Abstract
Members of the cyanobacterial genus Trichodesmium are well known for their substantial impact on nitrogen influx in ocean ecosystems and the enormous surface blooms they form in tropical and subtropical locations. However, the secondary metabolite composition of these complex environmental bloom events is not well known, nor the possibility of the production of potent toxins that have been observed in other bloom-forming marine and freshwater cyanobacteria species. In the present work, we aimed to characterize the metabolome of a Trichodesmium bloom utilizing MS/MS-based molecular networking. Furthermore, we integrated cytotoxicity assays in order to identify and ultimately isolate potential cyanotoxins from the bloom. These efforts led to the isolation and identification of several members of the smenamide family, including three new smenamide analogs (1–3) as well as the previously reported smenothiazole A-hybrid polyketide-peptide compounds. Two of these new smenamides possessed cytotoxicity to neuro-2A cells (1 and 3) and their presence elicits further questions as to their potential ecological roles. HPLC profiling and molecular networking of chromatography fractions from the bloom revealed an elaborate secondary metabolome, generating hypotheses with respect to the environmental role of these metabolites and the consistency of this chemical composition across genera, space and time.
Keywords: Trichodesmium, molecular networking, cyanotoxins, harmful algal blooms, metabolomics
Introduction
Blooms of toxin-producing cyanobacteria (harmful algal blooms, HABs) continue to be a threat to water resources in the U.S. and across the globe (Carmichael and Boyer, 2016). Research surrounding these bloom events with respect to cyanobacteria has generally focused on freshwater planktonic species and a suite of well-characterized toxins, including the anatoxins, saxitoxins and microcystins (Bláha et al., 2009). However, species of cyanobacteria in the marine realm have been a prolific source of exquisitely potent cytotoxic metabolites (Luesch et al., 2001; Taori et al., 2008; Pereira et al., 2012). Members of the bloom-forming genus Trichodesmium are an understudied group of marine cyanobacteria with respect to toxin production and environmental impact. With respect to new natural products, the cyclic peptide trichamide was characterized from a cultured strain of Trichodesmium erythraeum, although no significant cytotoxicity was observed against HCT-116 cells and CEM-TART cells when tested at 10 and 50 μg/mL, respectively (Sudek et al., 2006). The lipoamides, credneramides A and B were isolated and characterized from a field-collected benthic cyanobacterium identified as a new species of Trichodesmium (Malloy et al., 2012). These metabolites inhibited spontaneous calcium oscillations in murine cerebrocortical neurons (Malloy et al., 2012). Several known cyanotoxins, such as anatoxin, saxitoxin, microcystins and aplysiatoxins have been reported from Trichodesmium blooms collected from distinct geographic areas (Ramos et al., 2005; Detoni et al., 2016; Shunmugam et al., 2017).
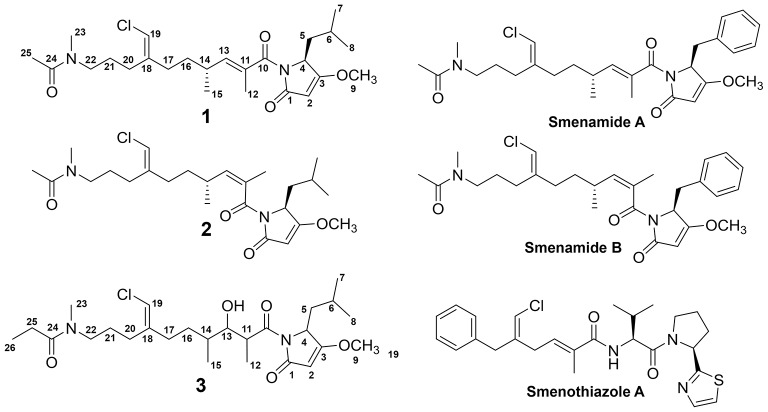
In the current report, we detail the comprehensive metabolic profiling of a Trichodesmium bloom collected from the western Gulf of Mexico utilizing MS/MS-based molecular networking (Watrous et al., 2012; Yang et al., 2013; Wang et al., 2016) and cytotoxicity assays. In our previous work on this Trichodesmium bloom, we have utilized cytotoxicity assays, NMR-guided isolation and MS-guided isolation independently to characterize chlorinated polyketides and hybrid polyketide peptides (Bertin et al., 2016, 2017a,b; Belisle et al., 2017). The current report attempts to describe the Trichodesmium bloom metabolome more completely, focusing on a networking tool to cluster molecules based on similarities in the MS/MS fragmentation patterns (Watrous et al., 2012). Our efforts ultimately led to the isolation and characterization of three new members of the smenamide family of molecules (1–3) and the previously reported smenothiazole A (Figure 1). Smenamides C and E demonstrated potent neurotoxicity (1 and 3).
Figure 1.
Structures of 1-3, smenamides A and B and smenothiazole A.
Materials and methods
General experimental procedures
Optical rotations were measured using a Jasco P-2000 polarimeter. UV spectra were measured using a Beckman Coulter DU-800 spectrophotometer. CD spectra were recorded using a Jasco J-1100 CD spectrometer. NMR spectra were collected using a Bruker 800 MHz NMR instrument. Additional NMR spectra were recorded on a Varian 500 MHz NMR instrument. HRESIMS analysis was performed using an AB SCIEX TripleTOF 4600 mass spectrometer with Analyst TF software. LC-MS/MS analysis was carried out using a ThermoFinnigan LCQ AdvantageMax mass spectrometer with an electrospray ionization (ESI) source. Semi-preparative HPLC was carried out using a Dionex UltiMate 3000 HPLC system and Agilent 1100 series system each equipped with a micro vacuum degasser, an autosampler and a diode-array detector.
Collection, extraction and fractionation of bloom material
Samples from a localized bloom of Trichodesmium were collected from Padre Island, Corpus Christi, TX during 9–11 May 2014 as described previously (Bertin et al., 2016, 2017a,b; Belisle et al., 2017). Briefly, bloom material was collected in 5-gallon buckets from ca. 0.5-meter water depth and concentrated by gentle filtration through an 18 μm mesh screen. A subsample of the cell mass was examined microscopically and identified using Komárek and Anagnostidis (2005) as being dominated by cyanobacteria of the genus Trichodesmium. The material was frozen and shipped for further chemical analysis. The biomass (ca. 14 g dry weight) was repeatedly extracted with 2:1 CH2Cl2:CH3OH and the extracts were combined and evaporated under reduced pressure (3.95 g). The extract was reconstituted in hexanes and applied to silica gel (300 mL) in a wide fritted column with a vacuum attachment. The extract was fractionated using a stepped gradient from 100% hexanes to 100% CH3OH resulting in nine fractions. Seven of the nine fractions (C-I) were further analyzed by means of cytotoxicity assays and MS/MS-based molecular networking. The first two fractions: 100% hexanes (A) and 90% hexanes in EtOAc (B) were intended to remove hydrocarbons and exceedingly lipophilic substances from the sample and were not analyzed further.
Molecular networking
Fractions C-I were subjected to LC-MS/MS analysis with data collection in data-dependent acquisition mode on a ThermoFinnigan LCQ AdvantageMax mass spectrometer with an electrospray ionization (ESI) source. A Kinetex 5 μm C18 column (100 × 4.6 mm) was used for separation of analytes. The LC method consisted of a linear gradient from 30 to 99% CH3CN in water + 0.1% formic acid over 17 minutes, followed by an isocratic period at 99% CH3CN of 3 minutes. The flow rate was held at 0.6 mL/min. The MS spray voltage was 5 kV with a capillary temperature of 400°C. For the MS/MS component, the CID isolation width was 2.0 and the collision energy was 35.0 eV. The raw data files were converted to mzXML format using MSConvert from the ProteoWizard suite (http://proteowizard.sourceforge.net/tools.shtml)1. The molecular network was generated using the online platform at Global Natural Products Social Molecular Networking website (gnps.ucsd.edu) using parameters detailed in Table S5. The network was visualized using the Browser Network Visualizer tool available on the gnps website.
Isolation of 1–3 and smenothiazole A
Fractions G (100% EtOAc, 104.0 mg) and H (75% EtOAc in CH3OH, 286.9 mg) were chosen for further purification based on the quantity of network ions in these fractions, the molecular features of these ions (ratio of M+ and M+2 isotope), and cytotoxicity results of the mixed chromatography fractions. Fractions G and H were combined based on similarities in LC-MS/MS profiles and 1H NMR resonances. The combined sample was further fractionated over a 2 g C18 SPE column eluting with 50% water in CH3CN (13.7 mg), 100% CH3CN (143.2 mg), 100% CH3OH (74.5 mg) and 100% EtOAc (54.5 mg). The fraction eluting with 100% CH3CN was subjected to reversed phase HPLC using a YMC 5 μm ODS column (250 × 10 mm); mobile phase: 65% CH3CN /35% water with 0.05% formic acid added to each solvent, flow 3 mL/min. Fractions were collected based on UV characteristics and HPLC fractions were analyzed by HRESIMS for ions of interest from the molecular network. Further purification was carried out using the YMC column mentioned above; mobile phase: 80% CH3CN in water with 0.05% formic acid added to each solvent, flow 3 mL/min resulted in the isolation of 7.0 mg of 1 (tR, 11.5 min). A mobile phase of 65% CH3CN in water with 0.05% formic acid added to each solvent, flow 3 mL/min was used to isolate 0.6 mg of 2 (tR, 26.0 min) and 0.3 mg of 3 (tR, 21.2 min). A final purification was carried out using a YMC 5 μm ODS column (250 × 10 mm); mobile phase: 80% CH3CN in water with 0.1% formic acid added to each solvent, flow 3 mL/min and 2.0 mg of smenothiazole A was isolated (tR, 5.0 min).
Smenamide C (1): colorless oil; [α]25D +38.2 (c 0.20, CH3OH); UV (CH3OH) λmax (log ε) 203 (4.2), 238 (4.0) nm; 1H NMR (800 MHz, DMSO-d6) and 13C NMR (200 MHz, DMSO-d6), see Table 1; HRESIMS m/z 467.2661 [M+H]+ (calcd for C25H40N2O4Cl, 467.2677) and m/z 489.2486 [M+Na]+ (calcd for C25H39N2O4ClNa, 489.2496).
Table 1.
NMR data for Smenamide C (1) Z-conformer (800 MHz for 1H NMR; 200 MHz for 13C NMR, DMSO).
| Position | δC, type | δH (J in Hz) | HMBC | COSY |
|---|---|---|---|---|
| 1 | 169.3, qC | |||
| 2 | 93.7, CH | 5.28, s | 1, 4, 10 | |
| 3 | 180.3, qC | |||
| 4 | 57.6, CH | 4.75, t (5.3) | 1, 2, 3, 5, 6, 10 | 5a, 5b |
| 5a | 38.6, CH2 | 1.72, m | 3, 4, 7, 8 | 4, 5b |
| 5b | 1.59, m | 3, 4, 7, 8 | 4, 5a | |
| 6 | 24.2, CH | 1.66, m | 4, 5, 7, 8 | 7, 8 |
| 7 | 24.0, CH3 | 0.85, d (6.6) | 5, 8 | 6 |
| 8 | 23.3, CH3 | 0.87, d (6.6) | 5, 7 | 6 |
| 9 | 59.7, CH3 | 3.87, s | 3 | |
| 10 | 170.6, qC | |||
| 11 | 131.7, qC | |||
| 12 | 13.9, CH3 | 1.77, s | 10, 11, 13 | |
| 13 | 141.9, CH | 5.59, d (9.8) | 10, 11, 12, 14, 15, 16 | 14 |
| 14 | 32.2, CH | 2.45, m | 11, 13, 15, 16, 17 | 13, 15, 16a, 16b |
| 15 | 20.4, CH3 | 0.94, d (6.6) | 14, 16, 18 | 14 |
| 16a | 34.9, CH2 | 1.49, m | 17, 18 | 14, 17a, 17b |
| 16b | 1.35, m | 17, 18 | 14, 17a, 17b | |
| 17a | 32.1, CH2 | 2.22, m | 14, 16, 18, 19, 20 | 16a, 16b, 17b |
| 17b | 2.10, ovlpa | 14, 16, 18, 19, 20 | 16a, 16b, 17a | |
| 18 | 142.8, qC | |||
| 19 | 112.6, CH | 6.04, s | 17, 18, 20 | |
| 20a | 27.4, CH2 | 2.12, m | 17, 18, 19, 21, 22 | 21 |
| 20b | 2.07, ovlp | 17, 18, 19, 21, 22 | 21 | |
| 21 | 24.8, CH2 | 1.56, m | 20, 22 | 20a, 20b, 22 |
| 22 | 46.7, CH2 | 3.26, m | 20, 21, 23, 24 | 21 |
| 23 | 35.9. CH3 | 2.94, s | 22, 24 | |
| 24 | 169.9, qC | |||
| 25 | 22.1, CH3 | 1.97, s | 23, 24 |
Overlapping signals.
Smenamide D (2): colorless oil; [α]25D +16.8 (c 0.10, CH3OH); UV (CH3OH) λmax (log ε) 203 (3.4), 240 (3.3) nm; 1H NMR (800 MHz, CDCl3) and 13C NMR (200 MHz, CDCl3), see Table S3; HRESIMS m/z 467.2693 [M+H]+ (calcd for C25H40N2O4Cl, 467.2677) and m/z 489.2492 [M+Na]+ (calcd for C25H39N2O4ClNa, 489.2496).
Smenamide E (3): colorless oil; [α]25D +21.9 (c 0.05, CH3OH); UV (CH3OH) λmax (log ε) 203 (4.1), 235 (3.8) nm; 1H NMR (800 MHz, DMSO-d6) and 13C NMR (200 MHz, DMSO-d6), see Table 2; HRESIMS m/z 499.2935 [M+H]+ (calcd for C26H44N2O5Cl, 499.2939).
Table 2.
NMR data for Smenamide E Z/E-conformersa (3) (800 MHz for 1H NMR; 200 MHz for 13C NMR, DMSO).
| Position | δC, type | δH (J in Hz) | HMBC | COSY |
|---|---|---|---|---|
| 1 | 170.3, qC | |||
| 2 | 94.2, CH | 5.31, s | 1, 3, 4 | |
| 3 | 180.8, qC | |||
| 4 | 58.1, CH | 4.63, m | 1, 2, 3, 5, 6, 10 | 5 |
| 5 | 39.2, CH2 | 1.72, m | 3, 4, 6, 7, 8 | 4 |
| 6 | 24.0, CH | 1.76, m | 4, 5, 7, 8 | 7, 8 |
| 7 | 24.3, CH3 | 0.84, d | 5, 6, 8 | 6 |
| 8 | 22.9, CH3 | 0.86, d | 5, 6, 7 | 6 |
| 9 | 59.5, CH3 | 3.86, s | 3 | |
| 10 | 175.8, qC | |||
| 11 | 42.7, CH | 3.95, m | 10, 12, 13 | 12, 13 |
| 12 | 14.4, CH3 | 0.90, d (6.8) | 10, 11, 13 | 11 |
| 13 | 74.3, CH | 3.73, m | 10, 11, 15 | 11, OH-13 |
| OH-13 | 4.40, m | 11, 13, 14 | 13 | |
| 14 | 34.3, CH | 1.48, ovlpb | 13, 15 | 15 |
| 15 | 12.9, CH3 | 0.81, d (6.2) | 13, 14, 16 | 14 |
| 16a | 31.5, CH2 | 1.48, ovlp | 13, 14, 15, 17 | 16b, 17a |
| 16b | 1.26, m | 13, 14, 15, 17 | 16a, 17b | |
| 17a | 28.1, CH2 | 2.25, m | 16, 18, 19 | 16a, 16b |
| 17b | 2.13, m | 16, 18, 19 | 16a, 16b | |
| 18 | 143.0 [142.8], qC | |||
| 19 | 112.3 [112.6], CH | 6.04 [6.06], s | 17, 18, 20 | 20 |
| 20 | 31.6 [31.4], CH2 | 2.02 [2.07], t (7.5) | 18, 19, 21, 22 | 21 |
| 21 | 25.4 [26.5], CH2 | 1.55 [1.63], m | 20, 22 | 20, 22 |
| 22 | 46.9 [48.9], CH2 | 3.24 [3.23], m | 20, 21, 23, 24 | 21 |
| 23 | 35.1 [33.2], CH3 | 2.94 [2.79], s | 22, 24 | |
| 24 | 173.0 [172.7], qC | |||
| 25 | 26.3 [25.7], CH2 | 2.27 [2.29], m | 24, 26 | 26 |
| 26 | 9.7 [10.1], CH3 | 0.97 [0.98], t (7.4) | 24, 25 | 25 |
E-conformer NMR values of 3 in brackets.
Overlapping signals.
Smenothiazole A: colorless oil; [α]23D−5.8 (c 0.10, CH3OH) UV (CH3OH) λmax (log ε) 202 (3.4) nm; 1H NMR (800 MHz, DMSO-d6) and 13C NMR (200 MHz, DMSO-d6), see Table S4; HRESIMS m/z 486.1984 [M+H]+ (calcd for C26H33ClN3O2S, 486.1982).
Cytotoxicity assay
Neuro-2A cells and HCT-116 cells were added to assay plates in 100 μl of Eagle's Minimum Essential Media (EMEM) or 100 μl of McCoy's 5A media respectively each supplemented with 10% FBS at a density of 5,000 cells/well. Cells were incubated overnight (37°C, 5% CO2) and examined microscopically to confirm confluence and adherence. Mixed chromatography fractions (C-I) were dissolved in DMSO (1% v/v) and tested at concentrations of 40 and 4 μg/mL with 10 μM doxorubicin used as a positive control. Compounds 1–3 were dissolved in DMSO (1% v/v) and added to the cells in the range of 100 to 0.1 μM in order to generate EC50 curves. Four technical replicates were prepared for each concentration and each assay was performed in triplicate. Doxorubicin was used as a positive control (EC50: 0.043 ± 0.032 μM for neuro-2A cells; EC50: 0.071 ± 0.004 μM for HCT-116 cells) and DMSO (1% v/v) was used as a negative control. Assays were resolved as previously reported (Bertin et al., 2017b) and EC50 curves were generated using Graphpad Prism software.
Results
trichodesmium bloom-cytotoxicity of chromatography fractions and molecular network
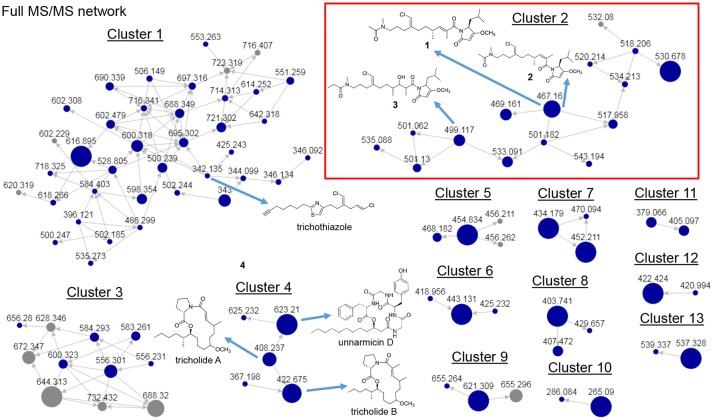
Several of the chromatography fractions (D–H) derived from the bloom material showed strong cytotoxicity against neuro-2A cells at 40 μg/mL (Figure S1). Fraction D showed the greatest potency at 4 μg/mL. Examination of the molecular network showed that compounds from cluster 3 were major ions in fraction D (Figure 2). However, these metabolites were not isolable following further purification procedures. The majority of the metabolites in the molecular network were found in fractions F–I. We identified Cluster 2 as a molecular cluster of interest due to the number of ions in the cluster and the M+ and M+2 ratio indicating a single chlorine atom in each of these metabolites (cf. Figures 2, 3). Additionally, fractions G and H showed potent cytotoxicity against neuro-2A cells; thus, our subsequent purification efforts centered on these two fractions. HPLC analysis indicated abundant metabolites in the combined G+H HPLC pre-fraction (Figure S2) and repeated chromatography resulted in the isolation of 1–3 as optically active colorless oils.
Figure 2.
Full MS/MS-based molecular network of Trichodesmium bloom. Previously identified molecules trichothiazole, tricholides A and B and unnarmicin D are noted. Cluster 2 (red box) shows new metabolites 1-3 (m/z 467.161, 467.161 and 499.117, respectively).
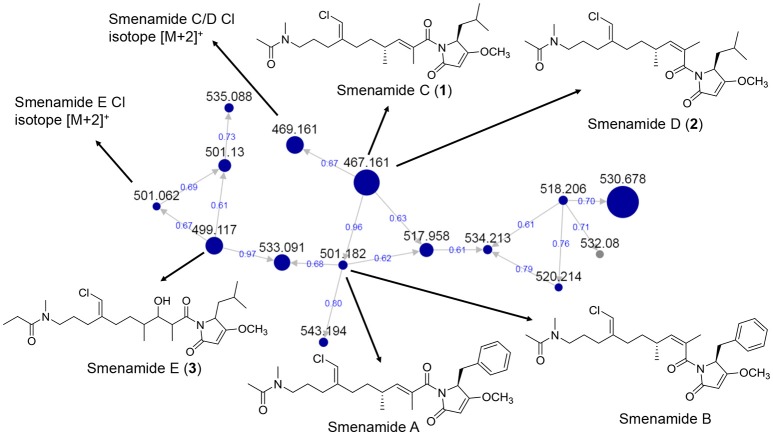
Figure 3.
MS/MS-based molecular networking cluster identifying 1–3 and smenamides A or B. Nodes are labeled with parent m/z values. Edges are labeled with cosine scores. Node size is relative to ion count.
Structure characterization of 1–3
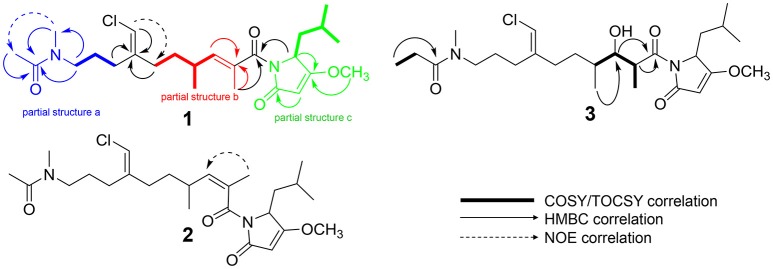
HRESIMS analysis of 1 gave an [M+H]+ of m/z 467.2661, suggesting a molecular formula of C25H39N2O4Cl and a requirement of 7 degrees of unsaturation. Examination of the 1H NMR spectrum of 1 showed several resonances with split signals in a 1:1 ratio, a phenomenon observed in several cyanobacteria metabolites with methylated tertiary amides such as smenamides A and B and kalkitoxin (Wu et al., 2000; Teta et al., 2013). The split signals were determined to be the result of two conformers in the E and Z configuration at the tertiary amide functionality in 1. This phenomenon was observed for all three new metabolites (1–3); in the structure characterization for each of these compounds, the data for the Z conformer is discussed. NMR data tables in the Supporting Information provide information on the E conformer. While the multiple conformers presented difficulties in NMR interpretation, three partial structures of 1 (a–c) were characterized initially based on 1H-1H COSY spin systems followed by HMBC correlation analysis (Figure 4). In the first partial structure (a), a moderately deshielded diastereotopic methylene group (H-20a, δH 2.12; H-20b, δH 2.07) was correlated by COSY to a second methylene group (H2-21, δH 1.56) which itself was correlated by COSY to a third methylene group (H2-22, δH 3.26). This latter deshielded methylene was correlated by HMBC to C-23 (δC 35.9) and the C-24 carbonyl (δC 169.9). The singlet methyl (H3-25, δH 1.97) showed an HMBC correlation to C-24 and characterized the western half of 1 with an N-methyl acetamide functionality. In the second partial structure of 1 (b), another moderately deshielded diastereotopic methylene group (H-17a, δH 2.22; H-17b, δH 2.10) showed COSY correlations to the H-16 methylene (H-16a, δH 1.49; H-16b, δH 1.35). H2-16 showed COSY correlations to the H-14 methine (δH 2.45), which itself showed COSY correlations to a doublet methyl (H3-15, δH 0.94) and olefinic proton (H-13, δH 5.59). A singlet methyl (H3-12, δH 1.77) and H-13 showed HMBC correlations to a quaternary carbon (C-11, δC 131.7) and the C-10 carbonyl (δC 170.6) extending the polyketide chain of 1. The C-11–C-13 olefin was assigned E geometry based on the 13C chemical shift of C-12 (δC 13.7) compared to δC 20.1 for the Z geometry (see below). The two sets of moderately deshielded methylenes (H2-20 and H2-17) showed HMBC correlations to the quaternary carbon at C-18 (δC 142.8). A deshielded methine singlet (H-19, δH 6.04) also showed an HMBC correlation to C-18, supporting an exomethylene vinyl chloride bridge connecting partial structures a and b. The configuration of the vinyl chloride was assigned as Z based on NOE correlations from H-19 to H2-17 and H2-16. The chemical shift of C-10 was consistent with that of an amide functionality and COSY correlations from H-4 to H-8 supported the assignment of a leucine residue in the third partial structure. However, the chemical shift at C-3 was somewhat deshielded for that of a standard amide or ester carbonyl (δC 180.3). An O-methyl singlet (H3-9, δH 3.87) was correlated to C-3 by HMBC supporting the presence of a methoxy functionality. Additionally, H-2 (δH 5.28) was correlated to C-3 and the C-1 carbonyl by HMBC. HMBC correlations from H-2 and H-4 to C-10 connected the third partial structure to the remainder of the molecule, establishing an isobutyl-methoxypyrrolinone moiety and satisfying the final three degrees of unsaturation required by the molecular formula. The structure of 1 was established as a highly functionalized linear polyketide-peptide of the smenamide family (Teta et al., 2013). While the correlations and chemical shifts described above relate to the Z conformer of 1, NMR data for the E conformer were also analyzed, and are listed in Table S1.
Figure 4.
Select 2D NMR correlations of compounds 1–3.
The absolute configuration of 1 (4S, 14R) was determined to be identical to that of smenamide A by comparison of the CD spectrum of 1 to that of naturally occurring smenamide A (Caso et al., 2017). The spectra were nearly identical in sign and magnitude.
HRESIMS analysis of 2 gave an [M+H]+ of m/z 467.2693, suggesting a molecular formula of C25H39N2O4Cl, identical to that of 1. Examination of 1H NMR, multiplicity-edited HSQC, and HMBC spectra of 1 and 2 showed that the two molecules were nearly identical (cf. Tables S2 and S3). 13C NMR differences were most pronounced at C-12 (δC 13.7 in 1; δC 20.1 in 2) and C-13 (δC 142.7 in 1; δC 135.9 in 2). These chemical shifts and the NOE correlations between H3-12 and H-13 in 2 supported the Z configuration of the C-11–C-13 olefin in 2 and established 2 as a geometric isomer of 1. The absolute stereochemistry of 2 is proposed to be identical to that of 1 based on similarity in optical rotation values.
HRESIMS analysis of 3 gave an [M+H]+ of m/z 499.2935, suggesting a molecular formula of C26H43N2O5Cl, and a requirement of 6 degrees of unsaturation. The examination of 1H and 13C NMR data and the placement of m/z 499 in the same molecular network cluster as smenamide C and D (1 and 2), suggested that 3 was a close structural analog. The reduction in degrees of unsaturation in 3 compared to 1 was due to the presence of a secondary alcohol at C-13 (H-13, δH 3.73; C-13, δC 74.3) in 3 and a methine at C-11 (H-11, δH 3.95; C-11, δC 42.7). The H-11 and H-13 methine protons were correlated by COSY and H-13 also showed a COSY correlation to H-14 (δH 1.48). Additionally, the C-25 methyl resonance of the acetyl group in 1 and 2 was not present in 3. COSY correlations between a methylene at H-25 (δH 2.27) and a methyl triplet at H-26 (δH 0.97) supported an N-methylpropanamide functionality in 3 and completed the planar structure of smenamide E (3). The secondary alcohol of 3 was resistant to acylation with Mosher's acid chloride and the configuration of this position could not be determined by chemical derivative formation. Therefore, in the current report, we report only the planar structure for this new metabolite.
During the attempt to isolate the compound with an m/z 530 from Cluster 2 in the network (Figure 3), we isolated a peptidic compound with spectrometric and spectroscopic characteristics consistent with that of the previously reported cytotoxin smenothiazole A (Esposito et al., 2015). Analysis of NMR data, optical rotation value, and CD spectra (negative Cotton effect at 234 nm, Figure S35) confirmed its identity.
Cytotoxicity of 1–3
Smenamides C and E (1 and 3) showed greater cytotoxicity to neuro-2A cells than to the human colon cancer cell line HCT-116. Smenamide E (3) showed the greatest potency to neuro-2A cells with an EC50 value of 4.8 ± 0.6 μM (EC50: 18.6 ± 1.8 μM against HCT-116 cells). Smenamide C (1) showed similar selective potency (EC50 neuro-2A: 7.2 ± 3.1 μM; EC50 HCT-116: 20.9 ± 2.1 μM). Interestingly, smenamide D (2), the geometric isomer of smenamide C (1), did not show cytotoxicity against either cell line.
Discussion
In our previous work on Trichodesmium blooms and their natural products, we have utilized cytotoxicity assays, NMR-guided isolation, and MS-guided isolation (Bertin et al., 2016, 2017a,b; Belisle et al., 2017). We have previously characterized the cytotoxic polyketide trichophycin A, the polyketides trichotoxins A and B, and the moderately cytotoxic polyketide-peptide trichothiazole (Bertin et al., 2016, 2017b; Belisle et al., 2017). In the current network, we did observe an m/z value consistent with trichothiazole (Figure 2). The node was in Cluster 1 (m/z 342) and showed an identical MS/MS fragmentation pattern to that of trichothiazole. However, we did not observe nodes for trichophycin A or trichotoxins A and B. It should be noted that the macrocyclic polyketide-peptides tricholides A and B and unnarmicin D (Bertin et al., 2017a) also clustered in the network (Figure 2, Cluster 4). It may be that some metabolites in the bloom metabolome do not ionize well by ESI+ or give informative fragments during MS/MS acquisition; this represents a limitation in the implementation of MS/MS-based networking to describe bloom metabolomes. Thus, the metabolite information gained in the network may be somewhat biased toward peptides and hybrid polyketide-peptides. Nevertheless, taking into account the limitations of this approach, molecular networking was a remarkable tool for visualizing a complex metabolome rich in metabolites with intriguing structural elements and cytotoxicity to neuro-2A cells. Analyzing fractions C-I using the networking procedure identified 93 nodes that were members of 13 clusters. This approach allowed us to isolate and characterize three new members of the smenamide family (1–3). Furthermore, within the smenamide cluster, we tentatively identified the known compound smenamide A or B (double bond isomers of each other at m/z 501). This later node in Cluster 2 (Figure 3) showed an identical MS/MS fragmentation pattern to that of smenamide A/B from published data and the HRESIMS analysis supported this identification (Figure S36) (Teta et al., 2013). Both of these known metabolites are very potent cytotoxins with IC50 values around 50 nM against Calu-1 cells (Teta et al., 2013). Smenamides C and E (1 and 3) were less potent cytotoxins than smenamide A and B, possibly due to the replacement of the phenylalanine amino acid unit with leucine (1, 3). Intriguingly, smenamide D (2) was not cytotoxic to either the neuro-2A and HCT-116 cell lines and we speculate that the cis configuration in the middle of the polyketide chain may affect binding of 2 to its molecular target. Smenothiazole A showed nanomolar levels against multiple cell lines and was previously isolated from a marine sponge; however, the authors indicate a likely cyanobacterial origin (Esposito et al., 2015). This is the first report of smenothiazole A from a bloom of Trichodesmium.
Overall, the networking procedure has identified new target molecules for isolation such as those in Cluster 1 (Figure 2). A comprehensive characterization of the chemical space within the bloom material is challenging as the metabolic composition of the sub-fractions we have generated are all nearly as complex as that of the sub-fraction from which 1–3 were isolated (Figure S2). The networking tool significantly improves the efficiency of our isolation and characterization workflow.
We did not identify anatoxins, saxitoxins or microcystins during the course of this analysis. This may be due to our focus on lipophilic metabolites, low abundance of these compounds in our samples, or a lack of informative MS/MS fragments. Trichodesmium blooms are complex events harboring a diverse array of microorganisms (Capone et al., 1997). Thus, the unequivocal identification of the producing organisms of the toxic metabolites described in this current work and other studies from environmental collections will ultimately require pure cultivation of producing organisms, the identification of biosynthetic gene clusters of toxic molecules, or the localization of metabolites to particular cell types (Simmons et al., 2008). In the original isolation and characterization of smenamides A and B, the authors suggest that a cyanobacterial symbiont is the true producer of the sponge-derived compounds (Teta et al., 2013). The present report supports this observation, as certain structural features such as the exomethylene vinyl chloride moiety are characteristic of cyanobacterial metabolism (Kan et al., 2000; Edwards et al., 2004; Nunnery et al., 2012). To the best of our knowledge, the N-methyl propanamide functionality in 3 has not previously been reported in a polyketide-peptide from cyanobacteria, and represents a biosynthetically intriguing unit because these organisms are not known to produce propionate. Conceivably, it may derive from an S-adenosylmethionine (SAM)-mediated methylation of an acetate precursor, the proposed first building block in the production of these smenamide-type natural products. This would be a similar biosynthetic transformation to that involved in producing the t-butyl group in apratoxin A which employs a combination of two SAM methyl transferases to incorporate these methyl groups (Grindberg et al., 2011; Skiba et al., 2017).
The identification of these neurotoxic metabolites (1 and 3) and the other more potent smenamides and smenothiazole A from a Trichodesmium bloom raises important questions as to their ecological role during these events. It will be important to characterize these bloom-associated metabolites in a longitudinal sense to evaluate their ongoing contribution to HABs.
Author contributions
MB was responsible for the study design. PZ and PM were involved in sample collection, organism identification and sample storage. CV and MB did the structure characterization of secondary metabolites. CV and SC carried out cytotoxicity studies. EG, WG, and MB did the molecular networking procedure. MB wrote the manuscript with editing help from all co-authors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Al Bach at the University of Rhode Island for assistance with certain spectroscopic measurements.
1ProteoWizardSoftware Foundation; http://proteowizard.sourceforge.net/tools.shtml
Funding. Acquisition of certain spectroscopic and spectrometric data in this publication was made possible by the use of equipment and services available through the RI-INBRE Centralized Research Core Facility at the University of Rhode Island, which is supported by the Institutional Development Award (IDeA) Network for Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103430. Certain NMR experiments were conducted at a research facility at the University of Rhode Island supported in part by the National Science Foundation EPSCoR Cooperative Agreement #EPS-1004057. We gratefully acknowledge the American Society of Pharmacognosy Starter Grant (MB).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2018.00316/full#supplementary-material
References
- Belisle R. S., Via C. W., Schock T. B., Villareal T. A., Zimba P. V., Beauchesne K. R., et al. (2017). Trichothiazole A, a dichlorinated polyketide containing an embedded thiazole isolated from Trichodesmium blooms. Tetrahedron Lett. 58, 4066–4068. 10.1016/j.tetlet.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J., Roduit A. F., Sun J., Alves G., Via C. W., Gonzalez M. A., et al. (2017a). Tricholides A and B and unnarmicin D: new hybrid PKS-NRPS macrocycles isolatedfrom an environmental collection of Trichodesmium thiebautii. Mar. Drugs 15:E206 10.3390/md15070206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J., Wahome P. G., Zimba P. V., He H., Moeller P. D. R. (2017b). Trichophycin A, a cytotoxic linear polyketide isolated from a Trichodesmium thiebautii bloom. Mar.Drugs 15:10. 10.3390/md15010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J., Zimba P. V., He H., Moeller P. D. R. (2016). Structure revision of trichotoxin, a chlorinated polyketide isolated from a Trichodesmium thiebautii bloom. Tetrahedron Lett. 57, 5864–5867. 10.1016/j.tetlet.2016.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bláha L., Babica P., Maršálek B. (2009). Toxins produced in cyanobacterial water blooms- toxicity and risks. Interdiscip. Toxicol. 2, 36–41. 10.2478/v10102-009-0006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone D. G., Zehr J. P., Paerl H. W., Bergman B., Carpenter E. J. (1997). Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229. 10.1126/science.276.5316.1221 [DOI] [Google Scholar]
- Carmichael W. W., Boyer G. L. (2016). Health impacts from cyanobacteria harmful algaeblooms: implications for the North American Great Lakes. Harmful Algae 54, 194–212. 10.1016/j.hal.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Caso A., Mangoni A., Piccialli G., Constantino V., Piccialli V. (2017). Studies toward thesynthesis of smenamide A, an antiproliferative metabolite from Smenospongia aurea:total synthesis of ent-smenamide A and 16-epi-smenamide A. ACS Omega 2, 1477–1488. 10.1021/acsomega.7b00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detoni A. M. S., Costa L. D. F., Pacheco L. A., Yunes J. S. (2016). Toxic Trichodesmium bloomoccurrence in the southwestern South Atlantic Ocean. Toxicon 110, 51–55. 10.1016/j.toxicon.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Edwards D. J., Marquez B. L., Nogle L. M., McPhail K., Goeger D. E., Roberts M. A, et al. (2004). Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptideneurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem. Biol. 11, 817–833. 10.1016/j.chembiol.2004.03.030 [DOI] [PubMed] [Google Scholar]
- Esposito G., Teta R., Miceli R., Ceccarelli L. S., Sala G. D., Camerlingo R., et al. (2015). Isolation and assessment of the in vitro anti-tumor activity of smenothiazole A and B, chlorinated thiazole-containing peptide/polyketides from the Caribbean sponge,Smenospongia aurea. Mar. Drugs 13, 444–459. 10.3390/md13010444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindberg R. V., Ishoey T., Brinza D., Esquenazi E., Coates R. C., Liu W. T., et al. (2011). Single cell genome amplification accelerates natural product pathway characterization from complex microbial assemblages. PLoS ONE 6:e18565 10.1371/journal.pone.0018565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y., Sakamoto B., Fujita T., Nagai H. (2000). New malyngamides from the Hawaiian cyanobacterium Lyngbya majuscula. J. Nat. Prod. 63, 1599–1602. 10.1021/np000250t [DOI] [PubMed] [Google Scholar]
- Komárek J., Anagnostidis K. (2005). Cyanoprokarota 19 Part 2: Oscillatoriales. Munchen: Elsevier. [Google Scholar]
- Luesch H., Yoshida W. Y., Moore R. E., Paul V. J., Corbett T. H. (2001). Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula. J. Am. Chem. Soc. 123, 5418–5423. 10.1021/ja010453j [DOI] [PubMed] [Google Scholar]
- Malloy K. L., Suyama T. L., Engene N., Debonsi H., Cao Z., Matainaho T., et al. (2012). Credneramides, A., and B: neuromodulatory phenethylamine and isopentylamine derivatives of a vinyl chloride-containing fatty acid from cf. Trichodesmium sp. nov. J. Nat. Prod. 75, 60–66. 10.1021/np200611f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnery J. K., Engene N., Byrum T., Cao Z., Jabba S. V., Pereira A. R., et al. (2012). Biosynthetically intriguing chlorinated lipophilic metabolites from geographically distant tropical marine cyanobacteria. J. Org. Chem. 77, 4198–4208. 10.1021/jo300160e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A. R., Kale A. J., Fenley A. T., Byrum T., Debonsi H. M., Gilson M. K., et al. (2012). The carmaphycins: new proteasomeinhibitors exhibiting an α,β-epoxyketone warhead from a marine cyanobacterium. Chembiochem. 13, 810–817. 10.1002/cbic.201200007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. G., Martel A., Codd G. A., Soler E., Coca J., Redondo A., et al. (2005). Bloom of the marine diazotrophic cyanobacterium Trichodesmium erythraeum in the northwest African upwelling. Mar Ecol. Prog. Ser. 301, 303–305. 10.3354/meps301303 [DOI] [Google Scholar]
- Shunmugam S., Gayathri M., Prasannabalaji N., Thajuddin N., Muralitharan G. (2017). Unraveling the presence of multi-class toxins from Trichodesmium bloom in the Gulf of Mannar region of the Bay of Bengal. Toxicon 135, 43–50. 10.1016/j.toxicon.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Simmons T. L., Coates R. C., Clark B. R., Engene N., Gonzalez D., Esquenazi E, et al. (2008). Biosynthetic origins of natural products isolatedfrom marine microorganism-invertebrate assemblages. Proc. Natl. Acad. Sci. U.S.A. 105, 4587–4594. 10.1073/pnas.0709851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiba M. A., Sikkema A. P., Moss N. A., Tran C. L., Sturgis R. M., Gerwick L., et al. (2017). Mononuclear iron-dependent methyltransferase catalyzes initial steps in assembly of the apratoxin A polyketide starter unit. ACS Chem. Biol. 12, 3039–3048. 10.1021/acschembio.7b00746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudek S., Haygood M. G., Youssef D. T. A., Schmidt E. W. (2006). Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl. Environ. Microbiol. 72, 4382–4387. 10.1128/AEM.00380-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taori K., Paul V. J., Luesch H. (2008). Structure and activity of largazole, a potentantiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J. Am.Chem. Soc. 130, 1806–1807. 10.1021/ja7110064 [DOI] [PubMed] [Google Scholar]
- Teta R., Irollo E., Sala G. D., Pirozzi G., Mangoni A., Costantino V. (2013). Smenamides, A., and B, chlorinated peptide/polyketide hybrids containing adolapyrrolidone unit from the Caribbean sponge Smenospongia aurea. Evaluation of theirrole as leads in antitumor drug research. Mar. Drugs 11, 4451–4463. 10.3390/md11114451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., et al. (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837. 10.1038/nbt.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J., Roach P., Alexandrov T., Heath B. S., Yang J. Y., Kersten R. D., et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U.S.A. 109, E1743–E1752. 10.1073/pnas.1203689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Okino T., Nogle L. M., Marquez B. L., Williamson R. T., Sitachitta N., et al. (2000). Structure, synthesis, and biological properties of kalkitoxin, a novel neurotoxin from themarine cyanobacterium Lyngbya majuscula. J. Am. Chem. Soc. 122, 12041–12042. 10.1021/ja005526y [DOI] [Google Scholar]
- Yang J. Y., Sanchez L. M., Rath C. M., Liu X., Boudreau P. D., Bruns N., et al. (2013). Molecular networking as a dereplication strategy. J. Nat. Prod. 76, 1686–1699. 10.1021/np400413s [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.