Abstract
The centromere is a chromosomal locus where a microtubule attachment site, termed kinetochore, is assembled in mitosis. In most eukaryotes, with the exception of holocentric species, each chromosome contains a single distinct centromere. A chromosome with an additional centromere undergoes successive rounds of anaphase bridge formation and breakage, or triggers a cell cycle arrest imposed by DNA damage and replication checkpoints. We report here a study in Schizosaccharomyces pombe to characterize a mutant (cnp3-1) in a gene encoding a homolog of mammalian centromere-specific protein, CENP-C. At the restrictive temperature 36°, the Cnp3-1 mutant protein loses its localization at the centromere. In the cnp3-1 mutant, the level of the Cnp1 (a homolog of a centromere-specific histone CENP-A) also decreases at the centromere. Interestingly, the cnp3-1 mutant is prone to promiscuous accumulation of Cnp1 at non-centromeric regions, when Cnp1 is present in excess. Unlike the wild type protein, Cnp3-1 mutant protein is found at the sites of promiscuous accumulation of Cnp1, suggesting that Cnp3-1 may stabilize or promote accumulation of Cnp1 at non-centromeric regions. From these results, we infer the role of Cnp3 in restricting the site of accumulation of Cnp1 and thus to prevent formation of de novo centromeres.
Keywords: centromere, CENP-A, CENP-C, and fission yeast
The centromere is a chromosomal locus on which a microtubule attachment site, termed kinetochore, is assembled in mitosis. It is therefore an essential component for faithful segregation of chromosomes. A variant of histone H3, CENP-A, is specifically incorporated into centromeric chromatin and plays an important role as the base of the kinetochore assembly. (Black and Cleveland 2011; Quénet and Dalal 2012; Catania and Allshire 2014). Distribution of CENP-A must be strictly regulated so that the centromere is positioned and organized appropriately for each species. Although it is generally accepted that centromeres are defined by a sequence-independent epigenetic manner (Allshire and Karpen 2008; Black and Cleveland 2011; Perpelescu and Fukagawa 2011), the underlying mechanisms to deposit and maintain CENP-A at the right place are still elusive.
In most eukaryotes, with the exception of holocentric species, each chromosome contains a single distinct centromere. It has been reported that a chromosome with an extra centromere (dicentric chromosome) causes deleterious problems such as chromosome breakage (Thrower and Bloom 2001; Thrower et al. 2003; Pobiega and Marcand 2010) and cell cycle arrest (Sato et al. 2012). A dicentric chromosome can also be generated by translocation of a centromere followed by meiotic recombination. To avoid these problems, each chromosome contains a single centromere whose position is maintained through generations. As CENP-A histone is a major component found in centromeric chromatin, distribution of CENP-A fundamentally influences formation and positioning of the centromere.
Recent studies have shown that CENP-A accumulation can be found in non-cetromeric chromatin as well. In chicken DT-40 cells, minor accumulation of CENP-A is detected around the native centromeres. When the native centromere is experimentally removed, a neocentromere is occasionally assembled by using the minor accumulation of CENP-A as a seed (Shang et al. 2013). In budding yeast, Cse4, a homolog of mammalian CENP-A, is enriched at promoters that contain histone H2A (Hildebrand and Biggins 2016). Psh1 is an E3 ubiquitin ligase (Hewawasam et al. 2010 ; Ranjitkar et al. 2010) that blocks stable Cse4 incorporation into these promoters by targeting mislocalized Cse4 for degradation (Collins et al. 2004). The ubiquitin system is a conserved pathway for preventing ectopic CENP-A accumulation in other organisms including fission yeast and fly (Gonzalez et al. 2014; Moreno-Moreno et al. 2006).
In fission yeast, overexpression of CENP-A causes the promiscuous incorporation of CENP-A near heterochromatic regions (Choi et al. 2012; Castillo et al. 2013; Gonzalez et al. 2014). Most of these loci with CENP-A accumulation do not transform into a neocentromere. It has been shown that the histone H2A variant (Pht1), a NAP family protein (Ccp1) and a histone chaperone FACT play a role in antagonizing CENP-A loading at non-centromeric regions (Choi et al. 2012; Ogiyama et al. 2013; Dong et al. 2016). These factors thus likely prevent formation of neocentromeres.
Interestingly, sub-telomeric regions in fission yeast can accommodate neocetromeres when the native centromere is disrupted (Ishii et al. 2008). Subsequent study has revealed that stable association of CENP-A chaperon, Scm3, facilitates neocetromere formation (Ogiyama et al. 2013). Nonetheless, neocentromere formation is a very rare event (several in ten thousand cells in fission yeast), suggesting that neocetromere formation is strongly suppressed likely by multiple mechanisms.
Having been interested in mechanisms to maintain appropriate distribution of CENP-A and prevent formation of de novo centromeres, we have taken a genetic approach in fission yeast model system. We assumed that a mutant unable to maintain proper distribution of CENP-A would be sensitive to expression of CENP-A at a high level. A strain ectopically expressing fission yeast CENP-A (Cnp1) tagged with GFP (GFP-Cnp1) from an inducible promoter, nmt1, was mutagenized and survivors were screened for mutants, which became temperature sensitive upon overexpression of GFP-Cnp1. We have previously reported a mutant (rpt3-1) identified through the screen (Kitagawa et al. 2014). Unlike the wild type fission yeast in which the localization of CENP-A is limited to the central region spanning 10–20 kb of the centromere, the rpt3-1 mutation allows spread of CENP-A localization at the centromere (40–70 kb). Likely due to abnormal distribution of CENP-A, the rpt3-1 mutant exhibits chromosome instability and enhanced gene silencing. In this study, we have characterized another mutant identified through the same screen. Our analyses have shown that the mutation in the cnp3+ gene encoding a fission yeast homolog of the mammalian CENP-C allows accumulation of Cnp1 at non-centromeric regions. We thus propose a role of the wild type Cnp3 in restricting the site of accumulation of Cnp1 to prevent formation of de novo centromeres.
Materials and Methods
Strain, culture media and strain construction
An S. pombe haploid wild-type strain SP6 (h- leu1-32) and its derivatives used in this study are listed in Table 1. The culture media were complete YES, minimal EMM2, and sporulation medium MEA and SPA (Moreno et al. 1991).Transformation was done using the lithium method (Ito et al. 1983). The cell number was measured using a hematology analyzer (Sysmex F-520P and Beckman multisizer 3 coulter counter).
Table 1. S. pombe strains used in this study.
| Strain | Genotype | Source and reference | |
|---|---|---|---|
| SP6 | h- | leu1-32 | Laboratory stock |
| MS01 | h- | leu1-32 transformed with pREP41-GFP-H3.3 | This work |
| MS02 | h- | leu1-32 transformed with pREP41-GFP-cnp1 | This work |
| MS03 | h- | leu1-32 cnp3-1 transformed with pREP41-H3.3 | This work |
| MS04 | h- | leu1-32 cnp3-1 transformed with pREP41-cnp3-1 | This work |
| MS168 | h+ | leu1-32::nmtGFP-cnp1-leu1+ cnp3::kanR ade6-M216 | This work |
| MS213 | h- | leu1-32 lys::nmtGFP-cnp1-lys1+ cnp3-mCherry-leu1+ | This work |
| MS216 | h+ | leu1-32::nmtGFP-cnp1-leu1+ ade6-M216 | This work |
| MS217 | h- | leu1-32 lys::nmtGFP-cnp1-lys1+ cnp3-1-mCherry-leu1+ | This work |
| MS262 | h+ | leu1-32::nmtGFP-cnp1-leu1+ cnp3-1 ade6-M216 | This work |
| MS343 | h- | leu1-32 GFP-cnp1-hph | Laboratory stock |
| MS344 | h- | leu1-32 GFP-cnp1-hph cnp3-1 | This work |
| MS413 | h- | leu1-32 cnp3-1 ade6-M210 ch16 | This work |
| MS468 | h- | leu1-32 cnp3-GFP-leu1+ | This work |
| MS497 | h- | leu1-32 GFP-cnp1-hph mis16-53 | This work |
| MS503 | h- | leu1-32 GFP-cnp1-hph mis18-818 | This work |
| MS508 | h | leu1-32 GFP-cnp1-hph mis6-302 | This work |
| MS525 | h- | leu1-32 lys::nmtGFP-cnp1-lys1+ cnp3-3HA 6his-leu1+ | This work |
| MS527 | h- | leu1-32 lys::nmtGFP-cnp1-lys1+ cnp3-1-3HA 6his-leu1+ | This work |
| MS532 | h- | leu1-32 GFP-cnp1-hph mis6-302 cnp3-1 | This work |
| MS533 | h- | leu1-32 GFP-cnp1-hph mis16-53 cnp3-1 | This wor |
| MS534 | h- | leu1-32 GFP-cnp1-hph cnp3-1 mis18-818 | This work |
| MS544 | h- | leu1-32 cnp3-1-GFP-leu1+ | This work |
| MS545 | h- | leu1-32 GFP-cnp1-hph cnp3-1mCherry-leu1+ mis18-818 | This work |
| MS546 | h- | leu1-32 GFP-cnp1-hph cnp3-mCherry-leu1+ mis18-818 | This work |
| MS548 | h- | leu1-32 GFP-cnp1-hph cnp3-mCherry-leu1+ | This work |
| MS549 | h- | leu1-32 GFP-cnp1-hph cnp3-1-mCherry-leu1+ | This work |
| MS568 | h- | leu1-32 Sad1-mCherry<<kanR transformed with pREP81-cnp3-GFP | This work |
| MS569 | h- | leu1-32 Sad1-mCherry<<kanR transformed with pREP81-cnp3-1-GFP(S508F) | This work |
| MS615 | h- | leu1-32 GFP-cnp1-hph mis12-mCherry-leu1+ | This work |
| MS616 | h- | leu1-32 GFP-cnp1-hph mis12-mCherry-leu1+ cnp3−1 | This work |
| MS617 | h+ | leu1-32 GFP-cnp1-hph mis12-mCherry-leu1+cnp3−1 mis18-818 | This work |
| MS618 * | leu1-32 GFP-cnp1-hph mis12-mCherry-leu1+ mis18-818 | This work | |
| MS620 | h- | leu1-32 GFP-cnp1-hph mis12-mCherry-leu1+ mis18-262 | This work |
| MS621 | leu1-32 GFP-cnp1-hph mis12-mCherry-leu1+ cnp3−1mis18-262 | This work | |
| TK5 | h- | leu1-32::nmt1GFP-cnp1-leu1+ | This work |
| TK85 | h- | leu1-32 ade6-M210 ch16 | Laboratory stock |
: The mating type was not determined.
For tagging GFP to the N-terminal of Cnp1, the cnp1+ gene was PCR-amplified with the primers, Cnp1-F and Cnp-R (Table 3), and cloned into pREP41-GFP(N-end tagging) after digestion with restriction enzymes, Sal I and Not I. The resulting plasmid was used to construct MS02 (Table 1), in which GFP-Cnp1 could be conditionally induced. Similarly, the cnp1+ gene was PCR-amplified with the primers, GFP-Cnp1-F and GFP-Cnp-R (Table 3), and cloned into pBR322-(leu1+) lacking the Bam HI site. The resulting plasmid was integrated at the leu1 locus to construct TK5 (Table 1).
Table 3. Plasmids and primers used in this study.
| *1 | pREP1-GFP-Cnp1 |
| *2 | pREP1-GFP-Cnp1-pUM1 |
| MS-1 | nmt1-GFP-cnp1-pBR322 |
| MS-2 | cnp3-mCherry-pBS |
| MS-3 | cnp3-1mCherry-pBS |
| MS-4 | cnp3-HA-pBS |
| MS-5 | cnp3-1HA-pBS |
| MS-6 | pREP81-cnp3-GFP |
| MS-7 | pREP81-cnp3-1-GFP |
| Cnp1-F | ATAGTCGACTATGGCAAAGAAATCTTTAATG |
| Cnp1-R | ATAGCGGCCGCTCAAGCACCACGAATCC |
| GFP-Cnp1-F | ATAGGATCCTATGAGTAAAGGAGAAGAAC |
| GFP-Cnp1-R | ATAGGATCCTCAAGCACCACGAATCCTC |
| Cnp3-F | ATACTCGAGATGTAGACGATGAATGAAACGTC |
| Cnp3-R | ATAGCGGCCGCGTCGTTCGTTTGGAAAATC |
| Rep81Cnp3-F | ATACATATGTTCTAGAGATGAAGATTCCG |
| Rep81-Cnp3-R | ATAGCGGCCGCGTCGYYCGTTTGGAAAATC |
1, 2 (Kitagawa et al. 2014)
To construct other plasmids and strains for expression of tagged Cnp1 and Cnp3, similar strategies were employed. The primers and plasmids used for these purposes were summarized in Table 3. All integrants were examined for their integrated locus by PCR as well as crossing.
Western blotting
Cell extracts were prepared from exponentially growing S.pombe cells. The cells were suspended in lysis buffer (25 mM HEPES-KOH at pH 7.5, containing 200mM NaCl, 10% glycerol, 0.2% NP-40 and protease inhibitors (Nacalai Tesque), 1 mM PMSF, and 1% Trasylol) were disrupted by Beads Smash 12 (WAKENYAKU) at 4°. After centrifugation the supernatants were used for SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The extracts were separated by SDS-PAGE gel and blotted to nitrocellulose membranes (Funakoshi). Antibodies for western blotting were diluted as follows: mouse anti-GFP (Roche) 1:1000, anti-HA (12CA5, Roche) 1:3000; and TAT1 anti-α-tubulin (gift from K. Gull, Oxford, UK) 1:5000. Blots were developed using ECL reagents, Substrate Plus (Pierce) and ECL prime (GE Healthcare). The intensity of each band was measured by Image J and indicated arbitrarily.
Microscopy
S. pombe strains were grown in YES or EMM2 medium. Images were acquired on a KEYENCE (BZ-9000) and Delta Vision (Applied Precision LLC, Issaquah, WA, USA), CH350L CCD camera (Photomettric, Tucson, AZ, USA). FISH analyses was performed as described (Funabiki et al. 1993).
Preparation of digoxigenin-labeled probe DNA
pRA140 (Chikashige et al. 1989) was used to visualized the centromere. The DNA fragments used to prepare the probes were digested by a mixture of restriction enzymes, AluI, DdeI, HaeIII, RsaI, and Sau3AI to yield an average fragment size of 300bp.These fragment were labeled by digoxigenin-dUTP using the random priming labeling kit(Takara 6045 random primer DNA labeling kit ver.2). Nonreacted nucleotides were removed by microspin G-25 column (GE healthcare).
Mini-chromosome stability assay
The stability of mini-chromosome was determined by a method developed by (Allshire et al. 1995). First, the cells were plated on YE (for Table 2) or EMM containing 10 μg/ml adenine to determine the initial percentage of the cells carrying the mini-chromosome. After incubation in the non-selective medium (Ade+), the final percentage of the cells carrying the mini-chromosome was determined. The rate of mini-chromosome loss per division was estimated using the formula: loss rate = 1-(F/I)1/N (F, the final percentage of cells carrying mini-chromosome; I, the initial percentage of cells carrying mini-chromosome; N, the number of generations between F and I).
Table 2. Stability of mini-chromosome. The stability of the mini-chromosome Ch16 was determined on YE media. (WT: TK85, cnp3-1: MS413).
| Loss rate | ||||
|---|---|---|---|---|
| Background | Per division | Percent division (%) | Fold increase in loss rate | |
| 26°C | WT | 1.4x10−4 | 0.014 | 1 |
| cnp3-1 | 3.4x10−2 | 3.4 | 243 | |
| 36°C | WT | 2.3x10−4 | 0.023 | 1 |
| cnp3-1 | 1.2x10−1 | 12 | 522 | |
Chromatin immunoprecipitation (Chip) assay
ChIP was performed as described previously (Kitagawa et al. 2014).
Isolation of the cnp3-1 mutant
A strain (TK5: h- leu1-32::nmt1-GFP-cnp1+ -leu1+) which was able to express fission yeast CENP-A, Cnp1, tagged with GFP from the nmt1 promoter, was chemically mutagenized as described (Matsumoto and Beach 1991). Exponentially growing cells suspended in TM buffer (50 mM Tris malate (pH 6.0) were treated with NTG (500 µg/ml) at 26° for 20 min and washed three times with TM buffer. Cells were grown in the YES liquid medium for 4h at 26° and washed with EMM2 medium containing thiamine. Cells were grown in the same medium thiamine and grown at 26°. The survivors were plated on the EMM2 plates containing thiamine and grown at 26°. They were subsequently transferred onto EMM2 plates with and without thiamine by replica and grown at 36°. The strains that exhibited temperature sensitivity only when GFP-Cnp1 was expressed were selected. They were then individually examined under a fluorescence microscope. We scored mutants with abnormal distribution of GFP-Cnp1 in the nucleus. Tetrad analysis indicated that five strains including the cnp3-1mutant carried a single mutation responsible for the phenotypes (temperature sensitivity and abnormal distribution of GFP-Cnp1).
Isolation of the cnp3+gene
By using the temperature sensitivity as a selection marker, a genomic DNA fragment complementing the temperature sensitivity was isolated from a fission yeast genomic DNA library. The integration mapping proved that this fragment originated from the cnp3-1 locus. A PCR-based strategy was used to identify the mutation site within the cnp3+ coding sequence. The cnp3-1 mutant gene contained mutation of S508.
Data availability
All strains and plasmids are available upon request. All data necessary for evaluating our conclusions are represented within this paper. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6618920.
Results
Isolation of cnp3-1
A strain (TK5: h- leu1-32::nmt1-GFP-cnp1+ -leu1+) which was able to express fission yeast CENP-A, Cnp1, tagged with GFP from the nmt1 promoter was chemically mutagenized and survivors were screened for mutants, which became temperature sensitive upon overexpression of GFP-Cnp1. In this study, we characterized one of the mutants identified through the screen.
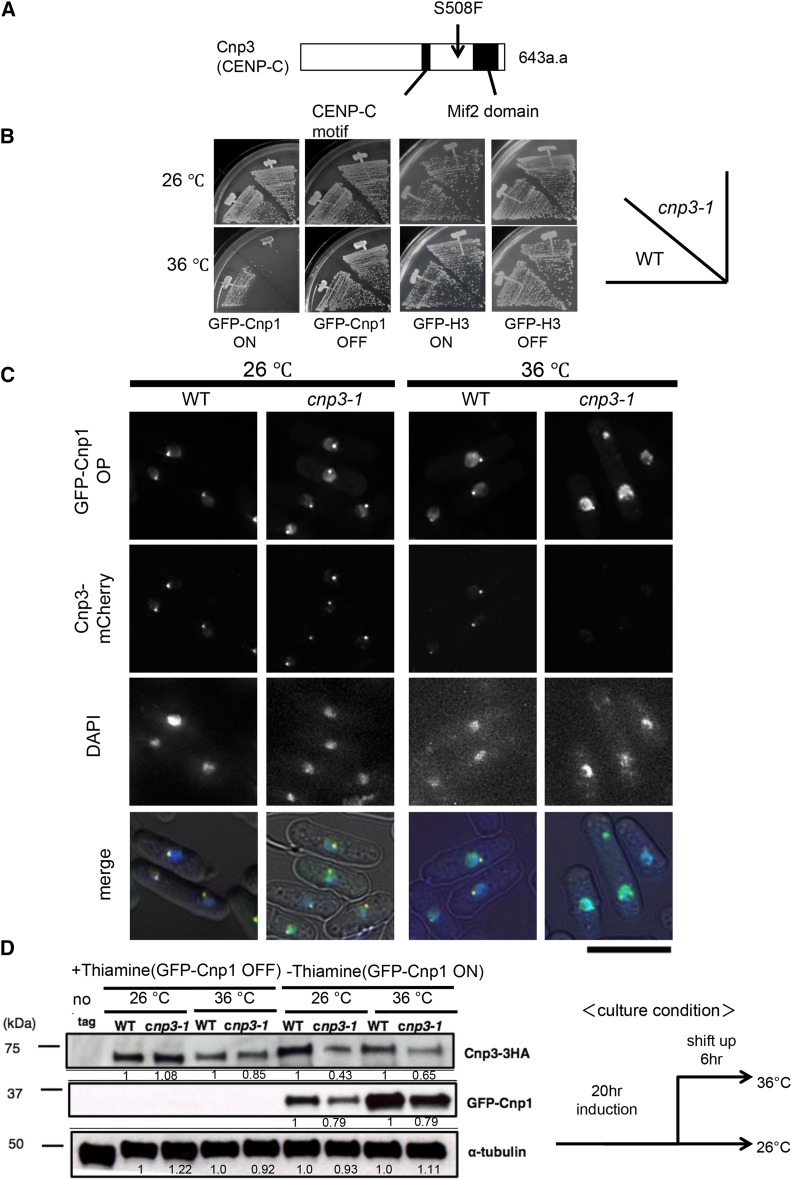
Genetic analysis and subsequent gene cloning revealed the temperature sensitivity upon overexpression of Cnp1 to be attributable to a mutation on the cnp3+ gene encoding a fission yeast homolog of CENP-C. The mutant is thus designated cnp3-1 hereafter. Sequencing of the cnp3-1 mutant gene identified a single point mutation changing TCT of the 508th codon to TTT, resulting in the change in the amino acid from serine to phenylalanine (Figure 1A). As shown in Figure 1B, the cnp3-1 mutant becomes temperature-sensitive upon overexpression of Cnp1, but not histone H3, demonstrating the specific sensitivity of the cnp3-1 mutant to Cnp1.
Figure 1.
Distribution of GFP-Cnp1. (A) Schematic illustration of the structure of Cnp3 protein and the position of the cnp3-1 mutation. (B) The wild type strain (WT) and the cnp3-1 mutant (cnp3-1) were transformed with pREP41-GFP- Cnp1 or GFP- H3. The resulting transformants (MS01, MS02, MS03 and MS04) were streaked as indicated and grown at 26°C or 36°C on EMM 2 media with thiamine for repression (GFP-Cnp1 OFF) or without thiamine for derepression (GFP-Cnp1 ON). (C) Localization of GFP-Cnp1 and Cnp3-mCherry were examined in the wild type (MS213) and cnp3-1 cells (MS217). They were grown at 26°C for 20 hr for induction of GFP-Cnp1 in absence of thiamine, and shifted to 36°C for 6 hr in absence of thiamine. The bar indicates 5 μm. (D) The levels of GFP-Cnp1and Cnp3-HA were examined by western blot in the wild type strain (WT: MS525) and the cnp3-1 mutant (cnp3-1: MS527). They were grown as (C) and the samples were taken 20 hr after the induction at 26 °C and 6 hr after the shift to 36 °C. α-tubulin was used as a loading control.
GFP-Cnp1 foci in the cnp3-1 mutant
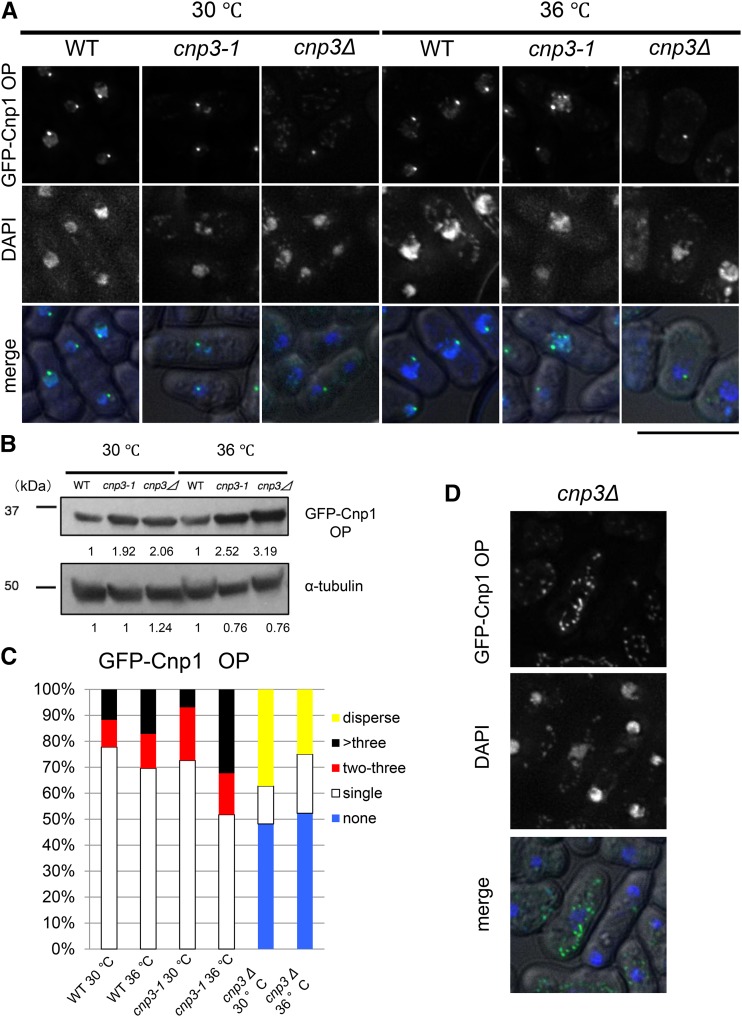
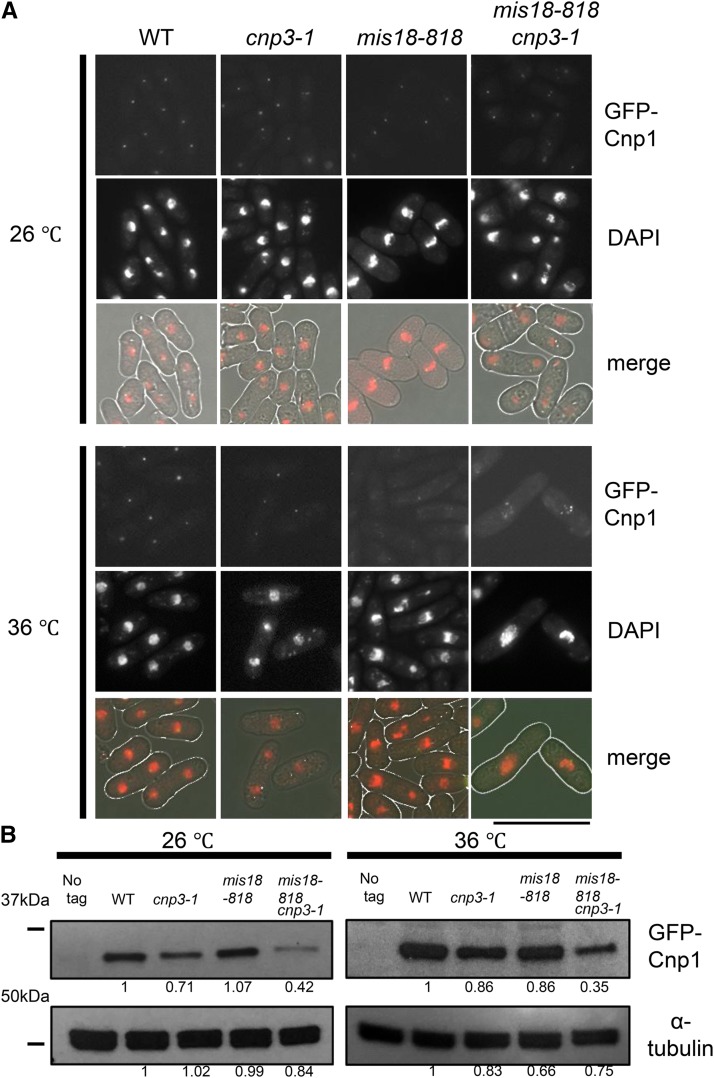
In order to examine distribution of ectopically expressed GFP-Cnp1 in the nucleus, we observed fission yeast cells overexpressing GFP-Cnp1 microscopically in various genetic background (Figure 1C and Figure 2A). We also examined the level of GFP-Cnp1 by immunoblot with an antibody to GFP. As shown in Figure 1D, GFP-Cnp1 was expressed in the cnp3-1 mutant at a level similar to that in the wild type strain at 26° and 36°. Likewise, it was expressed at comparable levels in the three strains (wild type, cnp3-1 mutant and deletion strain for the cnp3+ gene) at 30° and 36° (Figure 2B).
Figure 2.
Comparison between cnp3-1 and cnp3 Δ. (A and D) (A) Localization of GFP-Cnp1 were examined in WT (MS216), cnp3-1 (MS262) and cnp3Δ (MS168) cells. They were grown at 30 °C for 20 hr for induction of GFP-Cnp1 in absence of thiamine, and shifted to 36 °C for 6 hr in absence of thiamine. The GFP-Cnp1 appeared as cytoplasmic speckles in the cnp3Δ cells (MS168) was shown in (D). The bar indicates 5 μm. (B) The level of GFP-Cnp1 was examined by western-blot. α-tubulin was used as a loading control. The samples were taken 20 hr after the induction at 30 °C and 6 hr after the shift to 36 °C. (C) The statistic analysis of (A).
Because three centromeres of fission yeast cluster adjacent to the spindle pole body, SPB (Funabiki et al. 1993), the fluorescence signal of GFP-Cnp1 expressed from the native promoter appears as a discrete single speckle in each cell. Microscopic observation revealed that the signal of GFP-Cnp1 appeared as a single speck in most of the wild type cells (78% at 30° and 70% at 36°) even when GFP-Cnp1 was overexpressed (Figure 2A and 2C), suggesting that GFP-Cnp1 was incorporated into the native centromeres, but not into non-centromeric arm regions.
While distribution of the signal from GFP-Cnp1 in the cnp3-1 mutant at 26 or 30° appeared similarly to that in the wild type cells, it changed dramatically upon the shift to the restrictive temperature, 36° for 6 hr. As shown in Figure 1C, Figure 2A and 2C, the fluorescent signal from GFP appeared as multiple foci (48%), suggesting that the cnp3-1 mutant is prone to promiscuous accumulation of GFP-Cnp1. We also examined distribution of ectopically expressed GFP-Cnp1 in the deletion strain for the cnp3+ gene (cnp3Δ). The cnp3Δ strain is viable at 30°, but not at 36°. As shown in Figure 2A and C, no GFP-Cnp1 focus was observed in approximately 50% of the cnp3Δ cells. In the remaining 50% of the cells, GFP-Cnp1 appeared as a single speckle or cytoplasmic speckles (Figure 2D). Distribution of GFP-Cnp1 was thus strikingly different between the cnp3-1 mutant and cnp3Δ strain. We therefore speculate that the cnp3-1 mutation may not be a simple loss of function.
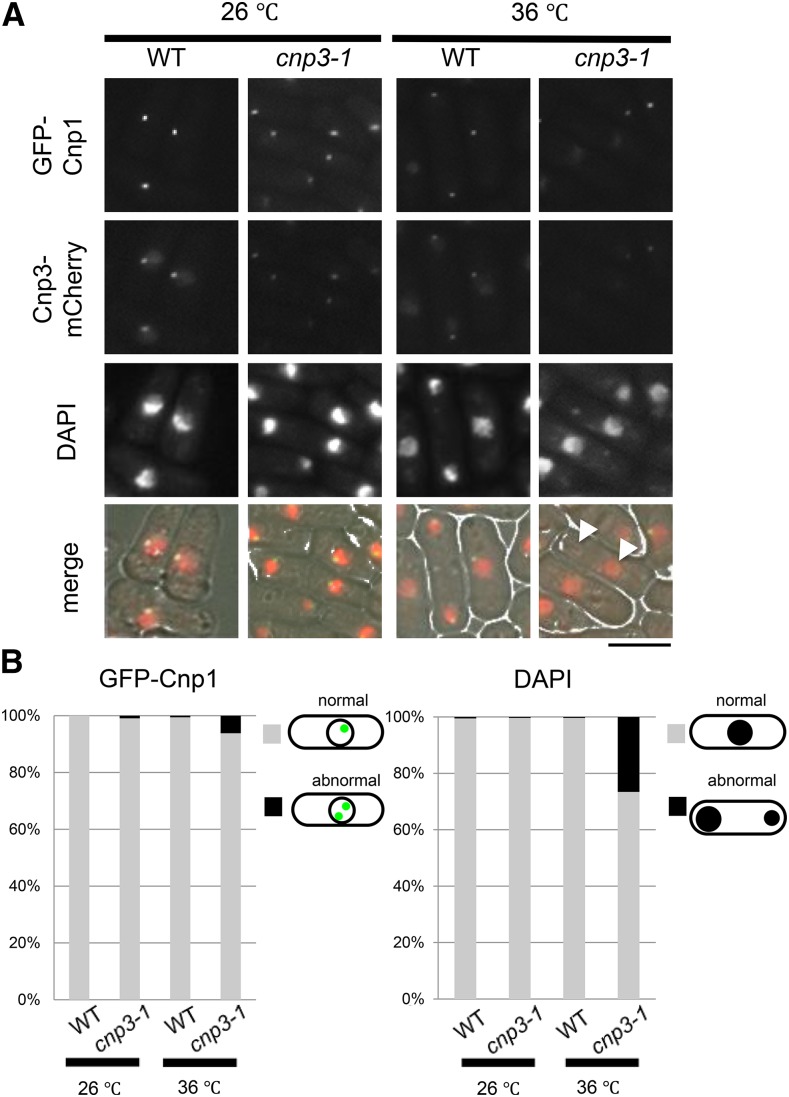
Characterization of Cnp3-1 mutant protein
We next attempted to investigate the cnp3-1 mutant under physiological conditions. The experiments described below were performed with cells expressing Cnp1 from the native promoter, unless otherwise stated. As described previously, tagging GFP to the N-terminal of Cnp1 does not affect growth of fission yeast (Takayama et al., 2008). To visualize the Cnp3 protein, mCherry was tagged to the C-terminus of Cnp3 and Cnp3-1 mutant protein, respectively. When Cnp1 is solely expressed from its native promoter, the cnp3-1 mutant is able to grow at 36°. Microscopic observation of the mutant at 36°, however, revealed that 1) although the signal from GFP-Cnp1 expressed from the native promoter appeared as a single speckle, the intensity of the fluorescent signal was reduced, 2) the level of Cnp3-1 mutant protein at the centromere significantly decreased, and 3) chromosomes segregated unequally in 20% of the cells (Figure 3A and B).
Figure 3.
Characterization of the cnp3-1 mutant. (A) The strains expressing GFP-tagged Cnp1 and mCherry-tagged Cnp3 (WT, MS548), Cnp3-1 mutant protein (cnp3-1, MS549) from the native promoter were grown at 26°C or 36°C for 24 hr and observed under a fluorescence microscope. The white arrow heads indicate mis-segregated chromatin mass scored as abnormal in (B). The bar indicates 10μm. (B) The statistic analysis of (A).
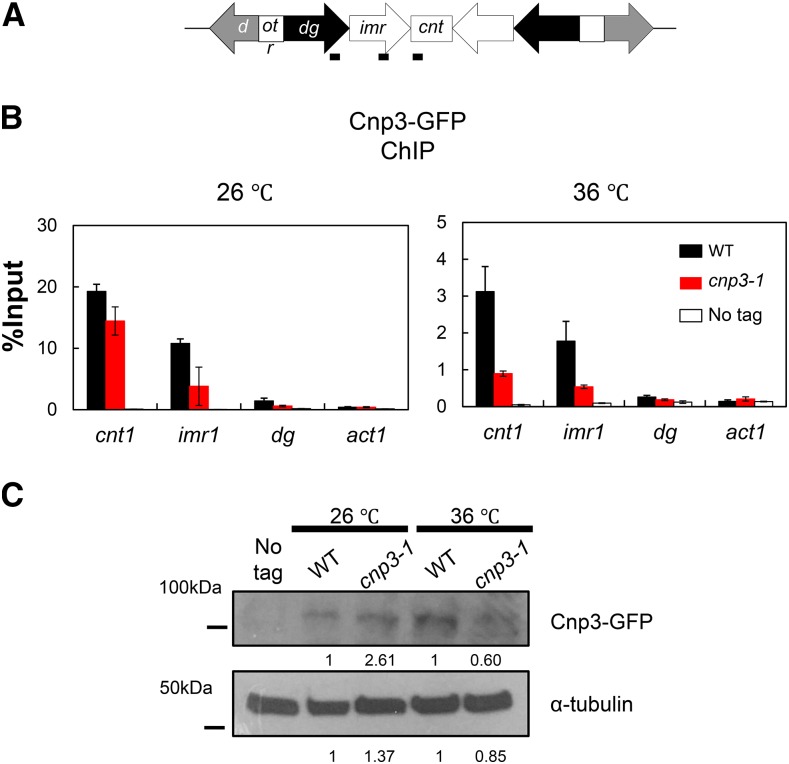
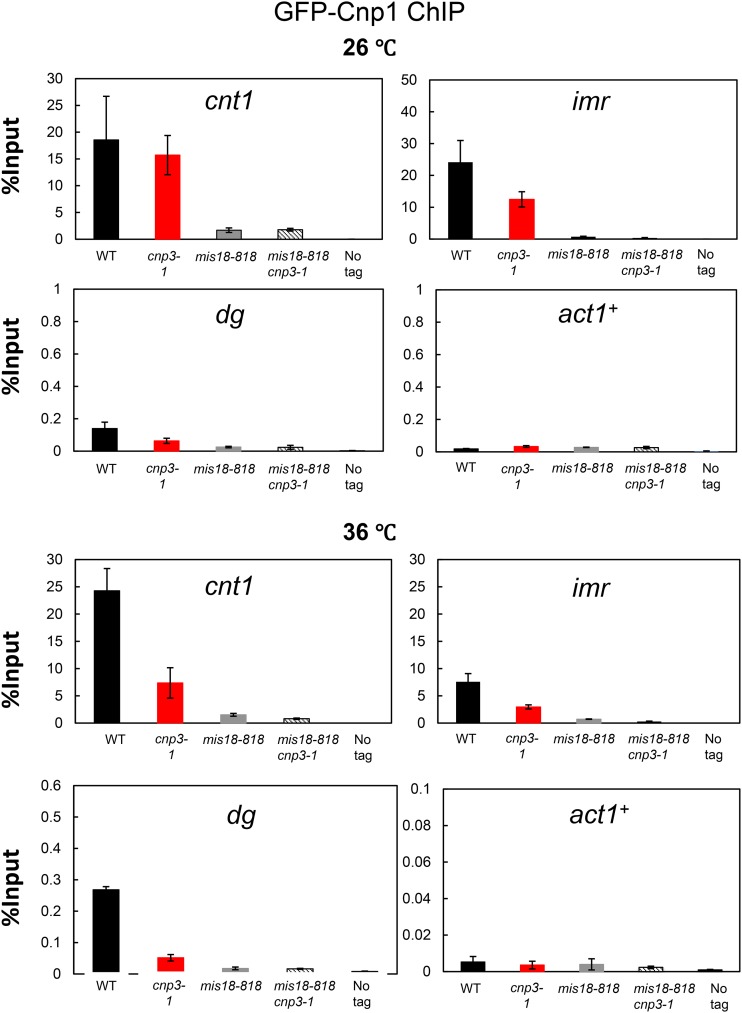
GFP was tagged to Cnp3 and Cnp3-1, respectively, and examined by immunoblot as well as ChIP (Figure 4A and C). As shown in Figure 4B and C, the level of Cnp3-1 mutant protein slightly reduced at 36°. ChIP analysis indicated that the amount of Cnp3-1 bound to centromere chromatin decreased upon the shift to the restrictive temperature (Figure 4A and B). ChIP analysis for Cnp1 also indicated that the amount of Cnp1 bound to centromere chromatin significantly decreased in the cnp3-1 mutant at 36° (Figure 5).
Figure 4.
Loss of Cnp3-1 from the centromere. (A) Schematic illustration of the centromere I (Cen I). Each centromere contains a central domain (cnt), flanked by innermost repeats (imr). This core domain is surrounded by arrays of outer repeats (otr). The black bars indicate the position of the primers used in the ChIP analysis. (B) Localization of Cnp3-GFP in Cen I was examined by ChIP analysis in the wild type stain (WT: MS468) and the cnp3-1 mutant (cnp3-1: MS544). The stains were grown at 26 °C and shifted to 36 °C for 6 hr. (C) The level of Cnp3-GFP was examined by western-blot in the wild type strain (WT: MS468) and the cnp3-1 mutant (cnp3-1: MS544) grown as in (B). α-tubulin was used as a loading control.
Figure 5.
Distribution of Cnp1 at the centromere. Distribution of GFP-Cnp1 at Cen I was examined by ChIP analysis in the indicated mutants (WT: MS343, cnp3-1: MS344, mis18-818; MS503 and cnp3-1 mis18-818: MS534) grown as in Fig. 4B.
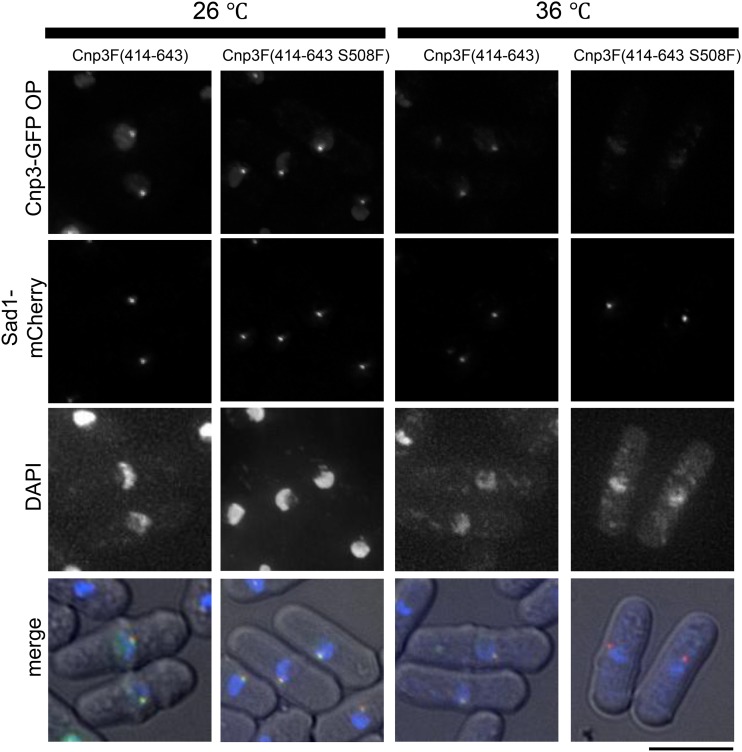
The centromere binding activity was mapped at a domain of Cnp3 spanning from 414 to 643 amino acids (Tanaka et al. 2009). This domain was fused with GFP, designated F(414-643)-GFP, and expressed in the wild type strain. As shown in Figure 6, F(414-643)-GFP was colocalized with SPB, a marker for the cluster of the three centromeres. When the mutation of cnp3-1 (S508F) was introduced in the domain, designated F(414-643 S508F)-GFP, it was colocallized with SPB at 26°, but not at 36°, indicating that F(414-643S508F)-GFP could recapture the phenotype of the full length Cnp3-1 mutant protein.
Figure 6.
S508 of Cnp3 is important for centromere-targeting. Localization of a partial fragment of Cnp3 spanning from 414 to 643 amino acids tagged with GFP were examined at 26°C or 36°C in the wild type strain (MS568 and MS569). Sad1 tagged with mCherry was used as a marker for the centromere cluster. These strain were grown as in Fig. 1C. The bar indicates 5 μm.
These results suggested that the Cnp3-1 mutant protein lost its binding activity to the centromere and allowed a partial loss of Cnp1 from the centromere at 36°, and prompted us to examine the function of the centromere in the mutant. As shown in Table 2, the stability of the mini-chromosome Ch16 was reduced in the cnp3-1 mutant, indicating a low activity of the centromere in the mutant.
Ectopic foci of Cnp1 in cnp3-1
As described above, the cnp3-1 mutant was prone to promiscuous accumulation of Cnp1 at 36° when Cnp1 was overexpressed. We speculated that when Cnp1 was present in excess, Cnp3-1 mutant proteins might interact with Cnp1 and promote promiscuous accumulation of Cnp1. In order to test this scenario under a physiological condition, we attempted to increase the level of free Cnp1 by disabling a system to incorporate Cnp1 in the centromere chromatin. It has been shown previously that Mis18 is responsible for incorporation of Cnp1 into centromeric chromatin (Hayashi et al. 2004; Williams et al. 2009). As shown in Figure 7A, the intensity of GFP-Cnp1 foci decreased at 36° in the temperature sensitive mis18-818 mutant. Remarkably, in the double mutant (mis18-818 cnp3-1), multiple foci of GFP-Cnp1, which were brighter than those in the single mis18-818 mutant, were observed (Figure 7). We speculated that foci of GFP-Cnp1 were formed ectopically in the cnp3-1 mis18-818 double mutant for the following three reasons; 1) ChIP analysis (Figure 5) indicated that the level of Cnp1 bound to the native centromere was extremely low, 2) the foci were formed even in the mis18-818 mutant defective in CENP-A deposition in the native centromere, and 3) the number of the foci in a nuclei occasionally exceeded three, the number of the fission yeast centromeres (Figure 7A).
Figure 7.
Multiple foci of GFP-Cnp1 in mis18 cnp3-1. (A) Cnp1 tagged with GFP was expressed from the native promoter. Each strain (WT: MS343, cnp3-1: MS344, mis18-818: MS503 and cnp3-1 mis18-818: MS534) was grown at 26 °C and shifted to 36 °C for 6 hr. The bar indicates 10 μm. (B) The level of Cnp1 in strains used in (A) was examined by immunoblot. α-tubulin was used as a loading control.
We next examined distribution of Mis12, a protein localized at centromeres in a Cnp1-independent manner (Goshima 2003). As shown in Figure S1, S2 and S3. Mis12 was observed as a discrete single speckle in the cnp3-1 mis18-818 and cnp3-1 mid18-262 double mutant at 26° (Figure S1) and 36° (Figure S2 and S3), suggesting that the centromere cluster was normally maintained. FISH analysis with a probe for the centromere also indicated that the centromere cluster was normally maintained (Figure S4). Furthermore, multiple foci of GFP-Cnp1 formed at 36° were found at positions different from the speckles of Mis12 (Figure S2), demonstrating that the foci of GFP-Cnp1 in the cnp3-1 mis18-818 double mutant were ectopic.
As shown in Figure S5 and S6, multiple foci were also observed when the cnp3-1 mutation was combined with mis6-302 or mis16-53, a mutation defective in deposition of Cnp1 into cenromeric chromatin.
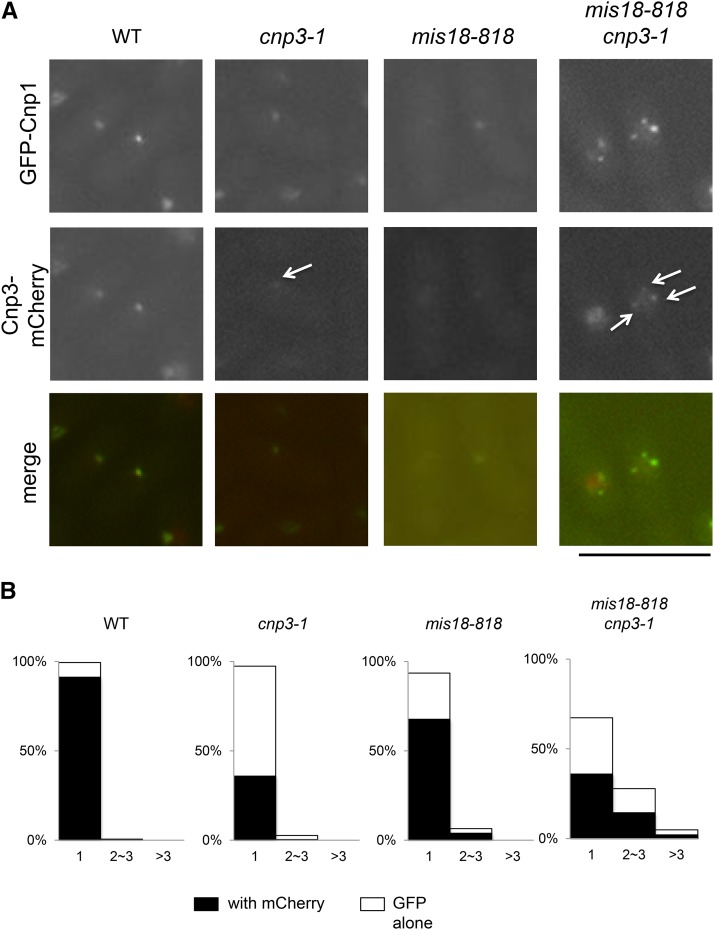
Finally, we examined direct contribution of Cnp3-1 mutant protein to ectopic formation of Cnp1 foci by localizing Cnp3-1 proteins. In the cnp3-1 mutant, weak fluorescent signal from Cnp3-1-mCherry was colocalized with GFP-Cnp1, which presumably represented the native centromere, indicating that the Cnp3-1 mutant protein lost the activity to bind to the native centromere (Figure 8A). In the double mutant (mis18-818 cnp3-1), the signal from Cnp3-1-mCherry, which was easily discernable, was colocalized with multiple GFP-Cnp1 foci, which were likely ectopic. As shown in Figure 8B, more than 33% of the double mutant cells exhibited multiple foci of GFP-Cnp1.
Figure 8.
Colocalization of Cnp1-foci and Cnp3-1 in mis18 cnp3-1. (A) Both Cnp1 tagged with GFP and Cnp3 (or Cnp3-1) tagged with mCherry were expressed from their native promoters in the four strains (WT: MS548, cnp3-1: MS549, mis18-818: MS546 and cnp3-1 mis18-818: MS545). They were grown as in Fig. 7A. The white arrows indicate the Cnp3 (Cnp3-1)-foci. The bar indicates 10 μm. (B) The statistic analysis of (A).
Based on the observation, we speculated that Cnp3-1 mutant protein preferentially recognized ectopically accumulating GFP-Cnp1 and might promote further accumulation.
Discussion
In this study we have characterized the mutant of cnp3+ gene (cnp3-1), which encodes a fission yeast homolog of mammalian CENP-C, with and without overexpression of Cnp1. The mutation site was mapped at S508, which is within the DNA binding domain of Cnp3 (Tanaka et al. 2009).
When Cnp1 is not overexpressed, the cnp3-1 mutant is viable from 26 to 36°. Both microscopic observation and ChIP analysis have revealed that the Cnp3-1 mutant protein looses the binding activity to the native centromeres at 36°. In addition, the level of Cnp1 bound to centromeres decreased. It has been shown by an in vitro study that human CENP-C stabilizes CENP-A nucleosomes on chromatin through physical interaction (Falk et al. 2015). We thus speculate that the reduction in the level of Cnp1 on centromeres is a direct consequence of a loss of the Cnp3-1 mutant protein from centromeric chromatin.
When Cnp1 is overexpressed, the cnp3-1 mutant is unable to grow at 36°. Microscopic observation revealed that multiple foci of GFP-Cnp1 were assembled in the mutant. Because we found cells (32%) with more than three foci of GFP-Cnp1(Figure 2C), some of the multiple foci, at least, are ectopically assembled. In a vast majority of the cnp3Δ strain overexpressing Cnp1, no focus of GFP-Cnp1 was observed, indicating that loss of function of the cnp3+ gene does not result in promiscuous accumulation of Cnp1. We therefore speculate that the single amino acid change, S508F, in Cnp3 causes a gain of function mutation.
The Mis18 protein forms a complex with Mis16 and is required for centromere localization of Scm3, a Cnp1-chaperone(Hayashi et al. 2004; Williams et al. 2009). Consistently with the function of Mis18, incorporation of GFP-Cnp1into centromeric chromatin was significantly reduced in a temperature sensitive mutant, mis18-818. Interestingly, when the mis18-818 mutation was combined with the cnp3-1 mutation, which is also defective in incorporation of Cnp1 into the native centromeres, multiple Cnp1-foci were observed. As Mis12, a protein localized at the centromere independently from Cnp1, appeared as a single speck, the cluster of the native centromeres is normally maintained in the double mutant, mis18 cnp3-1. Multiple Cnp1-foci therefore are ectopic. Furthermore, the Cnp3-1 mutant protein is associated with these foci, suggesting that it may contribute to ectopic accumulation of Cnp1.
It has been demonstrated that human CENP-A is overexpressed in some aggressive cancer cells, where it can be promiscuously incorporated in non-centromeric regions in the form of heterotypic nucleosomes containing H3.3 (Lacoste et al. 2014). The heterotypic nucleosomes is as stable as the CENP-A homo-nucleosome and can bind to CENP-C in vitro (Arimura et al. 2014). Furthermore, Cnp1 accumulation normally depends on Cnp3 (Tanaka et al., 2009). Taking these reports and our results together into consideration, we speculate that 1) the Cnp3-1 mutant has lost the specific binding activity to the native centromere, where the CENP-A homo-nucleosome is a major component in chromatin and 2) it binds to the heterotypic nucleosomes containing H3.3 more efficiently than the wild type Cnp3 protein, and finally 3) when Cnp1 is present in excess (in cells overexpressing Cnp1, or in the double mutant, mis18 cnp3-1, where deposition of Cnp1 into the native centromeres is prevented), the Cnp3-1 protein preferentially binds to and stabilizes the heterotypic nucleosomes promiscuously accumulated on non-centromeric regions. Alternatively it is possible that the Cnp3-1 protein may stabilize the CENP-A homo-nucleosomes promiscuously deposited at non-cetromeric regions.
With the specific binding activity to the native centromere, Cnp3 serves as a part of the epigenetic mechanism to restrict the position of CENP-A accumulation and prevents formation of de novo centromeres.
Acknowledgments
This work was supposed by JSPS KAKENHI (Grant Number JP26116508) to R.M., N.N. and M.Y. acknowledge the generous support of Okinawa Institute of Science and Technology. The Radiation Biology Center is a joint usage research center supported by the MEXT of Japan. A part of this work was performed in collaborative research projects in the Joint Usage Research Center.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6618920.
Communicating editor: C. Hoffman
Literature Cited
- Allshire R. C., Karpen G. H., 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9: 923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G., 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233. 10.1101/gad.9.2.218 [DOI] [PubMed] [Google Scholar]
- Arimura Y., Shirayama K., Horikoshi N., Fujita R., Taguchi H., et al. , 2014. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 4: 7115 10.1038/srep07115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Cleveland D. W., 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144: 471–479. 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A. G., Pidoux A. L., Catania S., Durand-Dubief M., Choi E. S., et al. , 2013. Telomeric repeats facilitate CENP-A(Cnp1) incorporation via telomere binding proteins. PLoS One 8: e69673 10.1371/journal.pone.0069673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania S., Allshire R. C., 2014. Anarchic centromeres: deciphering order from apparent chaos. Curr. Opin. Cell Biol. 26: 41–50. 10.1016/j.ceb.2013.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y. N. K., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M., 1989. composite motifs and repeat symmetry in S.pombe centromeres:direct analysis by integration of Not1 restriction site. Cell 57: 739–751. [DOI] [PubMed] [Google Scholar]
- Choi E. S., Stralfors A., Catania S., Castillo A. G., Svensson J. P., et al. , 2012. Factors that promote H3 chromatin integrity during ternscription prevent promiscuous deposition of CENP-A(cnp1) in fission yeast. PLoS Genet. 8: e1002985 10.1371/journal.pgen.1002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Furuyama S., Biggins S., 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972. 10.1016/j.cub.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Dong Q., Yin F. X., Gao F., Shen Y., Zhang F., et al. , 2016. Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading. Mol. Cell 64: 79–91. 10.1016/j.molcel.2016.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S. J., Guo L. Y., Sekulic N., Smoak E. M., Mani T., et al. , 2015. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348: 699–703. 10.1126/science.1259308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M., 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121: 961–976. 10.1083/jcb.121.5.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Iwasaki O., Obuse C., Yanagida M., 2003. The role of Ppe1/PP6 phosphatase for equal chromosome segregation in fission yeast kinetochore. EMBO J. 22: 2752–2763. 10.1093/emboj/cdg266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., He H., Dong Q., Sun S., Li F., 2014. Ectopic centromere nucleation by CENP–a in fission yeast. Genetics 198: 1433–1446. 10.1534/genetics.114.171173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., et al. , 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729. 10.1016/j.cell.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40: 444–454. 10.1016/j.molcel.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E. M., Biggins S., 2016. Regulation of Budding Yeast CENP-A levels Prevents Misincorporation at Promoter Nucleosomes and Transcriptional Defects. PLoS Genet. 12: e1005930 10.1371/journal.pgen.1005930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., et al. , 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091. 10.1126/science.1158699 [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A., 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa T., Ishii K., Takeda K., Matsumoto T., 2014. The 19S proteasome subunit Rpt3 regulates distribution of CENP-A by associating with centromeric chromatin. Nat. Commun. 5: 3597 10.1038/ncomms4597 [DOI] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A. V., Barth T., et al. , 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53: 631–644. 10.1016/j.molcel.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Matsumoto T., Beach D., 1991. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell 66: 347–360. 10.1016/0092-8674(91)90624-8 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O., Torras-Llort M., Azorin F., 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34: 6247–6255. 10.1093/nar/gkl902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiyama Y., Ohno Y., Kubota Y., Ishii K., 2013. Epigenetically induced paucity of histone H2A.Z stabilizes fission-yeast ectopic centromeres. Nat. Struct. Mol. Biol. 20: 1397–1406. 10.1038/nsmb.2697 [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T., 2011. The ABCs of CENPs. Chromosoma 120: 425–446. 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- Pobiega S., Marcand S., 2010. Dicentric breakage at telomere fusions. Genes Dev. 24: 720–733. 10.1101/gad.571510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quénet D., Dalal Y., 2012. The CENP-A nucleosome: a dynamic structure and role at the centromere. Chromosome Res. 20: 465–479. 10.1007/s10577-012-9301-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40: 455–464. 10.1016/j.molcel.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Masuda F., Takayama Y., Takahashi K., Saitoh S., 2012. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr. Biol. 22: 658–667. 10.1016/j.cub.2012.02.062 [DOI] [PubMed] [Google Scholar]
- Shang W. H., Hori T., Martins N. M., Toyoda A., Misu S., et al. , 2013. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell 24: 635–648. 10.1016/j.devcel.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y., Sato H., Saitoh S., Ogiyama Y., Masuda F., et al. , 2008. Biphasic incorporation of entromeric histone CENP-A in fission yeast. Mol. Biol. Cell 19: 682–690. 10.1091/mbc.e07-05-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H. L., Kagami A., Watanabe Y., 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17: 334–343. 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Thrower D. A., Bloom K., 2001. Dicentric chromosome stretching during anaphase reveals roles of Sir2/Ku in chromatin compaction in budding yeast. Mol. Biol. Cell 12: 2800–2812. 10.1091/mbc.12.9.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower D. A., Stemple J., Yeh E., Bloom K., 2003. Nuclear oscillations and nuclear filament formation accompany single-strand annealing repair of a dicentric chromosome in Saccharomyces cerevisiae. J. Cell Sci. 116: 561–569. 10.1242/jcs.00251 [DOI] [PubMed] [Google Scholar]
- Williams J. S., Hayashi T., Yanagida M., Russell P., 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33: 287–298. 10.1016/j.molcel.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and plasmids are available upon request. All data necessary for evaluating our conclusions are represented within this paper. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6618920.