Abstract
Autosomal recessive Stargardt disease (STGD1, MIM 248200) is caused by mutations in the ABCA4 gene. Complete sequencing of the ABCA4 locus in STGD1 patients identifies two expected disease-causing alleles in ∼75% of patients and only one mutation in ∼15% of patients. Recently, many possibly pathogenic variants in deep intronic sequences of ABCA4 have been identified in the latter group. We extended our analyses of deep intronic ABCA4 variants and determined that one of these, c.4253+43G>A (rs61754045), is present in 29/1155 (2.6%) of STGD1 patients. The variant is found at statistically significantly higher frequency in patients with only one pathogenic ABCA4 allele, 23/160 (14.38%), MAF = 0.072, compared to MAF = 0.013 in all STGD1 cases and MAF = 0.006 in the matching general population (P < 1 × 10−7). The variant, which is not predicted to have any effect on splicing, is the first reported intronic “extremely hypomorphic allele” in the ABCA4 locus; that is, it is pathogenic only when in trans with a loss-of-function ABCA4 allele. It results in a distinct clinical phenotype characterized by late onset of symptoms and foveal sparing. In ∼70% of cases the variant was allelic with the c.6006-609T>A (rs575968112) variant, which was deemed nonpathogenic. Another rare deep intronic variant, c.5196+1056A>G (rs886044749), found in 5/834 (0.6%) of STGD1 cases is, conversely, a severe allele. This study determines pathogenicity for three noncoding variants in STGD1 patients of European descent accounting for ∼3% of the disease. Defining disease-associated alleles in the noncoding sequences of the ABCA4 locus can be accomplished by integrated clinical and genetic analyses.
Keywords: macular dystrophy, macular flecks, progressive retinal degeneration, retinal flecks
INTRODUCTION
Mutations in the ABCA4 gene (STGD1, MIM #248200) (Allikmets et al. 1997) are the most frequent cause of Mendelian inherited recessive retinal dystrophies. Phenotypes of ABCA4 disease range from late-onset mild cases of central atrophy to very early-onset, RP-like, panretinal degeneration (Cremers et al. 1998; Martinez-Mir et al. 1998; Maugeri et al. 2000; Shroyer et al. 2001a; Lee et al. 2016; Ciccone et al. 2017; Tanaka et al. 2018). Phenotypic variability is caused by extensive ABCA4 disease-associated genetic variation, where more than 1000 definitely or possibly disease-causing variants have been determined in the coding sequences and splice sites of the ABCA4 gene (Cornelis et al. 2017) (J Zernant, and R Allikmets, unpubl. data). Recently, the understanding of disease-causing genetic variation in ABCA4 has substantially improved as a result of two major advances. First, many noncoding disease-associated ABCA4 alleles have been identified and proven pathogenic, mostly by affecting splicing (Zernant et al. 2011; Zernant et al. 2014; Bauwens et al. 2015; Bax et al. 2015; Sangermano et al. 2016, 2018; Albert et al. 2018). Second, we recently determined that some ABCA4 variants, which had been considered benign because of high minor allele frequency (MAF) in the general population, are in fact very mild conditional alleles, which result in disease expression only when in trans with a deleterious mutation, thereby called “extreme hypomorphs” (Zernant et al. 2017). Altogether, although these findings have increased the fraction of definitively diagnosed cases of ABCA4 disease substantially, 10%–15% of all STGD1 cases remain with only one proven disease-causing mutation.
It has been also demonstrated that large copy-number variants (CNVs) that elude detection by sequencing are exceptionally rare in the ABCA4 locus (Lee et al. 2016). Whole-exome sequencing (WES) of patients with phenotypes compatible with STGD1, but in which no ABCA4 mutations had been identified after complete sequencing of the ABCA4 locus (i.e., phenocopies), has uncovered causal genes in ∼70% of cases, including known retinal disease–associated genes (CRB1, CRX, etc.) (Tsang et al. 2014; Yamamoto et al. 2014) and new genes (RAB28, RDH11) (Xie et al. 2014; Riveiro-Alvarez et al. 2015). Therefore, the remaining “missing” alleles in monoallelic cases likely reside in the noncoding sequences of ABCA4 and are waiting to be assigned pathogenicity. In this study, we investigated three deep intronic variants (Table 1) for pathogenicity by an integrated analysis of clinical and genetic data in a large, familial cohort of patients with STGD1.
Table 1.
ABCA4 deep intronic variants investigated in this study
| Gene | Chromosome | Variant | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect on splicing | dbSNP | Genotype |
|---|---|---|---|---|---|---|---|---|
| ABCA4 | 1:94496509 | c.4253+43G>A | G | Noncoding | Substitution | None | rs61754045 | 100% heterozygous |
| ABCA4 | 1:94484082 | c.5196+1056A>G | A | Noncoding | Substitution | Activates cryptic donor site | rs886044749 | 100% heterozygous |
| ABCA4 | 1:94471747 | c.6006-609T>A | T | Noncoding | Substitution | None | rs575968112 | 100% heterozygous |
RESULTS
Determining Pathogenicity of Three Noncoding ABCA4 Variants
We had previously identified and described an intronic variant, c.4253+43G>A (rs61754045) in intron 28 of the ABCA4 gene (Zernant et al. 2011). At that time we noted that the variant was statistically significantly (P = 0.0003) more often found in STGD1 patients and also two times more frequently in cases with age-related macular degeneration than in matched general population controls (individuals >60 yr of age with no retinal pathology) (Zernant et al. 2011). We had suggested that the c.4253+43G>A variant was a good candidate for a disease-associated allele, but we were not able to make an unequivocal call of pathogenicity.
According to the updated data, the variant is present in 29/1155 STGD1 cases, almost exclusively in those with only one definite ABCA4 mutation (23/160, 14.38%; MAF = 0.072; Table 2), compared to MAF = 0.006 in non-Finnish European control cohort (gnomAD; Table 3). It was also present in 0.41% (3/725) of biallelic patients and in 1.1% (3/270) of cases with no other disease-causing ABCA4 alleles (Table 2). Therefore, the c.4253+43G>A variant is substantially enriched in STGD1 cases, specifically in those with one known pathogenic allele (P < 1 × 10−7; OR = 12.87 95% CI [8.37;19.79]). Family members were available for segregation analyses in ∼30% of cases, which always resulted in phasing the c.4253+43G>A allele in trans with the known ABCA4 mutation. All variants in trans from the c.4253+43G>A variant are severe, mostly confirmed loss-of-function alleles (Table 2), the situation exactly matching our recent observation for a frequent missense ABCA4 allele, c.5603A>T (p.Asn1868Ile; rs1801466) (Zernant et al. 2017).
Table 2.
STGD1 patients with deep intronic disease-associated ABCA4 variants
| Patient | Disease-associated ABCA4 variants in trans | Pathogenicity class | Deep-intronic disease-associated ABCA4 variants |
|---|---|---|---|
| 1 | c.161G>A (p.Cys54Tyr) | PS3 | c.[4253+43G>A;6006-609T>A] |
| 2 | c.161G>A (p.Cys54Tyr) | PS3 | c.[4253+43G>A;6006-609T>A] |
| 3 | c.247_250dup (p.Ser84Thrfs*15) | PVS1 | c.4253+43G>Aa |
| 4 | c.768G>T (p.Leu257Valfs*17) | PVS1 | c.4253+43G>Aa |
| 5 | c.768G>T (p.Leu257Valfs*17) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 6 | c.[1622T>C;3113C>T] (p.[Leu541Pro;Ala1038Val]) | PS3 | c.[4253+43G>A;6006-609T>A] |
| 7 | c.1819G>A (p.Gly607Arg) | PS4 | c.4253+43G>Aa |
| 8 | c.1819G>A (p.Gly607Arg) | PS4 | c.4253+43G>A |
| 9 | c.1988G>A (p.Trp663*) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 10 | c.3470T>G (p.Leu1157*) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 11 | c.3814-2A>G (p.?) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 12 | c.3898C>T (p.Arg1300*) | PVS1 | c.4253+43G>Aa |
| 13 | c.4248C>A (p.Phe1416Leu) | PP3 | c.4253+43G>A |
| 14 | c.4539+1G>T (p.?) | PVS1 | c.4253+43G>Aa |
| 15 | c.4539+1729G>T (p.?) | PP3 | c.[4253+43G>A;6006-609T>A] |
| 16 | c.4918C>T (p.Arg1640Trp) | PS3 | c.[4253+43G>A;6006-609T>A] |
| 17 | c.5312+2T>G (p.?) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 18 | c.5461-10T>C (p.Thr1821Valfs*13, p.Thr1821Aspfs*6) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 19 | c.5461-10T>C (p.Thr1821Valfs*13, p.Thr1821Aspfs*6) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 20 | c.5461-10T>C (p.Thr1821Valfs*13, p.Thr1821Aspfs*6) | PVS1 | c.[4253+43G>A;6006-609T>A] |
| 21 | c.5461-10T>C (p.Thr1821Valfs*13, p.Thr1821Aspfs*6) | PVS1 | c.4253+43G>A |
| 22 | c.5461-10T>C (p.Thr1821Valfs*13, p.Thr1821Aspfs*6) | PVS1 | c.4253+43G>A |
| 23 | c.5914G>A (p.Gly1972Arg) | PM2 | c.4253+43G>Aa |
| 24 | c.[70C>T(;)2041C>T] (p.[Arg24Cys(;)Arg681*]) | PP3, PVS1 | c.4253+43G>Aa |
| 25 | c.[4139C>T(;)5308T>G] (p.[Pro1380Leu(;)Tyr1770Asp]) | PS3, PM2 | c.[4253+43G>A;6006-609T>A] |
| 26 | c.6543_6578del (p.Leu2182_Phe2193del) homozygote | PVS1 | c.4253+43G>A |
| 27 | ND | c.[4253+43G>A;6006-609T>A] | |
| 28 | ND | c.4253+43G>A | |
| 29 | ND | c.4253+43G>A | |
| 30 | c.1726G>C (p.Asp576His) | PS4 | c.5196+1056A>G |
| 31 | c.3056C>T (p.Thr1019Met) | PS4 | c.5196+1056A>G |
| 32 | c.3413T>A (p.Leu1138His) | PM2 | c.5196+1056A>G |
| 33 | c.5882G>A (p.Gly1961Glu) | PS3 | c.5196+1056A>G |
| 34 | c.5882G>A (p.Gly1961Glu) | PS3 | c.5196+1056A>G |
Nucleotide positions and protein translation correspond to CCDS747.1 and NP_000341.2, respectively. Nucleotide numbering uses the A of the ATG translation initiation start site as nucleotide 1.
ND, not detected.
aNot screened for c.6006-609T>A.
Table 3.
Population frequency and in silico analysis of deep intronic ABCA4 variants
| Chr 1 position (hg19) | ABCA4 deep intronic variant | MAF in non-Finnish European controls | MAF in STGD1 cohort (n) | MAF in STGD1 with one ABCA4 mutation (n) | CADD score | Predicted effect on splicing | Associated with STGD1 |
|---|---|---|---|---|---|---|---|
| 94496509 | c.4253+43G>A | 0.006 | 0.013 (1155) | 0.069 (160) | 6.491 | No | Yes |
| 94484082 | c.5196+1056A>G | 0 | 0.003 (834) | N/A | 1.067 | Yes | Yes |
| 94471747 | c.6006-609T>A | 0.0047 | 0.009 (834) | 0.049 (132) | 1.848 | No | No |
Specifically, 26 unrelated patients harbor the c.4253+43G>A allele in trans from another ABCA4 mutation (Table 2). Of these, 16 patients carry ABCA4 variants, which are either LOF/null by definition (i.e., nonsense, frameshift, ±1 or 2 in splice sites) or as shown in functional studies, including c.5461-10T>C (p.Thr1821Valfs*13, p.|Thr1821Aspfs*6) (Sangermano et al. 2016). Of the missense alleles, several (e.g., c.[1622T>C;3113C>T] (p.[Leu541Pro;Ala1038Val]) (Zhang et al. 2015), c.161G>A (p.Cys54Tyr), c.4139C>T (p.Pro1380Leu) (Sun et al. 2000), and c.4918C>T (p.Arg1640Trp) (Shroyer et al. 2001b)) have direct functional proof. The c.5914G>A (p.Gly1972Arg) variant has been identified in a STGD1 patient in trans from other hypomorphic allele (Zernant et al. 2017). Therefore, the direct functional/clinical proof is currently vague for only three variants, c.1819G>A (p.Gly607Arg), c.4248C>A (p.Phe1416Leu), and c.4539+1729G>T (p.?). Altogether, these data allowed us to assign pathogenicity to the c.4253+43G>A variant; it is the second reported extremely hypomorphic ABCA4 allele, and first in noncoding sequences (i.e., it is causal only when in trans with a loss-of-function mutation).
Another deep intronic variant we had previously described (Zernant et al. 2014) and suggested as “possibly associated with the disease,” c.6006-609T>A (rs575968112), was seen in 15/834 (1.8%; MAF = 0.009) screened cases (MAF = 0.0047 in non-Finnish European control cohort, Table 3). Upon further analysis we determined that it forms a complex allele c.[4253+43G>A;6006-609T>A] (i.e., occurs on the same allele) in about 70% of cases in our cohort (Table 2). The c.6006-609T>A variant is always allelic with the c.4253+43G>A variant and has not been detected alone. According to the gnomAD database, the c.4253+43G>A variant is 1.2–2 times more frequent than c.6006-609T>A in every population except for Finnish Europeans, where the frequency of both variants is the same. Both variants are absent in East Asian population. Because both alleles, the c.4253+43G>A variant alone and the complex allele c.[4253+43G>A;6006-609T>A], are equally pathogenic based on genetic and clinical analyses, it is very likely that the hypomorphic c.4253+43G>A allele is pathogenic and the c.6006-609T>A is not, because the c.6006-609T>A allele does not add severity to the complex allele. Neither of these two variants are predicted to have any effect on splicing, whether on existing cryptic splice sites or on creating new sites, although c.4253+43G>A alters a splice silencer (ESS) and creates a splicing enhancer (ESE) SF2/ASF site (Supplemental Fig. 1A,B). Thus, an alternative splicing in photoreceptors caused by the c.4253+43G>A variant is likely. Recently, aberrant splicing caused by other ABCA4 intronic disease-associated variants, c.5461-10T>C, c.4539+2001G>A, and c.4539+2028C>T, was demonstrated in patient-derived photoreceptor precursor cells (PPCs), whereas no effect on splicing by these variants was predicted in silico (Sangermano et al. 2016; Albert et al. 2018).
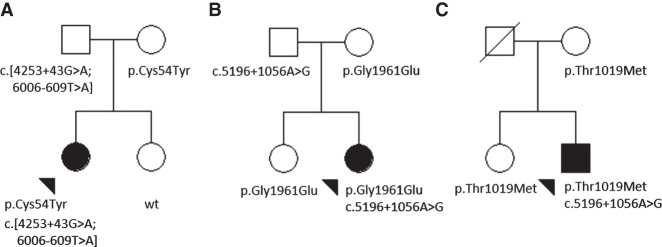
The third rare deep intronic variant, c.5196+1056A>G (rs886044749) (Braun et al. 2013; Zernant et al. 2014), is found in 5/834 (0.6%) of STGD1 cases (Tables 2, 3). Unlike the c.4253+43G>A and c.6006-609T>A variants, the c.5196+1056A>G variant is predicted to strongly activate a cryptic splice donor. Together with a strong cryptic splice acceptor downstream at c.5196+1318, splicing from these cryptic sites would result in a putative 262-bp pseudoexon inclusion and a frameshift in translation (Supplemental Fig. 1C). The variant is very rare, it is not described in any public databases (gnomAD, 1000 Genomes), and it was found exclusively in patients with one known ABCA4 mutation. All variants in trans from the c.5196+1056A>G are known disease-associated ABCA4 alleles (Table 2). In contrast to the c.4253+43G>A and the complex, c.[4253+43G>A;6006-609T>A] alleles, variants in trans from the c.5196+1056A>G variant do not include obvious loss-of-function alleles. For example, the frequent p.Gly1961Glu pathogenic variant in two families (Fig. 1B,C) consistently segregates with milder disease phenotypes similar to those observed in patients harboring p.Asn1868Ile (Burke et al. 2012, 2014; Zernant et al. 2017). Therefore, the aggregate evidence suggests that the c.5196+1056A>G variant is a rare, severe, deep intronic disease-associated allele.
Figure 1.
Examples of pedigrees showing the segregation of the deep intronic variants, c.[4253+ 43G>A;6006-609T>A] and c.5196+1056A>G, of ABCA4 in the families of (A) Patient 2, (B) Patient 33, and (C) Patient 31. Black arrowheads indicate the affected index patient in each family.
According to ENCODE analysis the three deep intronic variants are not located in predicted gene regulatory regions. The Combined Annotation Dependent Depletion (CADD) scores for these variants are also below the predicted pathogenic range of >20, but variants closer to coding sequences (c.4253+43G>A and c.6006-609T>A) score higher for their predicted possible deleterious effect (Table 3).
All three deep-intronic nucleotide positions are fully conserved, and adjacent sequences are also relatively conserved in nonhuman primates, suggesting an important functional role for these nucleotides—a crucial additional evidence for determining pathogenicity for a variant. All three alleles segregated with the disease in available families; that is, they were in trans from the proband's second pathogenic ABCA4 allele (examples in Fig. 1A–C).
Clinical Expression of the Three Noncoding ABCA4 Variants
Patients harboring the c.4253+43G>A variant alone or as a complex allele with c.6006-609T>A were predominantly Caucasian and relatively older in age with most cases presenting to the clinic in the fifth and sixth decade of life (median age at initial examination, 49.0 yr; range, 19–71 yr) (Supplemental Fig. 2). Most patients had relatively centralized fixation and visual acuity was correctable to 20/40 or better in at least one eye in 83% of patients. Reported ocular histories were consistent with a more recent or later onset of visual symptoms (median, 40.5 yr). Three patients reported symptomatic onset before the age of 20 yr; however, visual acuity in both eyes at the time was only marginally decreased, correctable between 20/25–20/40. Anterior segment examinations were unremarkable with the exception of mild nuclear sclerosis (NS+1) in a few cases. Dilated funduscopic examinations showed healthy optic discs, intact retinal vasculature, and otherwise normal peripheral retina. The most predominant features on funduscopy were retinal pigment epithelium (RPE) mottling, small patches of atrophy, through which large choroidal vessels were visible, and a reticular arrangement of yellow, pisciform flecks across the macula and mid-periphery (Fig. 2A,D). Disease changes were notably absent in the peripapillary region. The presence of dark pigment migration was only noted in the macula of one case (Patient 15), depositing in the foveola and along the temporal edge of the atrophic lesion in the right eye.
Figure 2.
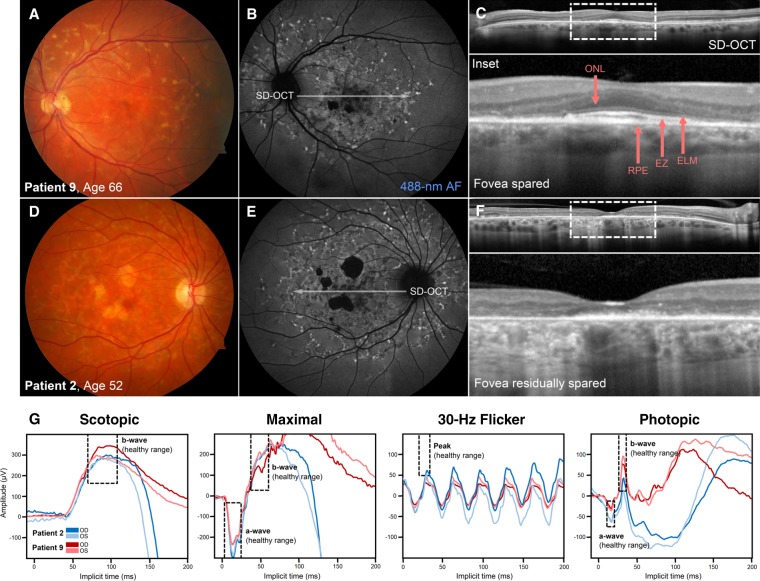
Clinical phenotype of patients harboring the deep intronic c.4253+43G>A variant of ABCA4 as a complex allele with c.6006-609T>A. (A) Color fundus photograph and (B) 488-nm autofluorescence (488-nm AF) image of the left eye of a 66-yr-old man (Patient 9, with c.1988G>A (p.Trp663*) in trans) exhibiting mottled atrophy and a distribution of autofluorescent flecks across the paramacular region of the fundus. (C) Spectral domain-optical coherence tomography (SD-OCT) revealed complete sparing of the photoreceptor-attributable layers, outer nuclear layer (ONL), external limiting membrane (ELM), ellipsoid zone (EZ), and RPE in the fovea resulting in preserved visual acuity of 20/20. (D) Color fundus photograph, (E) 488-nm AF image, and (F) SD-OCT scan of a 52-yr old woman (Patient 2, with c.161G>A (p.Cys54Tyr) in trans) revealing similar disease changes extending out into the midperiphery. Residual thickness of the foveal RPE layer can be noted, although the visible thinning of the ONL and absence of the ELM and EZ layers resulted in significantly decreased visual acuity (20/200) in this eye. (G) Full-field electroretinograms (ffERG) of Patient 2 and Patient 9 revealed relatively normal scotopic, maximal, and photopic and borderline 30-Hz flicker responses in both eyes. Areas delineated by dashed boxes indicate healthy amplitude and implicit time ranges.
Fundus autofluorescence (AF, 488-nm) abnormalities corresponding to changes observed on funduscopy were evident in all patients (Fig. 2B,E). Flecks were generally more hypoautofluorescent within the central macula and comparatively less confluent in the periphery indicating a centrifugal pattern of expansion. Lesions of discrete atrophy were homogenously hypoautofluorescent with uneven but well-demarcated edges. Very few patients demonstrated a complete loss of autofluorescence in the fovea. Remarkably, 83.3% of the cohort presented with structural and functional sparing of the fovea (VA 20/40 or better) in at least one eye at the initial examination—three patients (Patient 6, Patient 8, and Patient 9) exhibited unilateral sparing. SD-OCT through the fovea revealed intermittent disruptions of the reflective outer retinal bands, ELM, EZ, and interdigitation zone (IZ). Regions of discrete atrophy presented as areas of increased signal transmission into the underlying choroid because of the visible disruption of the photoreceptor layers and thinned or absent RPE. SD-OCT revealed visible preservation of all outer retinal bands (Fig. 2C, inset) in cases of complete foveal sparing such as Patient 9 (c.[4253+43G>A;6006-609T>A], p.Cys54Tyr*). Those with poor central acuity, such as Patient 2 (c.[4253+43G>A;6006-609T>A], p.Trp663*) (Fig. 2F, inset), were generally found to have ELM–EZ layer disruption but residual RPE reflectivity and thickness in the fovea on SD-OCT.
Virtually all patients for whom data were available had preserved generalized cone and rod function on full-field electroretinogram (ffERG) testing. Marginal decreases in 30-Hz flicker peak amplitudes were measured in five patients at the mean age of 57.6 yr (range, 42 to 71 yr). Representative scotopic, maximal, 30-Hz flicker and photopic waveforms from the right and left eyes of Patient 2 and Patient 7 are presented in Figure 2G. Follow-up examinations across periods of 1 to 29 yr revealed little indication of disease progression with respect to observable changes on funduscopy. Most patients also remained functionally stable at subsequent follow-up examinations, although a decrease in visual acuity, greater than two lines of vision, was evident in 25% of the cohort between the third and fifth decades of life. No significant distinctions between the clinical characteristics of patients harboring the c.4253+43G>A variant alone and patients harboring the c.4253+43G>A as a complex allele with c.6006-609T>A were noted.
DISCUSSION
Variation in the ABCA4 gene is extensive, and the complete understanding of genetic causality still remains obscure in at least 20% of all cases. The gene is expressed only in photoreceptors and, possibly, in the RPE, which renders RNA-based functional analyses challenging. This is specifically true for analyses of variants outside of coding regions for which protein-based tests are impossible.
Analyses of intronic variants have been attempted (Rivera et al. 2000; Braun et al. 2013) with the “illegitimate transcript” or “minigene” strategies to assess the effect of certain variants on splicing. These studies have limitations and the results, although suggestive, cannot be considered unequivocal (Sangermano et al. 2016). Most recently, a “midigene” approach was proposed, which is very effective for variants in canonical and noncanonical splice consensus sequences (Sangermano et al. 2018). Analysis of variants in deep intronic sequences of ABCA4, however, remains a tedious endeavor through iPSC-derived PPC (Albert et al. 2018). Although the PPC approach is gaining traction, the initial analysis for pathogenicity can be performed in very large, familial cohorts of STGD1 patients by a combination of in silico approaches including the assessment of variant frequencies in STGD1 patients and in the matched general populations, suggestions by predictive software programs, segregation analyses in families, and integration of clinical and genetic analysis. As demonstrated in this and in previous studies (Zernant et al. 2014, 2017), we have been able to assign pathogenicity to several variants in intronic sequences of ABCA4 by using these strategies prior to direct RNA- or protein-based functional studies. The latter, however, are still needed to determine the specific effect of variants on ABCA4 RNA and protein and would allow suggesting specific treatment modalities, such as AON-based correction of splicing (Albert et al. 2018).
The clinical phenotype of patients with the c.4253+43G>A allele is largely consistent with the described profile of STGD1 patients harboring the recently described hypomorphic c.5603A>T (p.Asn1868Ile) allele (Zernant et al. 2017). As compared to the more prevalent adolescent-onset form of STGD1, (Lambertus et al. 2015; Tanaka et al. 2018) patients harboring c.4253+43G>A or c.5603A>T (p.Asn1868Ile) progress through milder disease trajectories characterized by the confinement of disease changes to the macula, slower accumulation of flecks, and a significantly later onset of disease symptoms (fifth to sixth decade). The latter is likely a reflection of the enrichment of foveal sparing subphenotype in both the c.4253+43G>A (83.3%) and c.5603A>T (p.Asn1868Ile) (84.7%) groups. Several mechanisms for foveal sparing in ABCA4 disease and age-related macular degeneration (AMD) have been proposed, including the protective role of luteal macular pigment, anatomical arrangement, and morphology of foveal cones (Curcio et al. 1990) and the utilization of 11-cis retinol from Muller cells (Lee et al. 2014). However, our current findings support the hypothesis that slow disease progression is likely a major contributing factor for the functional persistence of cones in the fovea.
In summary, the analysis of three possibly disease-associated ABCA4 deep intronic variants revealed that two of these, the c.4253+43G>A and c.5196+1056A>G variants, are pathogenic, whereas the c.6006-609T>A variant is not. The c.5196+1056A>G variant is a highly penetrant, likely severe, allele, whereas the c.4253+43G>A variant is an “extreme hypomorph,” which is clinically expressed only when in trans with a loss-of-function ABCA4 allele. Functional studies involving ABCA4 RNA obtained from individual patients via iPS cells differentiated into PPCs would be the ultimate proof of specific functional outcome of each allele. However, these analyses are unlikely to change the clinical pathogenicity assessment obtained by integrated clinical and genetic analyses.
METHODS
Patients
STGD1 patients were recruited and clinically examined during a 20-yr period in different centers in the United States and in Europe. Ancestry of all patients was assessed from questionnaires and determined to be non-Finnish European. In total, 1155 patients were included in the analyses. Age of onset was defined as the age at which symptoms were first reported. Visual acuity was measured using the Early Treatment Diabetic Retinopathy Study Chart 1 or a Snellen acuity chart. Clinical examination, fundus photography, AF, and SD-OCT (Heidelberg Spectralis HRA+OCT) were performed using standard acquisition protocols following pupil dilation with Tropicamide 1% and Neosynephrine 2%.
Sequencing and Analysis of the ABCA4 Variants
Sequencing of the ABCA4 gene in 1155 patients was performed using 454 Titanium method (Roche) or Illumina TruSeq Custom Amplicon protocol (Illumina). (Zernant et al. 2011, 2014) Sequencing of the entire ABCA4 locus was performed in 217 patients using the RainDance target enrichment (RainDance Technologies) with Roche 454 sequencing or Illumina TruSeq Custom Amplicon protocol (Zernant et al. 2014). For genotyping of deep intronic variants in patient and control cohorts TaqMan Genotyping technology (Life Technologies) was applied. The variants’ segregation with the disease in available families was analyzed by Sanger sequencing.
All variants and their allele frequencies were compared to the 1000 Genomes database (Genomes Project et al. 2012) and to the gnomAD data set (Lek et al. 2016) (http://gnomad.broadinstitute.org; accessed April 2018). Frequent variants (MAF > 0.01) were considered nonpathogenic. Rare variants and variants identified with at least two times higher frequency in STGD1 patients than in the general control population were analyzed by a combination of predictive in silico methods and statistical analyses (Zernant et al. 2014). The possible effect of noncoding ABCA4 variants on splicing was assessed using five different algorithms (SpliceSiteFinder, MaxEntScan, NNSPLICE, GeneSplicer, and Human Splicing Finder) via Alamut software (http://www.interactive-biosoftware.com). The regulatory potential of the ABCA4 intronic variants was assessed by comparing their chromosome coordinates against the predicted regulatory regions from two ENCODE data sets: (1) combined DNase I hypersensitivity clusters from 125 cell types (“Digital DNase I Hypersensitivity Clusters in 125 cell types from ENCODE”) and (2) ChIP-seq clustered regions for 161 transcription factors in 91 cell types (“Transcription Factor ChIP-seq V4 (161 factors) with Factorbook motifs from ENCODE”). Evolutionary conservation of the variants was noted via UCSC Genome Browser (http://genome.ucsc.edu). The CADD algorithm (http://cadd.gs.washington.edu/score) was used to estimate combined predicted general deleteriousness of every variant. Combined data from MAF comparisons, predictive programs, determining the variants’ segregation with the disease were used to determine pathogenicity for the identified deep intronic variants. Statistical calculations using allele frequencies in cases and controls were performed with a 2 × 2 table. More detailed methods are provided in Supplement Materials.
ADDITIONAL INFORMATION
Data Deposition and Access
The variants described in the study were submitted to Leiden Open Variation Database (LOVD) under the accession numbers c.4253+43G>A (ABCA4_001024), c.6006-609T>A (ABCA4_000882), and c.5196+1056A>G (ABCA4_000028). The reference SNP (rs) accession numbers for the described variants are c.4253+43G>A (rs61754045), c.6006-609T>A (rs575968112), and c.5196+1056A>G (rs886044749). Raw data were not deposited to public access databases in the absence of patient consent but may be available through correspondence with the authors.
Ethics Statement
All study subjects provided full written consent prior to participating in the study under protocols #AAAB6560 and #AAAI9906 approved by the Columbia University Medical Center Institutional Review Board and protocol #20130770 by the Western Institutional Review Board at the Chicago Lighthouse. The study adhered to all tenets established in the Declaration of Helsinki.
Author Contributions
The study was conceived and designed by R.A., J.Z., and W.L. and sequenced by J.Z.; genetic analysis and interpretation were performed by R.A., J.Z., and T.N.; clinical data were acquired by W.L., F.T.C., G.A.F., M.B., T.R., P.R., and S.H.T.; clinical data were analyzed and interpreted by W.L., F.T.C., G.A.F., and S.H.T.; and the manuscript was drafted and revised by R.A., J.Z., and W.L. Study materials were provided by R.A.
Funding
This study was supported, in part, by grants from the National Eye Institute/National Institutes of Health (NIH) EY021163, EY019861, and EY019007 (Core Support for Vision Research) and by unrestricted funds from Research to Prevent Blindness (New York, NY) to the Department of Ophthalmology, Columbia University.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Albert S, Garanto A, Sangermano R, Khan M, Bax NM, Hoyng CB, Zernant J, Lee W, Allikmets R, Collin RWJ, et al. 2018. Identification and rescue of splice defects caused by two neighboring deep-intronic ABCA4 mutations underlying Stargardt disease. Am J Hum Genet 102: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, et al. 1997. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 15: 236–246. [DOI] [PubMed] [Google Scholar]
- Bauwens M, De Zaeytijd J, Weisschuh N, Kohl S, Meire F, Dahan K, Depasse F, De Jaegere S, De Ravel T, De Rademaeker M, et al. 2015. An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in Belgian Stargardt patients. Hum Mutat 36: 39–42. [DOI] [PubMed] [Google Scholar]
- Bax NM, Sangermano R, Roosing S, Thiadens AA, Hoefsloot LH, van den Born LI, Phan M, Klevering BJ, Westeneng-van Haaften C, Braun TA, et al. 2015. Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum Mutat 36: 43–47. [DOI] [PubMed] [Google Scholar]
- Braun TA, Mullins RF, Wagner AH, Andorf JL, Johnston RM, Bakall BB, Deluca AP, Fishman GA, Lam BL, Weleber RG, et al. 2013. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet 22: 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TR, Fishman GA, Zernant J, Schubert C, Tsang SH, Smith RT, Ayyagari R, Koenekoop RK, Umfress A, Ciccarelli ML, et al. 2012. Retinal phenotypes in patients homozygous for the G1961E mutation in the ABCA4 gene. Invest Ophthalmol Vis Sci 53: 4458–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TR, Duncker T, Woods RL, Greenberg JP, Zernant J, Tsang SH, Smith RT, Allikmets R, Sparrow JR, Delori FC. 2014. Quantitative fundus autofluorescence in recessive Stargardt disease. Invest Ophthalmol Vis Sci 55: 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone L, Lee W, Zernant J, Tanaka K, Schuerch K, Tsang SH, Allikmets R. 2017. Hyperreflective deposition in the background of advanced Stargardt disease. Retina 10.1097/IAE.0000000000001841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis SS, Bax NM, Zernant J, Allikmets R, Fritsche LG, den Dunnen JT, Ajmal M, Hoyng CB, Cremers FP. 2017. In silico functional meta-analysis of 5,962 ABCA4 variants in 3,928 retinal dystrophy cases. Hum Mutat 38: 400–408. [DOI] [PubMed] [Google Scholar]
- Cremers FP, van de Pol DJ, van Driel M, den Hollander AI, van Haren FJ, Knoers NV, Tijmes N, Bergen AA, Rohrschneider K, Blankenagel A, et al. 1998. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet 7: 355–362. [DOI] [PubMed] [Google Scholar]
- Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. 1990. Human photoreceptor topography. J Comp Neurol 292: 497–523. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertus S, van Huet RA, Bax NM, Hoefsloot LH, Cremers FP, Boon CJ, Klevering BJ, Hoyng CB. 2015. Early-onset Stargardt disease: phenotypic and genotypic characteristics. Ophthalmology 122: 335–344. [DOI] [PubMed] [Google Scholar]
- Lee W, Noupuu K, Oll M, Duncker T, Burke T, Zernant J, Bearelly S, Tsang SH, Sparrow JR, Allikmets R. 2014. The external limiting membrane in early-onset Stargardt disease. Invest Ophthalmol Vis Sci 55: 6139–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Xie Y, Zernant J, Yuan B, Bearelly S, Tsang SH, Lupski JR, Allikmets R. 2016. Complex inheritance of ABCA4 disease: four mutations in a family with multiple macular phenotypes. Hum Genet 135: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, Vilageliu L, Gonzalez-Duarte R, Balcells S. 1998. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet 18: 11–12. [DOI] [PubMed] [Google Scholar]
- Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. 2000. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet 67: 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveiro-Alvarez R, Xie YA, Lopez-Martinez MA, Gambin T, Perez-Carro R, Avila-Fernandez A, Lopez-Molina MI, Zernant J, Jhangiani S, Muzny D, et al. 2015. New mutations in the RAB28 gene in 2 Spanish families with cone-rod dystrophy. JAMA Ophthalmol 133: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, Jurklies B, Lorenz B, Scholl HP, Apfelstedt-Sylla E, et al. 2000. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet 67: 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangermano R, Bax NM, Bauwens M, van den Born LI, De Baere E, Garanto A, Collin RW, Goercharn-Ramlal AS, den Engelsman-van Dijk AH, Rohrschneider K, et al. 2016. Photoreceptor progenitor mRNA analysis reveals exon skipping resulting from the ABCA4 c.5461–10T→C mutation in Stargardt disease. Ophthalmology 123: 1375–1385. [DOI] [PubMed] [Google Scholar]
- Sangermano R, Khan M, Cornelis SS, Richelle V, Albert S, Garanto A, Elmelik D, Qamar R, Lugtenberg D, van den Born LI, et al. 2018. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res 28: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Lupski JR. 2001a. Null missense ABCR (ABCA4) mutations in a family with Stargardt disease and retinitis pigmentosa. Invest Ophthalmol Vis Sci 42: 2757–2761. [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Yatsenko AN, Wensel TG, Lupski JR. 2001b. Cosegregation and functional analysis of mutant ABCR (ABCA4) alleles in families that manifest both Stargardt disease and age-related macular degeneration. Hum Mol Genet 10: 2671–2678. [DOI] [PubMed] [Google Scholar]
- Sun H, Smallwood PM, Nathans J. 2000. Biochemical defects in ABCR protein variants associated with human retinopathies. Nat Genet 26: 242–246. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Lee W, Zernant J, Schuerch K, Ciccone L, Tsang SH, Sparrow JR, Allikmets R. 2018. The rapid-onset chorioretinopathy phenotype of ABCA4 disease. Ophthalmology 125: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang SH, Burke T, Oll M, Yzer S, Lee W, Xie YA, Allikmets R. 2014. Whole exome sequencing identifies CRB1 defect in an unusual maculopathy phenotype. Ophthalmology 121: 1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YA, Lee W, Cai C, Gambin T, Noupuu K, Sujirakul T, Ayuso C, Jhangiani S, Muzny D, Boerwinkle E, et al. 2014. New syndrome with retinitis pigmentosa is caused by nonsense mutations in retinol dehydrogenase RDH11. Hum Mol Genet 23: 5774–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. 2014. A Drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159: 200–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Schubert C, Im KM, Burke T, Brown CM, Fishman GA, Tsang SH, Gouras P, Dean M, Allikmets R. 2011. Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci 52: 8479–8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Xie YA, Ayuso C, Riveiro-Alvarez R, Lopez-Martinez MA, Simonelli F, Testa F, Gorin MB, Strom SP, Bertelsen M, et al. 2014. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum Mol Genet 23: 6797–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Lee W, Collison FT, Fishman GA, Sergeev YV, Schuerch K, Sparrow JR, Tsang SH, Allikmets R. 2017. Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J Med Genet 54: 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Tsybovsky Y, Kolesnikov AV, Rozanowska M, Swider M, Schwartz SB, Stone EM, Palczewska G, Maeda A, Kefalov VJ, et al. 2015. Protein misfolding and the pathogenesis of ABCA4-associated retinal degenerations. Hum Mol Genet 24: 3220–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.