Abstract
Congenital hemangiomas are tumor-like vascular malformations with poorly understood pathogenesis. We report the case of a neonate with a massive congenital scalp hemangioma that required urgent neurosurgical removal on the second day of life because of concern for high-flow arteriovenous shunting. Exome sequencing identified a rare damaging de novo germline mutation in MYH9 (c.5308C>T, p.[Arg1770Cys]), encoding the MYH9 nonmuscle myosin IIA. MYH9 has a probability of loss-of-function intolerance (pLI) score of >0.99 and is highly intolerant to missense variation (z score = 5.59). The p.(Arg1770Cys) mutation substitutes an evolutionarily conserved amino acid in the protein's critical myosin tail domain and is predicted to be highly deleterious by SIFT, PolyPhen-2, MetaSVM, and CADD. MYH9 is a known regulator of cytokinesis, VEGF-regulated angiogenesis, and p53-dependent tumorigenesis. These findings reveal a novel association of germline de novo MYH9 mutation with congenital hemangioma.
Keywords: hemangioma
INTRODUCTION
According to the International Society for the Study of Vascular Anomalies (ISSVA), congenital hemagiomas (CHs) belong to a subset of benign vascular tumors fully developed at birth, frequently symptomatic, and GLUT1-negative (Dasgupta and Fishman 2014). Most cases of CH affect the head and neck, followed by trunk and extremities (Dasgupta and Fishman 2014). Vascular anomalies such as CHs are treated with a variety of approaches including medical management, percutaneous sclerotherapy, chemotherapy, endovascular embolization, and surgical excision (Sadick et al. 2017). Targeted treatments for CHs are limited because of an incomplete understanding of disease pathogenesis, which is complicated by genetic and phenotypic heterogeneity.
Association with several well-described hereditary syndromes such as hereditary hemorrhagic telangiectasia (HHT) (OMIM# 187300) and CLOVE syndrome (OMIM# 612918) provides evidence of genetic determinants of CH pathogenesis. More recently, activating somatic mutations in the G-protein subunit-coding genes GNAQ and GNA11/14 have been detected in CHs by whole-exome sequencing (WES) and targeted sequencing (Funk et al. 2016; Lim et al. 2016). These findings have implicated MAPK/YAP signaling in the pathogenesis of CHs. Germline mutations in SMAD4 and VHL, among others, have been described in association with vascular anomalies in HHT, von Hippel–Lindau, and other syndromes, implicating germline mutations in the pathogenesis of congenital vascular anomalies (Glasker et al. 2013; Jelsig et al. 2016).
Herein, we report the case of a neonate that underwent neurosurgical excision of a large congenital scalp hemangioma. Exome sequencing of the patient's germline and hemangioma DNA and of parental germline DNA was performed to identify genomic variants.
RESULTS
Clinical Presentation
The patient is a female with a birth weight of 3540 g, born to a 31-yr-old G2P0010 mother at 39 wks 4 d of gestation via an elective Cesarean section. Prenatal ultrasound at 38 wks revealed a highly vascular mass anterior and superior to the left ear measuring 5.3 × 2.7 × 4.5 cm, along with mild cardiomegaly. These findings raised a concern for high blood flow within the lesion and the corresponding high rupture risk at delivery. Prenatal serologies for VDRL/RPR, HIV, rubella, hepatitis B, GBS, chlamydia, and GC were negative.
At the time of Cesarean section, the patient was neurologically intact, with APGAR score of 8 at 1 min and 9 at 5 min after delivery. She exhibited mild acrocyanosis but maintained oxygen saturations of >94% on room air. A 5 × 5 × 4-cm, soft, compressible, subcutaneous lesion was noted in the left temporal area anterior superior to the left external auditory canal, with moderate distortion of left ear architecture but patent ear canal. Transthoracic echocardiogram performed on patient's first day of life by pediatric cardiology demonstrated a small patent foramen ovale and patent ductus arteriosus with left-to-right flow and a holodiastolic flow reversal in the abdominal aorta. Dermatology formally evaluated patient and initially found no additional skin or mucosal abnormalities; no evidence of Kasabach–Merritt phenomenon (thrombocytopenia, consumptive coagulopathy with elevated D dimer, and low fibrinogen) was noted.
Initial bloodwork showed no abnormalities or evidence of consumptive coagulopathy, with normal hemoglobin, hematocrit, and platelet counts, mild elevation in PT/INR (12.3/1.21), reticulocyte counts of 5.15%, and direct/total bilirubin of 0.3/4.1 mg/dl not requiring phototherapy. Magnetic resonance (MR) imaging and MR angiography of the head showed a large left temporal soft tissue extradural lesion with avid enhancement, prominent extracranial arterial supply, and large arterial feeders from the superficial temporal and the occipital arteries concerning a high-flow lesion (Fig. 1). Radiographic differential diagnoses included a hemangioma, an arteriovenous malformation, or an arteriovenous fistula. Hemodynamic irregularity and a notable focus of plum red discoloration over the vertex of the lesion further supported concerns for a high-flow vascular lesion.
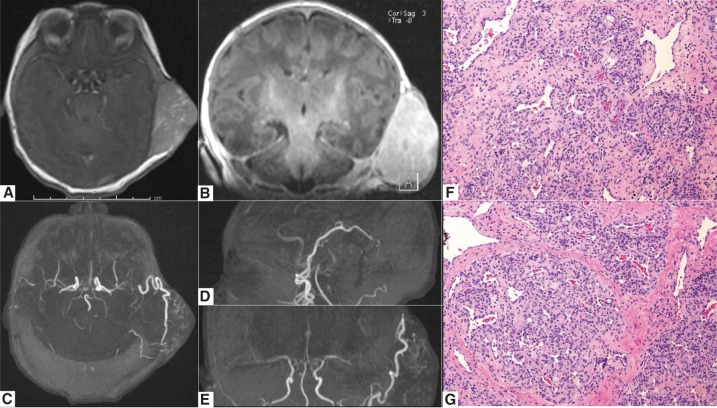
Figure 1.
Preoperative MRI/MRA imaging and tissue histology of a patient with a large congenital scalp hemangioma. (A,B) MR imaging of the brain with and without gadolinium, demonstrating a large extracranial contrast enhancing lesion in the left temporal area; axial and coronal T1-weighted post-gadolinium MR images shown; (C–E) axial, coronal, and sagittal reconstructions of the 3T time-of-flight MR angiogram images, demonstrating the arterial supply of patient's congenital scalp hemangioma arising from superior temporal and occipital arteries; (F,G) 20× images of H&E stains of congenital hemangioma showing dense fibrous stroma with large irregular vessels.
Given the size of the lesion and the potential for high cardiac failure, the patient was taken to the operating room on the second day of life for neurosurgical resection of the left temporal lesion, including ligation of the feeding superficial temporal artery and posterior auricular arteries. Intra-operative Doppler ultrasound confirmed a high-flow vascular lesion. The mass was superficial to the temporalis fascia and the underlying skull was intact. Frozen pathology was consistent with hemangioma, with no evidence of dysplastic or atypical features. Postoperative MR angiography/venography showed complete resection of the lesion. Permanent histologic analysis revealed lobular capillary proliferation within dense fibrous stroma with large irregular vessels. Immunohistochemical staining for Glut-1 staining was negative. These findings were consistent with congenital hemangioma (Fig. 1).
The patient had an uneventful postoperative clinical course and was discharged on postoperative day 5. Prior to discharge, the patient successfully passed a formal hearing screening with auditory brainstem responses in both right and left ears. Metabolic screen and cystic fibrosis screen were negative. No evidence of cataracts or nephropathy was noted during hospital stay or any of the postdischarge visits. Sutures were removed on postoperative day 21 with excellent wound healing. Postdischarge evaluation by a neonatologist noted few small hyperpigmented macules on the left calf and Mongolian spots on the buttocks. Evaluation by orthopedic surgery indicated no developmental pathology; however, the patient's lower extremities were mildly externally rotated. MR imaging of the brain at 4-mo follow-up showed no evidence of congenital hemangioma recurrence. The patient continues to meet all developmental milestones as expected, with her most recent visit in our clinic at 6 mo unremarkable.
Genomic Analyses
Whole-exome sequencing (WES) of the patient's lesion and germline DNA was performed as described in the Methods section. No confirmed pathogenic somatic variants with allele frequencies of >10% were identified; the list of mutations selected by our workflow, filter, and detection criteria is included in Supplemental Table 1. None of the previously described mutations identified as causal in patients with congenital hemangiomas and other vascular malformations were detected in our patient sample.
WES on patient–parent trio DNA was also performed as described; the identify-by-descent kinship analysis confirmed parent–offspring relationship in the trio, and principal component analysis suggested African–American descent (Supplemental Figure 1; Timberlake et al. 2016). A full 98.90% of targeted bases had eight or more independent reads, and 98.30% had 15 or more. The Genome Analysis Toolkit (GATK) Haplotype Caller was used to call variants (Van der Auwera et al. 2013). Allele frequencies were annotated in the Exome Aggregation Consortium (ExAC) and GnomAD databases (Lek et al. 2016). TrioDeNovo was used to identify de novo mutations (Wei et al. 2015). MetaSVM was used to infer the impact of missense mutations (Dong et al. 2015). Sanger sequencing of PCR amplicons containing the mutation verified mutations in genes of interest. A list of autosomal and X-linked recessive mutations, including recessive, missense, and loss of function mutations, and damaging inherited heterozygous variants are listed in Supplemental Tables 2,3. Whole-exome sequencing coverage data is included in Supplemental Table 4.
A rare damaging de novo germline mutation in MYH9 c.5308C>T, p.[Arg1770Cys], encoding the MYH9 nonmuscle myosin IIA, was identified (Table 1; Fig. 2). MYH9 has a probability of loss-of-function intolerance (pLI) score of >0.99 and is highly intolerant to missense variation (z score = 5.59). This mutation is rare; it is not reported in the ExAC database and is present in GnomAD database at a MAF of 4.08 × 10−6. The p.(Arg1770Cys) mutation substitutes an evolutionarily conserved amino acid in the protein's critical myosin tail domain and is predicted to be highly deleterious by SIFT, PolyPhen-2, MetaSVM, and CADD (Table 1). MYH9 c.5308C>T, p.[Arg1770Cys] mutation was confirmed by direct Sanger sequencing on patient and parent DNA, as shown in Figure 2. Although there are several 3D structures in PDB that share high homology with MYH9, the region around residue 1770 has not been solved; therefore there is no structural template to carry out in silico modeling (S Haider, pers. comm.).
Table 1.
Variant described in this study
| Gene | Chr-pos-ref-alt | Genotype | TranscriptID | HGVS protein | HGVS cDNA | Variant type | PolyPhen | MetaSVM | CADD | dbSNP | ExAC | pLI | mis_Z |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYH9 | 22-36681342-G-A | Heterozygous | NM_002473 | p.Arg1770Cys | c.5308C>T | missense | Damaging | Deleterious | 35 | N/A | N/A | 1 | 5.59 |
HGVS, Human Genome Variation Society; CADD, Combined Annotation Dependent Depletion; N/A, not available.
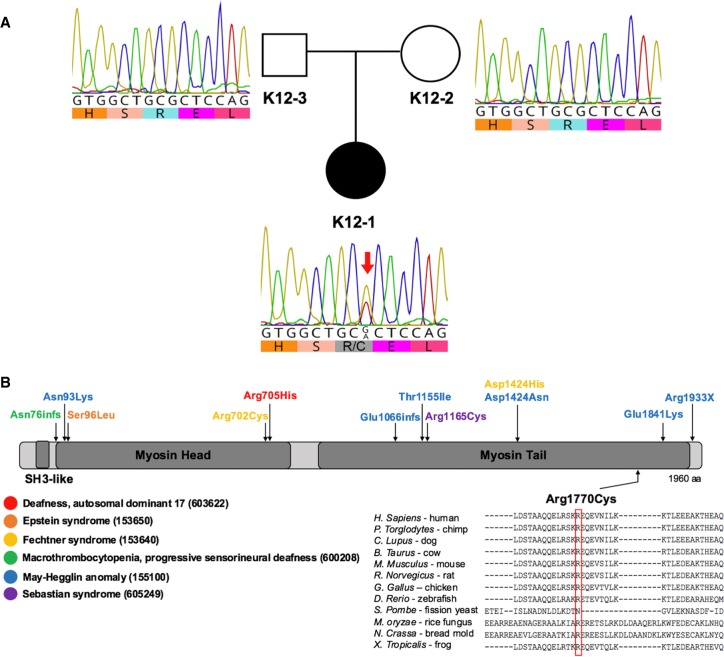
Figure 2.
Identification of MYH9 de novo damaging missense germline mutation in a patient harboring large congenital scalp hemangioma. (A) Pedigree structure and the Sanger chromatograms demonstrating the MYH9 c.5308C>T, p.[Arg1770Cys] mutation. The black circle represents the affected patient (K12-1); the unfilled square and circle represent patient's father (K12-3) and mother (K12-2), respectively, who do not have this phenotype. (B) Schematic representation of MYH9 protein structure and previously reported mutations affecting MYH9. Note the diversity of syndromes associated with the MYH9 mutations. Arrows depict the approximate location of previously described amino acid substitutions resulting from mutations in the MYH9. Each protein modification is color-coded and correlates with a key of all MYH9-associated syndromes. Dark rectangles represent functional domains. p.[Arg1770Cys] is depicted below the schematic; the amino acid sequence alignment for 12 different species is represented below, demonstrating phylogenetic conservation of Arg1770.
DISCUSSION
Here we report the first known damaging de novo missense mutation in MYH9 in a neonate presenting with a massive congenital scalp hemangioma. The relative intolerance of MYH9 to mutation, the evolutionary conservation and predicted deleteriousness of the involved residue in a critical domain of protein function, and the known role of MYH9 in cytokinesis, VEGF-regulated angiogenesis, and p53-dependent tumorigenesis support the pathogenicity of this mutation in congenital hemangioma formation.
MYH9 is located at 22q12.3, contains 41 exons, and encodes the 230-kDa conventional nonmuscle Myosin IIA (MYH9), shown to be important in phagocytosis, growth cone development, cytokinesis, cell shape maintenance, and cell adhesion (Berg et al. 2001). MYH9 is >70% identical to the heavy chain of smooth muscle myosin, containing an ATP-hydrolyzing catalytic motor domain that binds actin (exons 2–19) and a neck domain responsible for force transfer and binding to myosin α-helical light chains (exon 20). It also contains a coiled-coil tail domain critical in dimerization and myosin filament assembly (exons 21–40) and a regulatory phosphorylation domain (exon 41) (Sellers 2000). MYH9 is expressed in endothelial cells, macrophages, and fibroblasts in a variety of organs including lung, liver, spleen, kidney, and inner ear (Sellers 2000).
Single-nucleotide variants resulting in amino acid changes can have multiple detrimental effects by affecting the protein stability, cellular localization, cell signaling, or cell–cell interactions. Arg to Cys substitutions specifically can result in increased disulfide bonding between related proteins (Malfait et al. 2007). MYH9 c.5308C>T; p.[Arg1770Cys] mutation is located in the tail domain of myosin that forms the rod required for forming myosin minifilaments, which differ from thick filaments in the muscle. It is interesting to speculate that MYH9 c.5308C>T; p.[Arg1770Cys] mutation could result in dysfunctional unstable myosin monomers preventing proper assembly of minifilaments, with possibly dominant negative effects of mutant MYH9 explaining its pathogenicity in a heterozygous state. In vitro studies examined the effects of R1165C, D1424N, E1841K, and R1933Stop MYH9 mutations using electron microscopy and found that although wild-type tail fragments formed ordered arrays, mutants formed aberrant aggregates and acted in a dominant negative fashion with a wild-type protein (Franke et al. 2005). By the virtue of its location, MYH9 c.5308C>T; p.[Arg1770Cys] mutation could also affect the interactions between myosin tail and myosin head motor domains.
Germline mutations in MYH9 are associated with several dominantly inherited giant-platelet disorders characterized by thrombocytopenia and the presence of “Dohle bodies” (leukocyte inclusions), namely May–Hegglin anomaly, Sebastian syndrome, and Fechtner syndrome (Savioa and Pecci 2008). In addition to these features, Fechtner syndrome is characterized by cataracts, nephritis, and sensorineural deafness, similar to Alport syndrome (Savioa and Pecci 2008). Six MYH9 mutations were identified by the May–Hegglin/Fechtner Syndrome Consortium 2000; similar to our mutation, four of these occurred in the highly charged myosin tail domain essential for myosin filament assembly and were predicted to result in conformational changes and alterations in electrostatic properties of the MYH9 tail. Kim et al. reported several pathogenic MYH9 variants (c.3909C>A, p.[Phe1303Leu]; c.5188C>T, p.[Arg1730Cys]; c.5353C>T, p.[Arg1785Cys]) in patients with autosomal dominant nonsyndromic deafness (Kim et al. 2016), which may be associated with MYH9 tail domain mutations. Our patient's hearing and vision did not have any notable gross deficits, at least by 6-mo follow-up.
Recently, using insertional mutagenesis, MYH9 has been identified as a driver of novel oncogenic pathways in breast carcinoma (Kas et al. 2017). RNA-inhibition studies and mouse knockouts suggest MYH9 may induce formation of invasive squamous cell carcinomas, possibly by regulation of posttranscriptional stabilization of p53 (Schramek et al. 2014). Patients with malignant mesotheliomas were found to have several novel mutations associated with an integrin-linked kinase pathway including those in MYH9 (p.[Gln738*]; p.[Lys737Asn]; p.[Ala1197fs]) (De Rienzo et al. 2016). Moreover, higher expression levels of MYH9 in nonepithelioid tumors were associated with significantly worse survival (De Rienzo et al. 2016). Curiously, MYH9 was shown to interact with CXCR4 by immunoprecipitation, mediating nuclear translocation of CXCR4 and resulting in metastatic behavior of renal cell carcinoma via its effects on cell migration and invasion (Xu et al. 2016). Previous studies also support an active role of MYH9 in angiogenesis; it serves as a physical linker between the cytoskeleton and nucleolin, mediating translocation of nucleolin during VEGF-directed angiogenesis (Huang et al. 2006).
Given these compelling observations as well as MYH9’s association with congenital hemangioma pathogenesis, the effects of MYH9’s c.5308C>T; p.[Arg1770Cys] mutation on MYH9 protein structure and function merit further investigation in experimental models.
METHODS
This study was approved by Yale University's Human Investigations Committee and Human Research Protection Program. Written informed consent was obtained from patient's parents prior to sample collection. Tissue specimens were examined microscopically by a board-certified neuropathologist. Immunohistochemical staining was used for classification of the lesion as previously described per ISSVA criteria.
Kinship Analysis
Relationship between patient and parents was estimated using the pairwise identity-by-descent (IBD) calculation in PLINK (Purcell et al. 2007).
Principal Component Analysis
The EIGENSTRAT software was used to determine the ethnicity of each sample analysis as previously described (Lemaire et al. 2013). HapMap subjects were included in the analysis as reference panels.
Exome Sequencing and Data Analysis
Genomic DNA from tissue and blood was isolated using standard phenol–chloroform techniques and sequenced at the Yale Center for Genome Analysis (YCGA) per the Center's standard protocol. Coding sequences for each sample were captured using the xGEN Exome Research Panel v1.0 kit (Integrated DNA Technologies), followed by Illumina whole-exome sequencing, as previously described (Duran et al. 2016). Sequence reads were mapped to the reference genome (GRCh37/hg19) with BWA-MEM (version 0.7.10) (PMID 24974202) and further processed using the GATK Best Practices workflows (Van der Auwera et al. 2013), including duplication marking, insertions and deletions (indel) realignment, and base quality recalibration. Mean coverage of 95.6× and 191.5× was achieved for blood and tissue samples, respectively. Single-nucleotide variants (SNVs) and small indels were called with GATK HaplotypeCaller (PMIDs 20644199; 25431634). Variant annotation was performed using ANNOVAR (PMID 20601685), dSNP (v138), 1000 Genomes (August 2015) (Genomes Project et al. 2015), NHLBI Exome Variant Server (EVS), and ExAC (v3) (PMID 27535533).
Somatic SNVs and somatic indels were identified using Mutect (version 2.7.1) and Indelocator (version 36.3), respectively. Raw somatic SNVs were further filtered per GATK-Best Practice parameters. Called somatic variants were annotated using ANNOVAR (version 2015-12-14) (PMID 20601685) and VEP (version 83). Further filtering was performed based on the variant allele frequency in hemangioma or normal tissue and compared to the frequency of the variants in control databases such as ExAC, 1000 Genomes, and NHLBI (<1%). Copy-number variations (CNVs) were calculated using the ratio of hemangioma tissue coverage over normal coverage, normalized by total coverage variation, followed by segmentations performed with DNACopy package in R program. The MetaSVM algorithm (PMID 25552646) was used to predict deleteriousness of missense variants (“D-Mis,” or damaging missense) using software defaults.
De Novo Mutation Detection
De novo mutations were identified using the TrioDenovo (Wei et al. 2015) program as described previously (Duran et al. 2016). De novo calls were filtered based on the following hard filters: (1) minor allele frequency (MAF) ≤ 5 × 10−3 in ExAC; (2) pass GATK Variant Score Quality Recalibration (VSQR); (3) minimum 10 reads total, five alternate allele reads and a 20% alternate allele ratio in patient; (4) minimum depth of 10 reference reads and alternate allele ratio <3% in parents; (5) exonic or canonical splice-site variants; and (6) data quality score of ≥7 (suggested cutoff by authors of TrioDenovo). Read lengths were 74–99 bp; average 70.7 reads were obtained per sample, with median coverage of 70.3× and mean coverage of 74.2× at each base. Finally, in silico visualization was performed to remove false positives using Integrative Genomics Viewer11 (IGV) and BLAT search.
Data Confirmation
Candidate variant calls were confirmed by PCR amplification, utilizing custom primers (available upon request) followed by Sanger sequencing. Sanger read alignment and visualization was performed using Geneious R10 (Biomatters).
ADDITIONAL INFORMATION
Data Deposition and Access
The variant described in this paper has been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV000778433. Raw sequencing data were not deposited in the absence of patient consent but may be available by contacting the authors.
Ethics Statement
This study was approved by Yale University's Human Investigations Committee and Human Research Protection Program. Written informed consent was obtained from patient's parents prior to sample collection.
Funding
This study was supported by an initiative of the Office of Rare Diseases Research (RDCRN) and National Center for Advancing Translational Sciences (NCATS); neither of these organizations had a role in study design, data collection, interpretation, or analysis. K.T.K. is supported by the National Institutes of Health (NIH) Center for Mendelian Genomics.
Acknowledgments
We thank our patient and family for their agreement to participate in our study. We thank Yale Center for Genome Analysis for performing whole-exome sequencing.
Competing Interest Statement
The authors have declared no competing interest.
Author Contributions
E.I.F. and D.D. contributed to experimental design and data analysis, created figures, and wrote the manuscript; E.I.F. provided clinical information on the patient. S.C.J. and W.D. performed germline mutation analysis, E.Z.E.-O. performed somatic mutation analysis. P.A. assisted with manuscript revisions, submission, and adding patient data to ClinVar. A.A. and J.R.G. performed PCRs to validate mutations identified. A.H. assisted with analysis of pathologic specimens. E.I.F. and K.T.K. were a part of surgical team. K.T.K. guided experimental design and data analysis and performed critical review of the manuscript.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Berg JS, Powell BC, Cheney RE. 2001. A millennial myosin census. Mol Biol Cell 12: 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R, Fishman SJ. 2014. ISSVA classification. Semin Pediatr Surg 23: 158–161. [DOI] [PubMed] [Google Scholar]
- De Rienzo A, Archer MA, Yeap BY, Dao N, Sciaranghella D, Sideris AC, Zheng Y, Holman AG, Wang YE, Dal Cin PS, et al. 2016. Gender-specific molecular and clinical features underlie malignant pleural mesothelioma. Cancer Res 76: 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X. 2015. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 24: 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran D, Jin SC, DeSpenza T Jr, Nelson-Williams C, Cogal AG, Abrash EW, Harris PC, Lieske JC, Shimshak SJ, Mane S, et al. 2016. Digenic mutations of human OCRL paralogs in Dent's disease type 2 associated with Chiari I malformation. Hum Genome Var 3: 16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke JD, Dong F, Rickoll WL, Kelley MJ, Kiehart DP. 2005. Rod mutations associated with MYH9-related disorders disrupt nonmuscle myosin-IIA assembly. Blood 105: 161–169. [DOI] [PubMed] [Google Scholar]
- Funk T, Lim Y, Kulungowski AM, Prok L, Crombleholme TM, Choate K, Bruckner AL. 2016. Symptomatic congenital hemangioma and congenital hemangiomatosis associated with a somatic activating mutation in GNA11. JAMA Dermatol 152: 1015–1020. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasker S, Kruger MT, Klingler JH, Wlodarski M, Klompen J, Schatlo B, Hippchen B, Neumann HP, Van Velthoven V. 2013. Hemangioblastomas and neurogenic polyglobulia. Neurosurgery 72: 930–935; discussion 935. [DOI] [PubMed] [Google Scholar]
- Huang Y, Shi H, Zhou H, Song X, Yuan S, Luo Y. 2006. The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin. Blood 107: 3564–3571. [DOI] [PubMed] [Google Scholar]
- Jelsig AM, Torring PM, Kjeldsen AD, Qvist N, Bojesen A, Jensen UB, Andersen MK, Gerdes AM, Brusgaard K, Ousager LB. 2016. JP-HHT phenotype in Danish patients with SMAD4 mutations. Clin Genet 90: 55–62. [DOI] [PubMed] [Google Scholar]
- Kas SM, de Ruiter JR, Schipper K, Annunziato S, Schut E, Klarenbeek S, Drenth AP, van der Burg E, Klijn C, Ten Hoeve JJ, et al. 2017. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet 49: 1219–1230. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Lee S, Park HJ, Kang TH, Sagong B, Baek JI, Oh SK, Choi JY, Lee KY, Kim UK. 2016. Genetic association of MYH genes with hereditary hearing loss in Korea. Gene 591: 177–182. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire M, Fremeaux-Bacchi V, Schaefer F, Choi M, Tang WH, Le Quintrec M, Fakhouri F, Taque S, Nobili F, Martinez F, et al. 2013. Recessive mutations in DGKE cause atypical hemolytic-uremic syndrome. Nat Genet 45: 531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YH, Bacchiocchi A, Qiu J, Straub R, Bruckner A, Bercovitch L, Narayan D, Yale Center for Mendelian Genomics, McNiff J, Ko C, et al. 2016. GNA14 somatic mutation causes congenital and sporadic vascular tumors by MAPK activation. Am J Hum Genet 99: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfait F, Symoens S, De Backer J, Hermanns-Lê T, Sakalihasan N, Lapière CM, Coucke P, De Paepe A. 2007. Three arginine to cysteine substitutions in the pro-α (I)-collagen chain cause Ehlers–Danlos syndrome with a propensity to arterial rupture in early adulthood. Hum Mutat 28: 387–395. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadick M, Wohlgemuth WA, Huelse R, Lange B, Henzler T, Schoenberg SO, Sadick H. 2017. Interdisciplinary management of head and neck vascular anomalies: clinical presentation, diagnostic findings and minimalinvasive therapies. Eur J Radiol Open 4: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savioa A, Pecci A. 2008. MYH9-related disorders. In GeneReviews. University of Washington, Seattle, WA. [Google Scholar]
- Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. 2014. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343: 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. 2000. Myosins: a diverse superfamily. Biochim Biophys Acta 1496: 3–22. [DOI] [PubMed] [Google Scholar]
- Timberlake AT, Choi J, Zaidi S, Lu Q, Nelson-Williams C, Brooks ED, Bilguvar K, Tikhonova I, Mane S, Yang JF, et al. 2016. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. Elife 5: e20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11 10 11–11 10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Zhan X, Zhong X, Liu Y, Han Y, Chen W, Li B. 2015. A Bayesian framework for de novo mutation calling in parents–offspring trios. Bioinformatics 31: 1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Li P, Wei D, Wang Z, Bao Y, Sun J, Qu L, Wang L. 2016. NMMHC-IIA-dependent nuclear location of CXCR4 promotes migration and invasion in renal cell carcinoma. Oncol Rep 36: 2681–2688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.