase I/Ib study in children and adolescents with recurrent or refractory solid tumors and primary central nervous system tumors, with or without TRK, ROS1, or ALK fusions (recruiting, NCT02650401). |
| Larotrectinib (LOXO-101) | Phase II study in patients with advanced solid tumors harboring NTRK fusions (recruiting, NCT02576431) - A 35-year-old woman with recurrent multifocal GBM harboring in-frame EML4-NTRK3 fusion was treated with larotrectinib. The periventricular tumor shrank significantly (67 × 52 mm to 8 × 4 mm), while tumor at frontal and occipital lobes had no shrinkage. |
|
| MET fusion | PLB1001 | Phase I study in recurrent high-grade glioma patients with PTPRZ1-MET fusion gene (recruiting, NCT02978261) |
| Crizotinib | Single case experience. - Used in an 8-year-old GBM patient with PTPRZ1-MET fusion, led to substantial tumor shrinkage at 2 months, but treatment-resistant lesions were also detected with rapid progression that finally caused death of this patient. |
The revised 2016 WHO Classification of Tumors of the Central Nervous System combines biology-driven molecular marker diagnostics in addition to classical histology [88], formulating a concept for how glioma diagnoses should be structured in the molecular era and facilitating management of outcome prediction and treatment decisions. Different from many cancer-associated mutations with a variable and heterogeneous nature occurring in various morphologically and clinically distinct tumors, gene fusions, for the most part, are typically disease specific, which implicates its promising application in cancer diagnosis [42]. Recurrent gene fusions, such as ERG, ETV1, TFE3, NUT, POU5F1, NFIB, PLAG1, PAX8, and RELA, have been emerging as required diagnostic biomarkers in different type of cancers including ependymoma [13]. It is the inexorable trend to establish more gene fusions as clinical biomarkers across different subtypes of gliomas to identify patients with specific promising therapeutic targets.
Taken together, data-intensive studies are playing the key role in detecting gene fusions, while further functional characterization of these fusions should be a high priority. A more detailed understanding of its diverse regulatory pathway and involvement in cellular processes will provide valuable insight into malignant glioma biology. The integration of diagnosis, stratification, and targeted treatment based on gene fusion will direct the future avenues of next-generation personalized treatment for glioma patients.
Conflicts of Interests
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81101907, 81472354), “Young Talented Physician” Project of Shanghai, and “Pu Jiang Talent” Project of Shanghai (PJ[2014]0002617). The authors also would like to thank Dr. Peng Yang for his assistance with all figures.
References
- 1.Campregher PV, de Oliveira Pereira W, Lisboa B, Puga R, Deolinda ER, Helman R, Marti LC, Guerra JC, Manola KN, Petroni RC. A novel mechanism of NPM1 cytoplasmic localization in acute myeloid leukemia: the recurrent gene fusion NPM1-HAUS1. Haematologica. 2016;101:e287–290. doi: 10.3324/haematol.2015.137364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masetti R, Togni M, Astolfi A, Pigazzi M, Manara E, Indio V, Rizzari C, Rutella S, Basso G, Pession A. DHH-RHEBL1 fusion transcript: a novel recurrent feature in the new landscape of pediatric CBFA2T3-GLIS2–positive acute myeloid leukemia. Oncotarget. 2013;4:1712–1720. doi: 10.18632/oncotarget.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Y, Zhang Y, Li Y, Hu H, Wang L, Li H, Wang R, Ye T, Luo X, Zhang Y. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer. 2014;84:121–126. doi: 10.1016/j.lungcan.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Kim HP, Cho GA, Han SW, Shin JY, Jeong EG, Song SH, Lee WC, Lee KH, Bang D, Seo JS. Novel fusion transcripts in human gastric cancer revealed by transcriptome analysis. Oncogene. 2014;33:5434–5441. doi: 10.1038/onc.2013.490. [DOI] [PubMed] [Google Scholar]
- 5.Parker BC, Engels M, Annala M, Zhang W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J Pathol. 2014;232:4–15. doi: 10.1002/path.4297. [DOI] [PubMed] [Google Scholar]
- 6.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 7.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 8.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RG. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon MP, Pedeutour F, Sirvent N, Grosgeorge J, Minoletti F, Coindre JM, Terrier-Lacombe MJ, Mandahl N, Craver RD, Blin N. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- 10.Rutkowski P, Van Glabbeke M, Rankin CJ, Ruka W, Rubin BP, Debiec-Rychter M, Lazar A, Gelderblom H, Sciot R, Lopez-Terrada D. Imatinib mesylate in advanced dermatofibrosarcoma protuberans: pooled analysis of two phase II clinical trials. J Clin Oncol. 2010;28:1772–1779. doi: 10.1200/JCO.2009.25.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non–small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 12.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 13.Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129–156. doi: 10.1186/s13073-015-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 15.Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Ding Q, Chen Y, Ouyang Q, Jiang L, Dai J, Lu Y, Wu X, Liang Q, Wang H. Clinical features and molecular basis of 102 Chinese patients with congenital dysfibrinogenemia. Blood Cells Mol Dis. 2015;55:308–315. doi: 10.1016/j.bcmd.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340:192–200. doi: 10.1016/j.canlet.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gainor JF, Shaw AT. Novel targets in non–small cell lung cancer: ROS1 and RET fusions. Oncologist. 2013;18:865–875. doi: 10.1634/theoncologist.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mino-Kenudson M, Chirieac LR, Law K, Hornick JL, Lindeman N, Mark EJ, Cohen DW, Johnson BE, Janne PA, Iafrate AJ. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, Zielenska M, Squire JA. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang R. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, Kimura Y, Drilon A, Guha U, Rusch V. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–6608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, Luo X, Wang L, Li H, Zhang Y. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 26.Jabber Al-Obaidi MS, Martineau M, Bennett CF, Franklin IM, Goldstone AH, Harewood L, Jalali GR, Prentice HG, Richards SM, Roberts K. ETV6/AML1 fusion by FISH in adult acute lymphoblastic leukemia. Leukemia. 2002;16:669–674. doi: 10.1038/sj.leu.2402435. [DOI] [PubMed] [Google Scholar]
- 27.Oliva E, de Leval L, Soslow RA, Herens C. High frequency of JAZF1-JJAZ1 gene fusion in endometrial stromal tumors with smooth muscle differentiation by interphase FISH detection. Am J Surg Pathol. 2007;31:1277–1284. doi: 10.1097/PAS.0b013e318031f012. [DOI] [PubMed] [Google Scholar]
- 28.Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L, Ilouze M, Dvir A, Boyle T, Wynes M, Miller VA, Lipson D, Palmer GA. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist. 2015;20:316–322. doi: 10.1634/theoncologist.2014-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum Mol Genet. 2003;12(2):R145–R152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- 30.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;(37 Suppl):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 31.Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, Buhlmann P, Niggli FK, Schafer BW. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Motoi T, Khanin R, Olshen A, Mertens F, Bridge J, Dal Cin P, Antonescu CR, Singer S, Hameed M. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosom Cancer. 2012;51:127–139. doi: 10.1002/gcc.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertens F, Tayebwa J. Evolving techniques for gene fusion detection in soft tissue tumours. Histopathology. 2014;64:151–162. doi: 10.1111/his.12272. [DOI] [PubMed] [Google Scholar]
- 34.Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- 36.Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, Johnson NA, Zhao Y, Telenius A, Neriah SB. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha KC, Lalonde E, Li L, Cavallone L, Natrajan R, Lambros MB, Mitsopoulos C, Hakas J, Kozarewa I, Fenwick K. Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines. BMC Med Genet. 2011;4:75–87. doi: 10.1186/1755-8794-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH, Anderson S, Haegert A, Brahmbhatt S, Shukin R. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J Pathol. 2012;227:53–61. doi: 10.1002/path.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Xia J, Jia P, Pao W, Zhao Z. Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief Bioinform. 2013;14:506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305:1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Razzaq SK, Vo AD, Gautam M, Li H. Identifying fusion transcripts using next generation sequencing. Wiley Interdiscip Rev RNA. 2016;7:811–823. doi: 10.1002/wrna.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veeraraghavan J, Ma J, Hu Y, Wang XS. Recurrent and pathological gene fusions in breast cancer: current advances in genomic discovery and clinical implications. Breast Cancer Res Treat. 2016;158:219–232. doi: 10.1007/s10549-016-3876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think! Genome Biol. 2011;12:125–134. doi: 10.1186/gb-2011-12-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro-Oncology. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spina R, Voss DM, Asnaghi L, Sloan A, Bar EE. Atracurium besylate and other neuromuscular blocking agents promote astroglial differentiation and deplete glioblastoma stem cells. Oncotarget. 2016;7:459–472. doi: 10.18632/oncotarget.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics. 2013;14:818–832. doi: 10.1186/1471-2164-14-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science (New York, NY) 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–388. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Hood FE, Royle SJ. Pulling it together: the mitotic function of TACC3. BioArchitecture. 2011;1:105–109. doi: 10.4161/bioa.1.3.16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duncan CG, Killela PJ, Payne CA, Lampson B, Chen WC, Liu J, Solomon D, Waldman T, Towers AJ, Gregory SG. Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget. 2010;1:265–277. doi: 10.18632/oncotarget.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao R, Natsume Y, Saiki Y, Shioya H, Takeuchi K, Yamori T, Toki H, Aoki I, Saga T, Noda T. Disruption of Tacc3 function leads to in vivo tumor regression. Oncogene. 2012;31:135–148. doi: 10.1038/onc.2011.235. [DOI] [PubMed] [Google Scholar]
- 54.Costa R, Carneiro BA, Taxter T, Tavora FA, Kalyan A, Pai SA, Chae YK, Giles FJ. FGFR3-TACC3 fusion in solid tumors: mini review. Oncotarget. 2016;7:55924–55938. doi: 10.18632/oncotarget.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier B, Giry M. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5 doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lasorella A, Sanson M, Iavarone A. FGFR-TACC gene fusions in human glioma. Neuro-Oncology. 2016;19:475–483. doi: 10.1093/neuonc/now240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Zhou Q, Gao G, Wang Y, Fang Z, Li G, Yu M, Kong L, Xing Y, Gao X. The effects of ponatinib, a multi-targeted tyrosine kinase inhibitor, against human U87 malignant glioblastoma cells. Onco Targets Ther. 2014;7:2013–2019. doi: 10.2147/OTT.S67556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tabernero J, Bahleda R, Dienstmann R, Infante JR, Mita A, Italiano A, Calvo E, Moreno V, Adamo B, Gazzah A. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2015;33:3401–3408. doi: 10.1200/JCO.2014.60.7341. [DOI] [PubMed] [Google Scholar]
- 62.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 63.Addeo R, Zappavigna S, Parlato C, Caraglia M. Erlotinib: early clinical development in brain cancer. Expert Opin Investig Drugs. 2014;23:1027–1037. doi: 10.1517/13543784.2014.918950. [DOI] [PubMed] [Google Scholar]
- 64.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maire CL, Ligon KL. Molecular pathologic diagnosis of epidermal growth factor receptor. Neuro-Oncology. 2014;16(Suppl 8):viii1–viii6. doi: 10.1093/neuonc/nou294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karpel-Massler G, Schmidt U, Unterberg A, Halatsch ME. Therapeutic inhibition of the epidermal growth factor receptor in high-grade gliomas: where do we stand? Mol Cancer Res. 2009;7:1000–1012. doi: 10.1158/1541-7786.MCR-08-0479. [DOI] [PubMed] [Google Scholar]
- 67.Kreisl TN, Lassman AB, Mischel PS, Rosen N, Scher HI, Teruya-Feldstein J, Shaffer D, Lis E, Abrey LE. A pilot study of everolimus and gefitinib in the treatment of recurrent glioblastoma (GBM) J Neuro-Oncol. 2009;92:99–105. doi: 10.1007/s11060-008-9741-z. [DOI] [PubMed] [Google Scholar]
- 68.Reardon DA, Desjardins A, Vredenburgh JJ, Gururangan S, Friedman AH, Herndon JE, 2nd, Marcello J, Norfleet JA, McLendon RE, Sampson JH. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J Neuro-Oncol. 2010;96:219–230. doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, Kabuubi P, Ayers-Ringler J, Rabbitt J, Page M. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27:579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee EQ, Kaley TJ, Duda DG, Schiff D, Lassman AB, Wong ET, Mikkelsen T, Purow BW, Muzikansky A, Ancukiewicz M. A multicenter, phase II, randomized, noncomparative clinical trial of radiation and temozolomide with or without vandetanib in newly diagnosed glioblastoma patients. Clin Cancer Res. 2015;21:3610–3618. doi: 10.1158/1078-0432.CCR-14-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5:25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J, Lee Y, Cho HJ, Lee YE, An J, Cho GH, Ko YH, Joo KM, Nam DH. NTRK1 fusion in glioblastoma multiforme. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, Chen J, Robinson HE, Shim HS, Chmielecki J. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 74.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, Bauer TM, Farago AF, Wheler JJ, Liu SV. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schram AM, Taylor BS, Hechtman JF, Benayed R, Wang L, Hanusch B, Young R, Grommes C, Ku N, Hyman DM. AACR Annual Meeting 2017 Online Proceedings and Itinerary Planner Homesection 34. 2017. LB-302/10 — potential role of larotrectinib (LOXO-101), a selective pan-TRK inhibitor, in NTRK fusion-positive recurrent glioblastoma. [Google Scholar]
- 76.Charest A, Lane K, McMahon K, Park J, Preisinger E, Conroy H, Housman D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosom Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 77.Chen HM, Yu K, Tang XY, Bao ZS, Jiang T, Fan XL, Chen XW, Su XD. Enhanced expression and phosphorylation of the MET oncoprotein by glioma-specific PTPRZ1-MET fusions. FEBS Lett. 2015;589:1437–1443. doi: 10.1016/j.febslet.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 78.Gottardo NG, Gajjar A. Chemotherapy for malignant brain tumors of childhood. J Child Neurol. 2008;23:1149–1159. doi: 10.1177/0883073808321765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hipp SJ, Steffen-Smith E, Hammoud D, Shih JH, Bent R, Warren KE. Predicting outcome of children with diffuse intrinsic pontine gliomas using multiparametric imaging. Neuro-Oncology. 2011;13:904–909. doi: 10.1093/neuonc/nor076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.International Cancer Genome Consortium PedBrain Tumor P Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22:1314–1320. doi: 10.1038/nm.4204. [DOI] [PubMed] [Google Scholar]
- 83.Carvalho D, Mackay A, Bjerke L, Grundy RG, Lopes C, Reis RM, Jones C. The prognostic role of intragenic copy number breakpoints and identification of novel fusion genes in paediatric high grade glioma. Acta Neuropathol Commun. 2014;2:23–34. doi: 10.1186/2051-5960-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nanmbirajan A, Malgulwar PB, Pathak P, Faruq M, Rajeshwari M, Gupta RK, Suri V, Sarkar C, Singh M, Sharma MC. C11orf95-Rela fusion positive pediatric supratentorial ependymomas are an aggressive subset with increased expression of stem cell marker nestin and vascular endothelial derived growth factor. Neuro-Oncology. 2016;18:iii30–iii31. [Google Scholar]
- 87.Korshunov A, Witt H, Hielscher T, Benner A, Remke M, Ryzhova M, Milde T, Bender S, Wittmann A, Schottler A. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010;28:3182–3190. doi: 10.1200/JCO.2009.27.3359. [DOI] [PubMed] [Google Scholar]
- 88.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 89.Milinkovic V, Bankovic J, Rakic M, Milosevic N, Stankovic T, Jokovic M, Milosevic Z, Skender-Gazibara M, Podolski-Renic A, Pesic M. Genomic instability and p53 alterations in patients with malignant glioma. Exp Mol Pathol. 2012;93:200–206. doi: 10.1016/j.yexmp.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 90.Latysheva NS, Babu MM. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 2016;44:4487–4503. doi: 10.1093/nar/gkw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shugay M, Ortiz de Mendibil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29:2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 92.Abate F, Zairis S, Ficarra E, Acquaviva A, Wiggins CH, Frattini V, Lasorella A, Iavarone A, Inghirami G, Rabadan R. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;8:97–110. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davare MA, Tognon CE. Detecting and targetting oncogenic fusion proteins in the genomic era. Biol Cell. 2015;107:111–129. doi: 10.1111/boc.201400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subramaniam DS, Xiu J, Mehta S, Gatalica Z, Swensen J, Sanai N, Heimberger AB. RNA-Seq analysis of glioma tumors to reveal targetable gene fusions. J Clin Oncol. 2017;35:2019. [Google Scholar]
- 95.Chmielecki J, Peifer M, Jia P, Socci ND, Hutchinson K, Viale A, Zhao Z, Thomas RK, Pao W. Targeted next-generation sequencing of DNA regions proximal to a conserved GXGXXG signaling motif enables systematic discovery of tyrosine kinase fusions in cancer. Nucleic Acids Res. 2010;38:6985–6996. doi: 10.1093/nar/gkq579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]