Abstract
Multi-parental recombinant inbred populations, such as the Collaborative Cross (CC) mouse genetic reference population, are increasingly being used for analysis of quantitative trait loci (QTL). However specialized analytic software for these complex populations is typically built in R that works only on command-line, which limits the utility of these powerful resources for many users. To overcome analytic limitations, we developed gQTL, a web accessible, simple graphical user interface application based on the DOQTL platform in R to perform QTL mapping using data from CC mice.
Keywords: collaborative cross, qtl, software
The utility of model organisms for genetic analysis of biological systems has dramatically increased with the establishment of genetic reference populations. Modern, multi-parental populations specifically designed for quantitative trait locus (QTL) and systems genetics analyses originated with the Collaborative Cross (CC) mouse genetic reference population (Threadgill et al. 2002; Threadgill and Churchill 2012). The CC population is derived from eight founder strains, A/J, C57BL/6J, 129S1Sv/ImJ, NOD/ShiLtJ, NZO/H1LtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ, representing the three major Mus musculus subspecies (M. m. musculus, M. m. domesticus, and M. m. castaneus) and which captures 90% of the genetic variation in laboratory mice (Roberts et al. 2007). Although the CC has an organized genetic structure (Churchill et al. 2004) and is increasingly being used to identify genetic factors controlling a variety of phenotypes from infectious disease and cancer to molecular circuitry (Rasmussen et al. 2014; Dorman et al. 2016; Venkatratnam et al. 2018), genetic analysis of phenotypes using the CC can be challenging due to the multi-allelic structure of the population and complex analytic tools needed to perform analyses (Aylor et al. 2011).
Although not a replicable population like the CC, the Diversity Outbred (DO) population was derived from the CC population to increase the recombination load in order to improve mapping resolution for QTL analysis (Svenson et al. 2012). To support genetic analysis using the DO population, DOQTL was developed (Gatti et al. 2014), which also is increasingly being used for analysis of CC data. DOQTL is an R-based program developed to overcome several analytic challenges of multi-parental populations by implementing an integrated pipeline for haplotype reconstruction, regression modeling to account for kinship, significance thresholds through permutation analysis, and combined association mapping and parental allele-specific tests. Although DOQTL has become the predominant analytic platform for analysis of CC data, it presents a substantial barrier for most biologists with limited computer programming background. Exploiting recent advancements in web framework technologies in R programming, we developed gQTL, which is web application to simplify genetic analyses using data collected from CC mice that will greatly extend the utility of the CC model for a much broader user base.
Methods
gQTL was implemented using the R Shiny framework (Chang et al. 2016), which provides necessary tools for rapid prototyping of interactive web applications. gQTL relies on functions from the DOQTL R package to perform QTL mapping (Gatti et al. 2014). Since the CC population has a fixed genetic architecture, associated genotypes and haplotype probabilities for each CC line are stored and loaded into memory in the backend when gQTL is launched. The genotype probabilities for each CC and founder strain were obtained from UNC Systems Genetics data repository (http://csbio.unc.edu/CCstatus/index.py), while the MegaMUGA and GigaMUGA marker set from which the genotypes are determined in the CC was obtained from The Jackson Laboratory data repository (ftp://ftp.jax.org/MUGA/). The user has the ability to choose between either of these marker sets during the submission of the analysis.
Data availability
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its figures. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6453092.
Results and Discussion
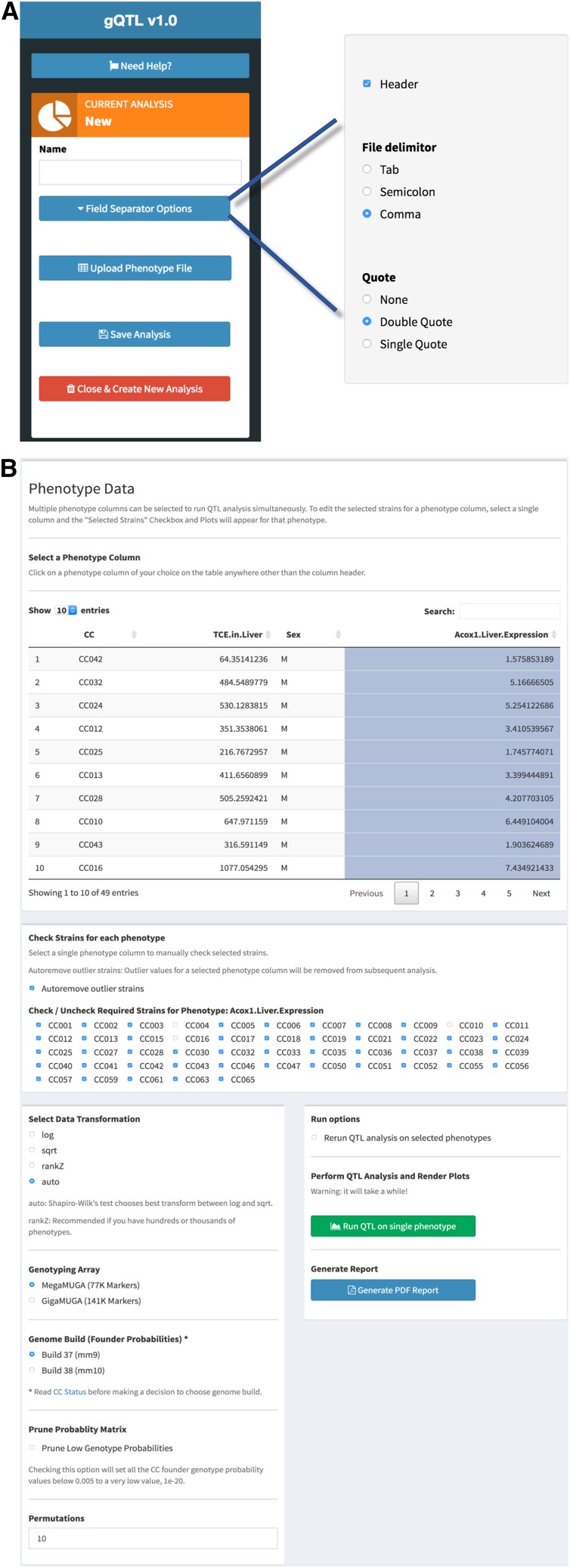
After creating a user account, data can be uploaded into a server-side deployment of gQTL, which accepts simple tab delimited or comma separated text files containing a sex identifier and multiple phenotype columns from individual or strain pooled CC data (Figure 1A). At least 3 columns containing Strain (CC), Sex and Phenotype values are mandatory. The CC column can be official or alias names (Supplementary Material, Table S1). Each row can be a line mean or individual mice, sex column should contain M or F, and multiple phenotype columns can be used. In a recent toxicology study, we used the CC population to evaluate the inter-strain variability in oxidative metabolism of trichloroethylene (TCE) and found several QTL controlling tissue TCE levels and expression of specific genes using DOQTL (Venkatratnam et al. 2017); datasets from this project are used here to illustrate simplicity of gQTL (Supplemental Material, Table S2). After uploading the data file, users can remove outliers, normalize the data and perform QTL mapping. Uploaded data are presented as a table, wherein specified phenotype columns can be selected for analysis (Figure 1B). Data from specific CC strains for each phenotype can be manually removed using simple check boxes, or automatic outlier removal can be selected. Trait outliers are detected using the standard boxplot outlier rule, 1.5 × interquartile range (IQR) (Tukey 1977). Multiple data transformation choices (log, sqrt, rankZ) are available for user selection, or an automated transformation selection feature can be specified that uses the Shapiro-Wilk test of normality to determine the optimal transformation between log and sqrt (Shapiro and Wilk 1965). For a selected phenotype column, data quality plots, including raw and normalized histogram and QQ plots, are displayed (Figure 2). Finally, individual or multiple phenotype data columns can be submitted to the server for QTL mapping. Significance thresholds are determined through permutation analysis using a user-specified number of permutations (Churchill and Doerge 1994). QTL mapping with 1000 permutations typically takes about 5 hr to finish due to the fact that DOQTL runs on a single core; future implementations will transition to multiple cores. E-mail notifications keep the user informed on the current state of the job(s) running on the server. Each user account can store up to seven different analyses for later revisiting and re-submission of QTL mapping jobs with different parameters.
Figure 1.
Screen shots of data entry and initial processing. (A) Data loading and file type selection. (B) Uploaded data visualization, outlier selection, and normalization options.
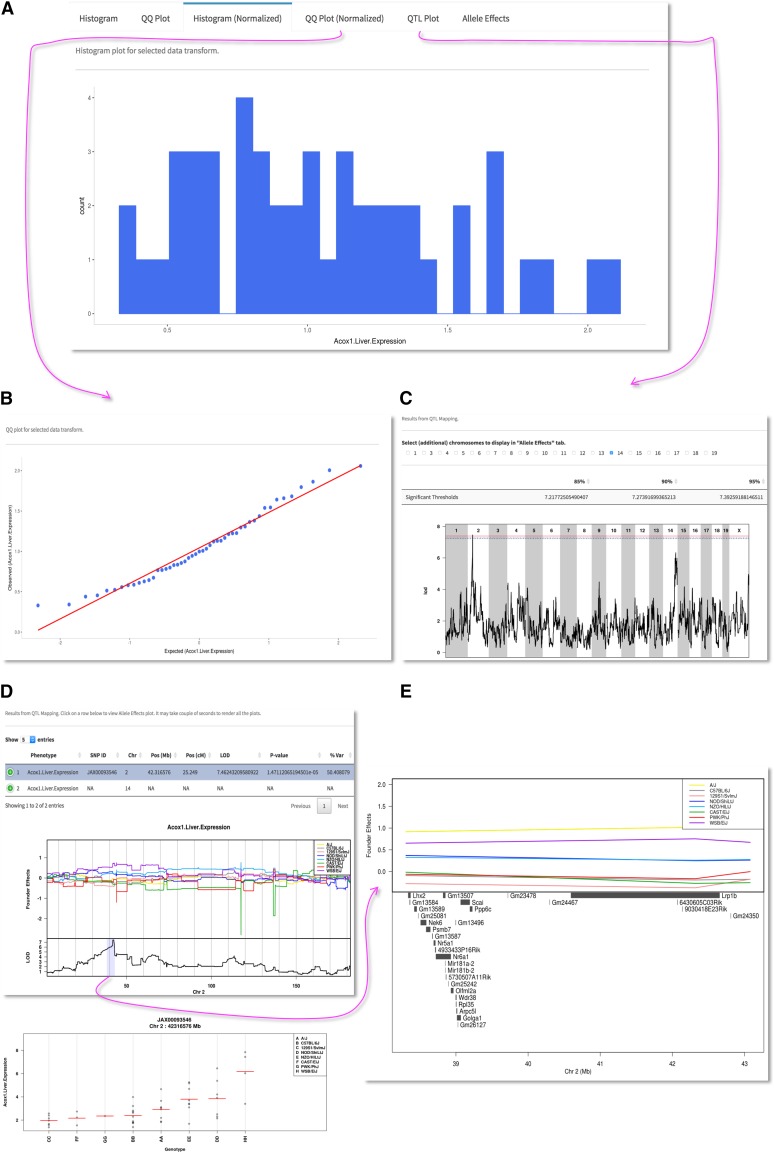
Figure 2.
Screen shots of QTL analysis results. (A) Options for data visualization with normalized histogram. (B) QQ plot. (C) QTL plot with threshold levels and locations of significant markers. (D) Allele effect and genotype-phenotype plots. (E) A zoomed version of the significant QTL interval.
After the analyses are complete, QTL results can be explored using the web application (Figure 2; Gatti et al. 2014). Linkage plots are displayed along with permutation determined LOD scores for the 85, 90 and 95% significance threshold levels. Chromosome-wide, CC founder strain-specific allele effect plots are automatically generated for any locus reaching significance that shows the marker ID with the maximal LOD and its location in cM and Mb coordinates on Build 37 (mm9) or Build 38 (mm10) depending on marker set selected, as well as Mb coordinates of the confidence interval based on a 95% Bayesian credible interval (Sen and Churchill 2001). Higher resolution images of the 95% intervals can be selected that show underlying gene annotations. Other chromosomes that may contain regions of interest but not reach at least 85% significance can be manually selected to generate additional chromosome-specific allele effect plots. For those loci reaching at least 85% significance thresholds, phenotypes for each CC sample is also plotted by genotype to visualize those genotypes driving the QTL signal. A comprehensive PDF report is automatically generated for archiving (Supplemental Material, Figure S1). Additionally, a ZIP archive containing the PDF report along with publication quality PNG figures at 600 dpi can be downloaded.
gQTL v1.0 provides an easy to use graphical user interface for QTL mapping analyses of studies in CC mice with the upload of quantitative phenotype data collected in CC mice being the only input required from users. We plan to extend the application to include the ability to use phenotypes from CC Recombinant Inbred Intercrosses (CC-RIX) in subsequent version releases (Zou et al. 2005).
Web Resources
The web application is freely available at: https://genomics.tamu.edu/gqtl. A built-in help menu exists on gQTL with instructions on setting up user accounts, uploading phenotype data files, inspecting phenotype data, running QTL analysis, viewing QTL analysis results and generating reports of QTL results. The source code, from the original developers (Gatti et al. 2014), for the underlying DOQTL package is available at GitHub (https://github.com/dmgatti/DOQTL).
Acknowledgments
The authors thank members of the Threadgill and Rusyn labs for providing user feedback during development. Development of gQTL was supported by the Texas A&M Institute for Genome Sciences and Society and, in part, by grants from the U.S. EPA (STAR RD-83516602 and RD 83580201), Department of Defense (D17AP00004), and the National Institutes of Health (P42 ES027704, P30 ES023512, P42 ES004911, RM1 HG008529) to I.R. and D.W.T. Its contents are solely the responsibility of the grantees and do not necessarily represent the official views of the U.S. EPA or NIH. Further, the U.S. EPA and NIH do not endorse any products or services mentioned in the publication U.S. EPA or NIH.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6453092.
Communicating editor: D. J. de Koning
Literature Cited
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222. 10.1101/gr.111310.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W., J. Cheng, J. Allaire, Y. Xie and J. McPherson, 2016 shiny: Web application framework for R, http://shiny.rstudio.com.
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. 10.1038/ng1104-1133 [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman A., Baer D., Tomlinson I., Mott R., Iraqi F. A., 2016. Genetic analysis of intestinal polyp development in Collaborative Cross mice carrying the Apc (Min/+) mutation. BMC Genet. 17: 46 (erratum: BMC Genet. 17: 147) 10.1186/s12863-016-0349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative Trait Locus Mapping Methods for Diversity Outbred Mice. G3-Genes Genomes Genetics 4: 1623–1633. 10.1534/g3.114.013748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A. L., Okumura A., Ferris M. T., Green R., Feldmann F., et al. , 2014. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346: 987–991. 10.1126/science.1259595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pardo-Manuel de Villena F., Wang W., McMillan L., Threadgill D. W., 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome 18: 473–481. 10.1007/s00335-007-9045-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S., Churchill G. A., 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro S. S., Wilk M. B., 1965. An analysis of variance test for normality (complete samples). Biometrika 52: 591–611. 10.1093/biomet/52.3-4.591 [DOI] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447. 10.1534/genetics.111.132597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Churchill G. A., 2012. Ten years of the Collaborative Cross. Genetics 190: 291–294. 10.1534/genetics.111.138032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Hunter K. W., Williams R. W., 2002. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm. Genome 13: 175–178. 10.1007/s00335-001-4001-y [DOI] [PubMed] [Google Scholar]
- Tukey J. W., 1977. Box-and-Whisker Plots, pp. 39–43 in Exploratory Data Analysis. Addison-Wesley, Reading (Sunderland). [Google Scholar]
- Venkatratnam A., Furuya S., Kosyk O., Gold A., Bodnar W., et al. , 2017. Collaborative Cross Mouse Population Enables Refinements to Characterization of the Variability in Toxicokinetics of Trichloroethylene and Provides Genetic Evidence for the Role of PPAR Pathway in Its Oxidative Metabolism. Toxicol. Sci. 158: 48–62. 10.1093/toxsci/kfx065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatratnam A., House J. S., Konganti K., McKenney C., Threadgill D. W., et al. , 2018. Population-based dose-response analysis of liver transcriptional response to trichloroethylene in mouse. Mamm. Genome 29: 168–181. 10.1007/s00335-018-9734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Gelfond J. A., Airey D. C., Lu L., Manly K. F., et al. , 2005. Quantitative trait locus analysis using recombinant inbred intercrosses: theoretical and empirical considerations. Genetics 170: 1299–1311. 10.1534/genetics.104.035709 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its figures. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6453092.