Abstract
Prenatal exposure to ethanol causes a wide range of adverse physiological, behavioral and cognitive consequences. However, identifying allelic variants and genetic networks associated with variation in susceptibility to prenatal alcohol exposure is challenging in human populations, since time and frequency of exposure and effective dose cannot be determined quantitatively and phenotypic manifestations are diverse. Here, we harnessed the power of natural variation in the Drosophila melanogaster Genetic Reference Panel (DGRP) to identify genes and genetic networks associated with variation in sensitivity to developmental alcohol exposure. We measured development time from egg to adult and viability of 201 DGRP lines reared on regular or ethanol- supplemented medium and identified polymorphisms associated with variation in susceptibility to developmental ethanol exposure. We also documented genotype-dependent variation in sensorimotor behavior after developmental exposure to ethanol using the startle response assay in a subset of 39 DGRP lines. Genes associated with development, including development of the nervous system, featured prominently among genes that harbored variants associated with differential sensitivity to developmental ethanol exposure. Many of them have human orthologs and mutational analyses and RNAi targeting functionally validated a high percentage of candidate genes. Analysis of genetic interaction networks identified Cyclin E (CycE) as a central, highly interconnected hub gene. Cyclin E encodes a protein kinase associated with cell cycle regulation and is prominently expressed in ovaries. Thus, exposure to ethanol during development of Drosophila melanogaster might serve as a genetic model for translational studies on fetal alcohol spectrum disorder.
Keywords: genome-wide association, genetic networks, ethanol sensitivity, Drosophila Genetic Reference Panel, mutational analysis
In humans, prenatal exposure to ethanol causes a wide range of adverse physiological, behavioral and cognitive consequences, including growth deficiency, developmental delay, reduced brain size, and fetal death. These conditions are known as fetal alcohol effects (FAE) or fetal alcohol spectrum disorder (FASD) (Hoyme et al. 2005; Manning and Eugene Hoyme 2007; Memo et al. 2013).
Rodent models have been used to examine morphological and neurological changes that occur following alcohol exposure, but the mechanisms of those effects are still unclear (Kleiber et al. 2011; Kleiber et al. 2012; Schambra et al. 2015; Marquardt and Brigman 2016; Saito et al. 2016). Damage to the heart, brain and skeleton in response to prenatal alcohol exposure has been documented in animal models (Cavieres and Smith 2000; Debelak and Smith 2000; Su et al. 2001; Smith et al. 2014; Sarmah and Marrs 2017). Studies on chicken embryos and cell lines revealed altered expression of genes related to ribosome biogenesis, mRNA splicing and protein processing, as well as energy metabolism and oxidative phosphorylation (Downing et al. 2012; Garic et al. 2014; Rogic et al. 2016). The nervous system is especially susceptible to developmental alcohol exposure, with widespread transcript abundance changes among genes associated with cell adhesion, synaptogenesis and synaptic signaling (Tyler and Allan 2014; Halder et al. 2015; Mandal et al. 2015). However, comprehensive population level studies that can accurately assess genotype by exposure effects are impractical for studies in vertebrate animal models.
Identifying allelic variants and genetic networks associated with variation in susceptibility to prenatal alcohol exposure is especially challenging in human populations, due to incomplete or unreliable maternal drinking histories, and the diversity of phenotypic manifestations, some of which may appear after a time lag. Drosophila melanogaster has been proposed as a model for FASD (McClure et al. 2011; Logan-Garbisch et al. 2014), since developmental ethanol exposure leads to reduced viability and developmental delay. Altered expression of insulin-like peptides and their receptors in the brain (McClure et al. 2011) as well as oxidative stress (Logan-Garbisch et al. 2014) have been implicated as possible mechanisms. Previous studies on the effects of developmental ethanol exposure were, however limited to a few genotypes, focused on selected pathways, and did not provide insights in the genetic underpinnings that determine individual variation in sensitivity to developmental ethanol exposure.
We took advantage of the Drosophila melanogaster Genetic Reference Panel (DGRP; Mackay et al. 2012; Huang et al. 2014) to perform a genome wide association (GWA) analysis to infer candidate genes associated with variation in development time and viability upon ethanol exposure. The DGRP represents a population of fully sequenced, wild-derived, inbred lines with well-annotated genomes. We found extensive variation in viability and development time among DGRP lines grown on regular and on ethanol-supplemented food, with flies developing on average slower when exposed to ethanol. They also showed reduced viability and impaired sensorimotor integration as measured through locomotor reactivity. Analysis of candidate genes revealed a genetic interaction network with Cyclin E (CycE) as a central hub gene. CycE encodes a serine-threonine protein kinase which plays a regulatory role in development and is highly expressed in ovaries (Richardson et al. 1993; Richardson et al. 1995; Sauer et al. 1995). Mutational analyses and RNAi interference experiments provide causal validation for CycE and associated genes as developmental targets for ethanol exposure.
Materials and Methods
Drosophila stocks
We used 201 DGRP lines (Mackay et al. 2012; Huang et al. 2014) reared on cornmeal-molasses-yeast medium (hereafter referred to as standard or regular medium) at 25° and 70% humidity under a 12 hr light-dark cycle (lights on at 6:00 am) to measure viability and development time, and a subset of 39 DGRP lines (Ayroles et al. 2009) to measure locomotor reactivity. The DGRP consists of 205 lines derived from a natural population from North Carolina by 20 generations of full-sib inbreeding followed by whole genome sequencing to high coverage (Mackay et al. 2012). For functional validation seven P{MiET1} mutants and their co-isogenic control w1118iso; 2iso; 3iso (Bellen et al. 2011) were obtained from the Bloomington Drosophila stock center (bab1, CG17150, CG42820, CG43729, CG6024, Nek2, nuf and SKIP; Bloomington, IN). In addition, we obtained 18 RNAi transgenic fly strains of the phiC31 (KK) RNAi library (CG1440, CG32264, CCG34351, CG34370, CG43894, CycE, dve, Egfr, fd59A, ft, fz, Lim1, mam, msn, pbl, sgg, ZnT41F and zormin), together with the corresponding progenitor line (60010) from the Vienna Drosophila RNAi Center (VDRC; Dietzl et al. 2007). These lines and the appropriate progenitor controls were crossed to a weak Ubiquitin-GAL4 driver to suppress the expression of the gene of interest in hybrid F1 offspring (Garlapow et al. 2015).
Viability and development time
To measure viability and development time we allowed parents to lay eggs overnight. The next day we collected eggs from the parental vials and placed 50 eggs per replicate per line on standard medium or on medium supplemented with 10% (v/v) ethanol. We used five replicate vials per line and per medium for measurement of development time and viability. To account for ethanol evaporation all the vials were kept in behavioral chambers with controlled humidity and temperature and all DGRP lines were exposed to the same condition. In addition experimental replicates were done with a randomized design to avoid batch effects. Flies were not exposed to CO2 anesthesia for at least 24 hr prior to the assay. To measure development time we collected and counted eclosing adult flies every morning between 9:00 and 10:00 am. We used the mean eclosion day across all flies as a measurement of development time for each line. We used the fraction of surviving adults out of 50 eggs as a measurement of viability. Sensitivity to ethanol was determined as the difference in viability or development time between flies grown on ethanol-supplemented and regular food.
Locomotion
Locomotor reactivity was measured as startle behavior for 39 DGRP lines, as described previously (Yamamoto et al. 2008). A single three- to five-day-old fly grown on ethanol or regular food was placed in a vial and subjected to a mechanical disturbance by quickly tapping the vial twice on the bench top. The vial was placed horizontally, and the locomotor score was recorded as the amount of time the fly remained mobile within a 60 s period immediately following the disturbance. This assay was performed with 20 replicate measurements per line per sex and per condition.
Quantitative genetic analyses
We used mixed model factorial analysis of variance (ANOVA) to partition variance in replicate means of development time and viability to ethanol exposure among the DGRP lines, according to the model Y = μ + T + L + LxT + ε, where μ is the overall mean, T is the fixed effect of treatment (ethanol or regular), L represents line (random), LxT is the interaction term (random) and ε is the within line (error) variance. We used a reduced ANOVA of form Y = μ + L + ε for each growth condition separately. We estimated variance components from the full model using the restricted maximum likelihood method and calculated broad sense heritability as H2 = σ2G/σ2P, where σ2G is the total genetic variation (σ2L + σ2LT) and σ2P is the total phenotypic variation (σ2L + σ2LT + σ2ε).
For locomotor behavior we obtained data for both sexes; therefore, we partitioned the variance using the ANOVA model: Y = μ + T + S + L + TxS + LxS + LxT + LxTxS + ε, where μ is the overall mean, T is the fixed effect of treatment (ethanol or regular), L represents line (random), S is the fixed effect of sex (females or males), TxS, LxS, LxT and LxTxS are the interaction terms and ε is the within line (error) variance. We used a reduced ANOVA of form Y = μ + S + L + LxS + ε for each growth condition separately. We calculated the broad-sense heritability of locomotor reactivity from the full model as H2 = σ2G/σ2P = (σ2L+ σ2LT + σ2LS + σ2LTS) / (σ2L+ σ2LT + σ2LS + σ2LTS+ σ2ε).
Genome-wide association analysis
We performed GWA analyses for development time and viability on each rearing medium, as well as sensitivity, using a mixed linear model implemented using the pipeline available at http://dgrp2.gnets.ncsu.edu. GWA analyses were performed on line means using 1,876,781 variants that are present at minor allele frequencies of at least 0.05. The effects of Wolbachia pipientis infection, common polymorphic inversions, and polygenic relatedness, were taken into account, as described previously (Mackay et al. 2012, Huang et al. 2014).
Bioinformatics analyses
We annotated DNA variants using the gene models in Flybase release r5.57 (McQuilton et al. 2012). We downloaded the complete genetic interaction networks from FlyBase (release r5.57). The genes in the networks are represented as nodes, whereas edges between the nodes represent interactions. We mapped candidate genes significant at P < 5x10−5 from GWA analyses for viability, development time, and all candidate genes combined from both traits to the graphical interface of genetic networks using the igraph package in R (R Core Team 2016). We then extracted subnetworks from the global networks whose edges were either a direct connection between candidate genes or bridged by only one gene not among the candidate gene list. We evaluated the significance of the size of the largest cluster among the subnetworks by a randomization test in which we randomly extracted subnetworks with the same number of input genes. The P-value was determined by dividing the number of instances where the size of the largest cluster exceeds the observed largest size by the total number of randomizations (α=0.05) (Antonov et al. 2008; Carbone et al. 2016; Fochler et al. 2017).
We performed Gene Ontology enrichment analyses using DAVID software (Huang et al. 2009). Human orthologs were obtained using the DRSC Integrative Ortholog Prediction Tool (DIOPT, version 5.4; http://www.flyrnai.org/diopt; Hu et al. 2011). A gene interaction network for human orthologs was constructed using R-Spider (http://www.bioprofiling.de; Antonov 2011).
Functional analyses of candidate genes
Viability and development time:
We selected nine (CG1440, CG34370, CG42820, CG43729, CG43894, CG6024, msn, nuf and SKIP) and ten (bab1, CG17150, CG32264, CG34351, dve, fd59A, Lim1, Nek2, ZnT41F and zormin) genes for association with alcohol-dependent variation in viability and development time at P < 10−6, respectively. In addition, we functionally tested the six most connected genes in the network associated with phenotypic variation (CycE, ft, fz, mam, msn and sgg) along with two computationally predicted genes (Egfr and pbl). Viability and development time were measured for all genotypes as described above for the DGRP lines, but with 10 replicate vials per genotype.
Locomotion:
Locomotor behaviors were quantified using two different assays, startle induced locomotor reactivity (Yamamoto et al. 2008) and negative geotaxis, for the subset of highly interconnected candidate genes (CycE, ft, fz, and sgg). We analyzed negative geotaxis based on the countercurrent apparatus designed by Benzer (Benzer 1967). We collected 50 3-5 day old flies of the same sex per replicate and performed five replicate assays for mutant lines and their control, reared on ethanol or regular medium. Flies were allowed to recover overnight from CO2 anesthesia. To begin the assay, flies were tapped to the bottom of the first start tube, and the apparatus was positioned vertically. The flies were given 15 s to reach the distal tube. This procedure was repeated seven times, such that flies could choose to go upward a maximum of eight times. At the end of the trial, all eight start tubes containing flies were removed and frozen at −20° before manually counting the flies in each tube. Each individual fly is assigned a score from 0 (did not move up) to 7 (moved up every trial) (Carbone et al. 2016).
Alcohol sensitivity:
Alcohol sensitivity was measured for a CycE –targeted RNAi knockdown line compared to its control as 50% sedation by collecting 10 replicates of 8 flies (3-7 days old) of the same sex per replicate and adding 1 ml of ethanol solution onto a vial plug. The number of intoxicated flies, which lost postural control, was recorded every 2 min. The observation time ended when at least 4 flies became intoxicated (Maples and Rothenfluh 2011).
Data analyses:
We analyzed sensitivity to ethanol exposure by comparing differences in development time, viability, startle-induced locomotor reactivity, negative geotaxis and alcohol sensitivity between ethanol supplemented and regular medium and between mutant lines and appropriate controls, using a mixed model ANOVA of form Y = μ + T + L + T×L + ε, where T indicates the fixed effect of treatment (ethanol vs. regular medium), L indicates mutant or control genotypes (fixed) and ε is the residual variance. For startle-induced locomotor reactivity, negative geotaxis, and alcohol sensitivity we performed data analyses for sexes separately. Significance of the T×L interaction term indicates an effect of the mutation on the sensitivity to ethanol exposure for a given trait.
Data availability
DGRP lines are publicly available from the Bloomington stock center, IN. Raw phenotypic data for line means are presented in Table S1. Supplemental material is available at Figshare: https://doi.org/10.25387/g3.6213629.
Results
Variation in development time and viability of dgrp lines upon ethanol exposure
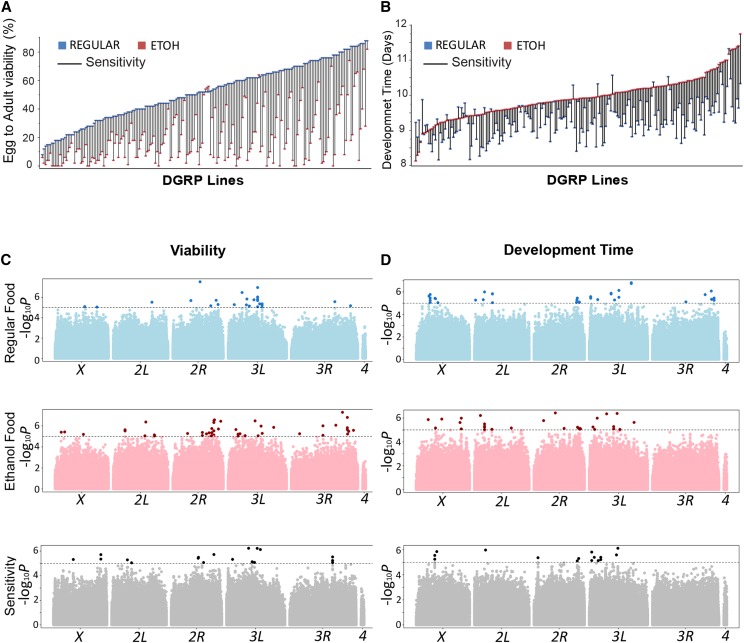
We measured development time and egg-to-adult survival of 201 DGRP lines reared on standard medium and medium supplemented with 10% ethanol. Viability was measured as the fraction of eggs that hatched and developed to adults. We observed extensive variation in viability across the lines, with overall reduced viability when flies were reared on ethanol (Figure 1A). Analyses of variance show significant Line by Treatment interactions, indicating that sensitivity to ethanol exposure as measured by viability or development time is dependent on genetic background (Tables S2 and S3). Broad sense heritability estimates were 0.54 for growth conditions combined and 0.46 for growth on ethanol-supplemented as well as regular medium, showing a substantial genetic contribution to the observed variation (Table S2). However, differences between viability on regular food and ethanol-supplemented food varied greatly for each individual line, indicating substantial variation in sensitivity to developmental alcohol exposure (Figure 1A). The mean viability for flies was 52% reared on standard medium and 20% on ethanol supplemented medium. Sensitivity to ethanol, estimated as the difference of survival between growth on ethanol-supplemented food vs. standard food, ranged from 0 to 100%, with a mean of 37%.
Figure 1.
Phenotypic variation and genome-wide associations for viability, development time and sensitivity to ethanol exposure among 201 DGRP lines. (A) Distribution histogram for variation in viability. (B) Distribution histogram for variation in development time. Blue symbols in panels A and B indicate growth on standard medium and red symbols indicate growth on medium supplemented with 10% ethanol. The differences between the two growth conditions, illustrated by the black connecting lines, represent the sensitivity to ethanol exposure. (C) GWA analysis for viability for flies reared on regular food (top panel), ethanol-supplemented medium (middle panel) and the difference, reflecting sensitivity (lower panel). (D) GWA analysis for development time for flies reared on regular food (top panel), ethanol-supplemented medium (middle panel) and the difference, reflecting sensitivity (lower panel). Sensitivity to ethanol was determined as the difference in viability or development time between flies grown on ethanol-supplemented and regular food. The X-axes in (C) and (D) indicate chromosomal locations. The dashed lines correspond to the P < 10−5 statistical threshold. Darker dots above the line indicate SNPs that pass the statistical threshold.
Development time was measured as the number of days it takes for an egg to develop to adult for 198 of the 201 lines that survived on both ethanol-supplemented and standard medium. Mean eclosion time was 9.3 days for flies reared on standard food and 9.9 days (one half day later) for flies reared on ethanol (Figure 1B). Surprisingly, while on average flies grown on ethanol developed more slowly than those reared on standard medium, some lines developed faster on ethanol-supplemented food (Figure 1B). Heritability estimates were ∼0.3 for growth on ethanol-supplemented as well as regular medium (Table S3). Again, there was wide variation in sensitivity to ethanol exposure, i.e., the difference between development time on ethanol-supplemented food and regular medium. Whereas some lines appeared marginally or not at all affected by this concentration of ethanol, three of the lines were not viable. These lines were excluded from subsequent analyses. Sensitivity of development time, calculated as the difference in mean development time on ethanol-supplemented vs. standard food, ranged from -1.1 to 2.1 days, with an average sensitivity of 0.6 days (Figure 1B).
The correlation between sensitivity to ethanol exposure for viability and development time was not statistically significant (r = 0.08, P = 0.25). However, we found significant positive correlations between development time (r = 0.47; P < 0.0001) and viability (r = 0.48; P < 0.0001) on ethanol-supplemented vs. regular media (Figure S1).
Phenotypic variation in startle-induced locomotor activity after developmental ethanol exposure
To assess the effect of developmental alcohol exposure on sensorimotor behavior, we measured startle-induced locomotor activity in a subset of 39 DGRP lines. On average, startle-induced locomotor activity was higher when flies were reared on regular food and reduced when reared on ethanol-supplemented medium, indicating compromised sensorimotor function (Figure 2). We did not observe significant differences between males and females on average, although there was significant genetic variation in sexual dimorphism of locomotor behavior (Table S4). The heritability estimate for the two growth conditions combined was 0.64 (Table S4).
Figure 2.
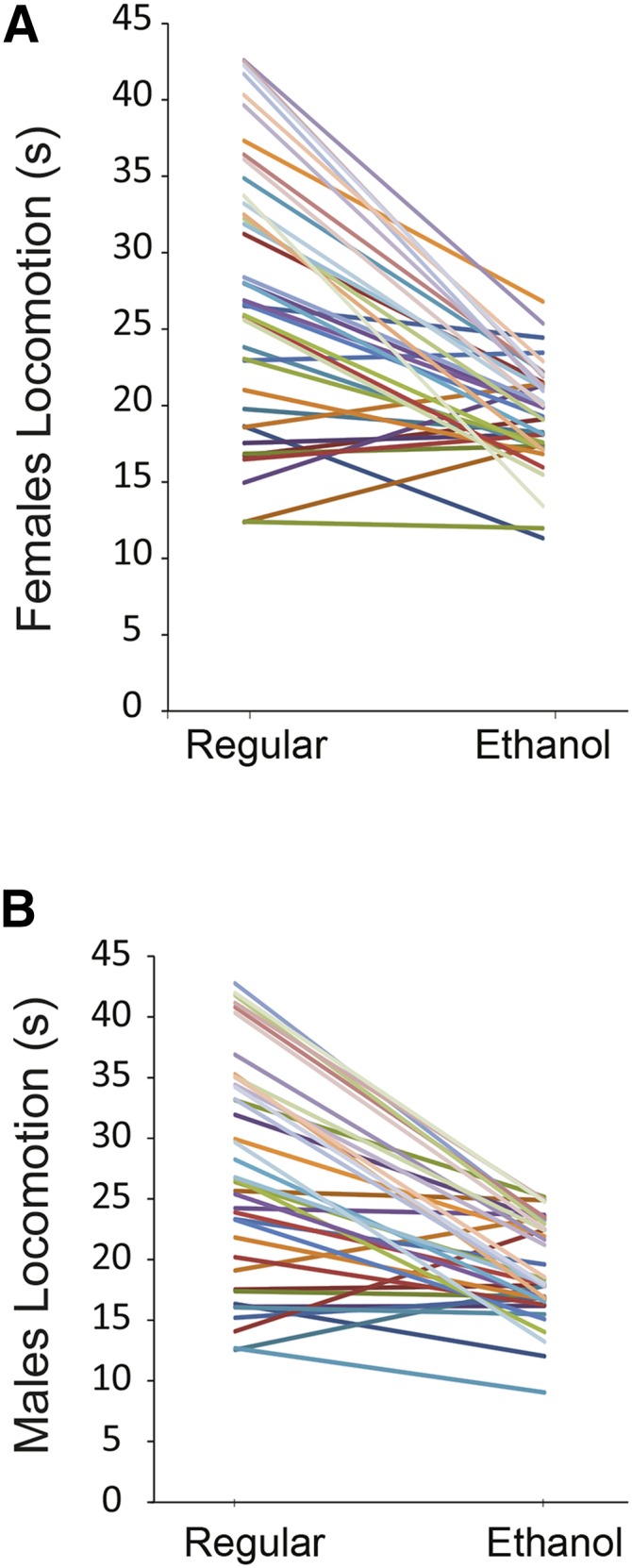
Reaction norms that illustrate variation in startle-induced locomotor activity among 39 DGRP lines grown on regular and ethanol supplemented media for females (A) and males (B). Colors represent the different DGRP lines.
Gwa analyses for development time and viability
We performed single marker genotype-phenotype association analyses using line means for viability and development time on regular and ethanol-supplemented food, and for sensitivity for these traits using the DGRP web portal (http://dgrp2.gnets.ncsu.edu). We tested 1,891,456 variants with minor allele frequencies greater than 0.05, after accounting for effects of Wolbachia infection, common polymorphic inversions, and polygenic relatedness, and identified alleles associated with alcohol sensitivity for development time and viability at a nominal threshold of P < 10−5. Quantile-quantile plots showed deviations of observed from expected values at this significance level (Figure 1, C and D, Figure S2).
We identified 26 polymorphisms in 19 genes associated with viability on regular food, 47 variants located in or near 33 genes that were associated with phenotypic variation in viability on ethanol supplemented food, and 19 polymorphisms in or near 12 genes that contribute to sensitivity of viability (Figure 1C, Table S5). There was no overlap between these SNPs. Candidate genes associated with variation in viability for all conditions represent Gene Ontology categories associated with cell morphogenesis, axonogenesis, and neuron development. At a relaxed P value < 5 x10−5, the number of associated SNPs increased to 284, located in or near 194 genes, and with more input genes Gene Ontology enrichment analysis revealed neuron development, nervous system development, chemotaxis, and organ morphogenesis as significant categories (Table S6).
We identified 37 variants in 27 genes associated with variation for development time when flies were reared on regular food, 30 polymorphisms located in or near 23 genes that were associated with variation for development time on ethanol supplemented food, and 15 polymorphisms in or near 14 genes that contributed to variation in sensitivity of development time (Figure 1D, Table S7). There was no overlap between these SNPs. At P < 5x10−5 we identified 198 SNPs located in or near 138 genes. Gene Ontology enrichment analysis revealed Wnt signaling, signal transduction, tissue morphogenesis and neuron development, associated with variation in developmental time on ethanol-supplemented food (Table S8).
Functional analysis of candidate genes from the gwa analyses
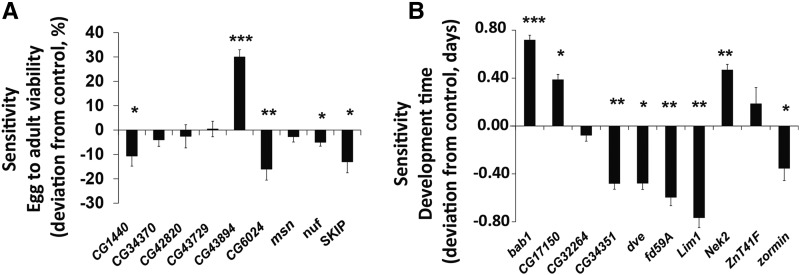
We conducted functional analyses of candidate genes using either RNAi knockdown of gene expression or P{MiET1} insertional mutants corresponding to candidate genes that harbor SNPs associated with sensitivity to ethanol exposure for development time or viability at P < 10−6 (Tables S5 and S7). To separate the effects of developmental mutations from mutations affecting sensitivity to ethanol exposure, we measured viability and development time on regular and ethanol-supplemented media for the mutants and their corresponding controls. Among the nine mutants tested, five (56%) showed a significant difference in viability from the control (Figure 3A; Table S9A). Mutations in CG1440, CG6024, nuf and SKIP caused a decrease in viability, while the CG43894 mutant significantly increased viability when grown on ethanol. This indicates that the wild type CG43894 allele limits viability on exposure to ethanol. CG1440 is predicted to be involved in proteolysis and response to toxic substance (Mi et al. 2010); nuf is associated with actin cytoskeleton reorganization (Riggs et al. 2003; Cao et al. 2008); CG6024 and CG43894 have unknown functions. SKIP has been implicated in sensory perception of smell (Tunstall et al. 2012).
Figure 3.
Functional validation of candidate genes associated with variation in sensitivity to ethanol exposure for viability (A) and development time (B). Data are shown as differences between viability or development time on ethanol-supplemented food vs. regular food and presented as deviation from the appropriate control ± SE * P < 0.05; ** P < 0.001; *** P < 0.0001.
We tested ten mutants/RNAi lines for effects of ethanol exposure on development time, and found that eight of them showed a significant difference between growth on ethanol and regular medium (Figure 3B; Table S9B). bab1, CG17150 and Nek2 mutants showed an increase in development time, while CG34351, dve, fd59A, Lim1 and zormin developed faster on ethanol. These genes are involved in a wide range of biological processes. bab1 is associated with cuticle pigmentation (Kopp et al. 2000; Dembeck et al. 2015); dve is involved in midgut (Nakagawa et al. 2011) and reproductive structure development (Minami et al. 2012); fd59A plays role in controlling egg-laying behavior (Lacin et al. 2014); Lim1 is associated with eye development and regulation of transcription and gene expression (Kojima et al. 2005; Roignant et al. 2010); CG17150 plays a role in sperm competition (Karak et al. 2015); Nek2 contributes to regulation of mitotic nuclear division (Prigent et al. 2005) and protein phosphorylation (Schertel et al. 2013); CG34351 and zormin have unknown functions.
A Genetic interaction network for sensitivity to developmental alcohol exposure
We asked to what extent the 254 genes (Tables S5 and S7) associated with variation in viability on regular or ethanol-supplemented media as well as sensitivity to ethanol at P < 5x10−5 participate in known gene-gene interactions. We identified a network comprised of 42 interacting candidate genes and 82 computationally recruited genes (Figure S3A). Gene ontology analysis of 124 interacting genes showed significant enrichment of organ and tissue morphogenesis, and cell development, including nervous system and sensory organ development (Table S10). CycE, fz, mam, msn and sgg were the most connected candidate genes.
Similarly, we assessed to what extent the 224 (Tables S3 and S5) genes associated with variation in development time on regular or ethanol-supplemented media as well as sensitivity to ethanol (i.e., the difference in development time between the two growth conditions) at P < 5x105, participate in known gene-gene interactions. Here, we identified a network of 39 interacting candidate and 44 computationally recruited genes (Figure S3). Gene ontology analysis of 83 interacting genes showed significant enrichment of genes associated with cell development and nervous system development, organ and tissue morphogenesis, and regulation of signal transduction (Table S11). Bx, ft, kuz and sgg were the most highly connected candidate genes.
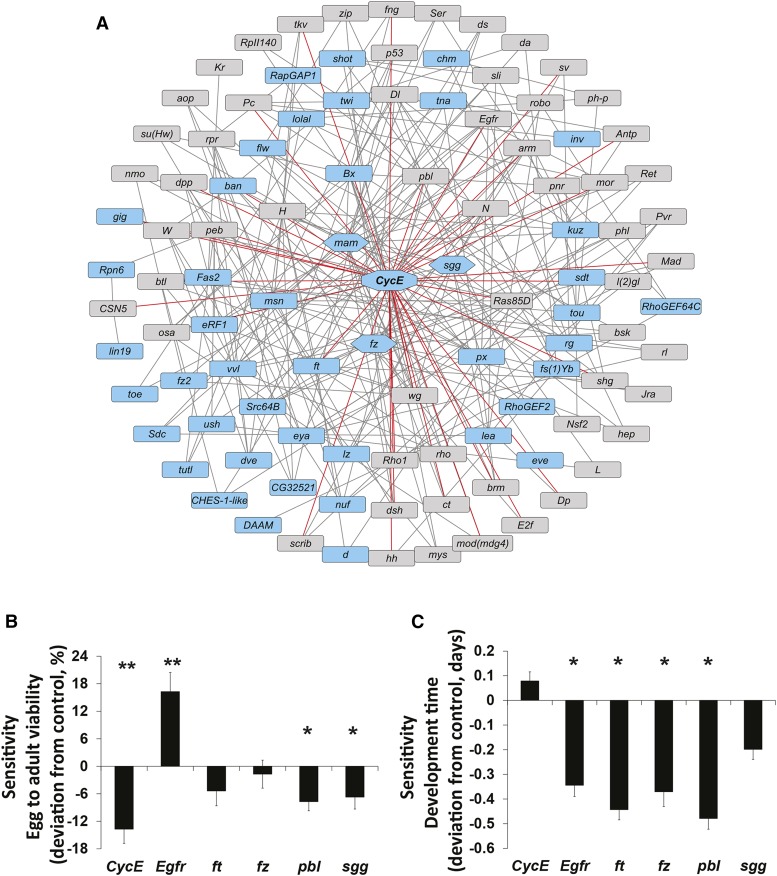
The genetic interaction networks for viability and development time (Figures S7 and S8) did not reach statistical significance individually. However, since viability and development time are related traits, we combined the candidate genes from both GWA analyses and constructed a network using 461 input genes with a significance threshold of P < 5x10−5 (Figure 4). This network was significant compared to the probability of obtaining the same size network by chance (P < 0.001) and consisted of 184 interconnected genes with 78 candidate genes and 106 computationally recruited genes. For clarity, the network depicted in Figure 4 shows only genes with three or more known genetic interactions.
Figure 4.
A genetic interaction network for ethanol-dependent variation in viability and development time. (A) The network consists of candidate genes identified by the GWA analysis at P < 5 × 10−5. We identified 184 interconnected genes (P < 0.001) with 78 candidate genes and 106 computationally recruited genes, i.e., genes that were not identified in our study, but are known to interact with our candidate genes based on published data curated in FlyBase. The network shows only genes with three or more known genetic interactions. The octagon shape in the center indicates the most highly connected CycE gene with red lines highlighting its 54 nodal connections. Hexagons indicate the additional highly connected genes mam, sgg and fz. Blue rectangular boxes indicate candidate genes identified by GWA analyses and gray boxes represent computationally recruited genes. (B) Functional validation of highly connected candidate genes from the network analysis for viability. (C). Functional validation of highly connected candidate genes from the network analysis for development time. * P < 0.05 and ** P < 0.001 indicates a significant difference between sensitivity in RNAi knockdown lines grown on ethanol-supplemented food vs. regular food. Data are shown as deviation from the appropriate control ± SE.
CycE, fz, ft, mam and msn were the most connected candidate genes. CycE stands out with 49 genetic interactions. CycE is associated with variation in viability of flies grown on ethanol-supplemented food, and in the network for development it appears as a computationally recruited gene (Figure 4A). This gene has previously been identified in a network for variation in alcohol sensitivity of adult flies (Morozova et al. 2015). Gene ontology analysis of 184 interacting genes from this combined analysis revealed similar enrichment of developmental genes, including development of sensory organs and oogenesis (Table S12).
Functional analysis of hub genes from the network
Next we assessed the effects on alcohol sensitivity of RNAi knockdown of hub genes in the combined network analysis (Figure 4, B and C; Table S9C and S9D). We measured both viability and development time for eight mutants of highly connected genes in the network (CycE, Egfr, ft, fz, mam, msn, pbl and sgg). We assessed the effect of RNAi knockdown for the hub genes CycE, sgg, mam and fz using a weak Ubiquitin-GAL4 driver by qRT-PCR and found significant reduction (10–30%) in expression levels in the RNAi knockdown background compared to the control. Reduction in msn expression has been confirmed previously (Fochler et al. 2017).
We determined sensitivity to ethanol as the differences in viability or development time between flies grown on ethanol-supplemented food and regular food (Figure 4). We confirmed functional associations for sensitivity to ethanol for viability for CycE, Egfr, pbl and sgg, with Egfr mutants showing increased viability when flies were grown on ethanol containing food compared to regular medium. In contrast, sensitivity to ethanol for development time increased for Egfr, ft, fz and pbl mutants, which developed more slowly on ethanol food compared to regular medium. We did not observe significant differences for either trait for mam and msn mutants.
Finally, we used the same RNAi lines to evaluate pleiotropic effects of the hub candidate genes on locomotion and adult ethanol sensitivity. Adult flies exhibit altered behavioral responses when grown on ethanol-supplemented medium, including changes in locomotor behavior and sensitivity to ethanol vapors. We used two different assays to measure locomotor behaviors, startle-induced locomotor reactivity and negative geotaxis (Figure 5). Startle-induced locomotion showed antagonistic sexual dimorphism with tendencies to decrease in males and increase in females. However, in female mutants an increase in the startle response was only significant for fz, whereas in male mutants statistically significant declines in startle behavior were observed for CycE and ft (Figure 5A). In contrast to startle behavior, negative geotaxis was strongly affected in at least one sex of all mutant lines (Figure 5B). With the exception of fz, the mutants became more active when grown on ethanol. Knock-down time as a measure of alcohol sensitivity was altered only in the CycE mutant males (Figure 5C).
Figure 5.
Pleiotropic effects of the hub candidate genes. (A) Locomotor reactivity. (B) Locomotion. (C) Alcohol sensitivity. Black bars indicate females, white bars indicate males. Data are shown as deviation from the appropriate control ± SE * P < 0.05; ** P < 0.001; *** P < 0.0001.
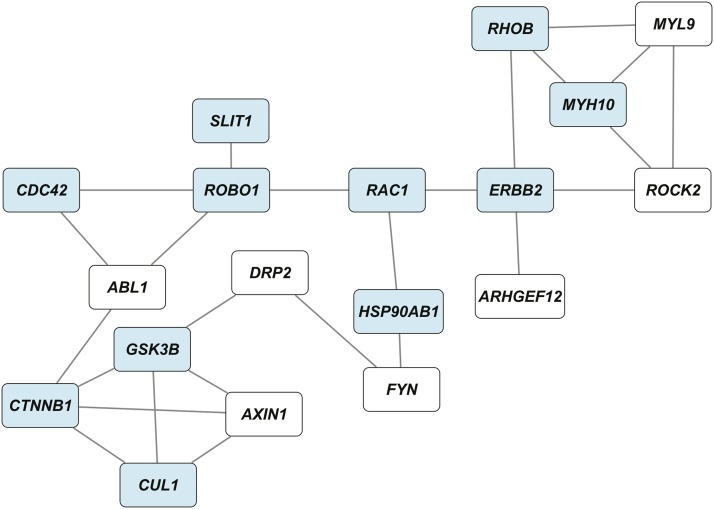
Among candidate genes from the network depicted in Figure 4A, 92% have human orthologs and 18 of them formed a network of interconnected candidate genes (P < 0.005; Figure 6). This network is enriched for genes associated with signal transduction, regulation of protein metabolic processes, and central nervous system development, including brain development (Table S13).
Figure 6.
A genetic interaction network of human orthologs corresponding to Drosophila candidate genes. Human orthologs were identified as counterparts to Drosophila genes depicted in Figure 4A. Blue rectangles indicate genes previously implicated in alcohol-related phenotypes in model systems and humans. The probability to obtain this network by chance is P < 0.005.
Discussion
We harnessed the power of natural variation captured by the Drosophila melanogaster Genetic Reference Panel to identify genes and genetic networks associated with sensitivity to developmental alcohol exposure. Despite the limited size of the DGRP, we were able to identify allelic variants at a lenient P-value. We could then use mutational analyses using transposon-tagged mutants and RNAi to assess whether genes that harbor these allelic variants affect the phenotype.
A previous study also reported that flies grown on ethanol show delayed development, decreased viability, reduced adult size, increased locomotion, and resistance to ethanol vapors (McClure et al. 2011). Developmental delay and reduced viability in this study were attributed to ethanol-mediated inhibition of the insulin signaling pathway. Whereas we were able to document phenotypic variation in locomotor behaviors that depend on sensorimotor integration among 39 DGRP lines, this sample did not provide enough power for GWA analysis. Our study also did not identify variants in genes of the insulin signaling pathway associated with variation in ethanol sensitivity in the DGRP, but instead identified a wide range of developmental genes, in line with previous cell-based or animal model studies (Mandal et al. 2015; Schambra et al. 2015; Fish et al. 2017).
Examination of the genetic interaction network associated with variation in developmental alcohol sensitivity identifies Cyclin E as a central hub gene. The gene product of this highly interconnected gene is a protein kinase associated with cell cycle regulation and oogenesis (Richardson et al. 1993; Richardson et al. 1995; Sauer et al. 1995). CycE regulation is crucial for proper S to G phase transition (Shcherbata et al. 2004). Expression of CycE is controlled by Myc, a transcriptional regulator implicated in the biosynthesis of ribosomes and important for growth and proliferation during normal development (Gallant 2009; Bellosta and Gallant 2010; Quinn et al. 2013). Thus, altered CycE transcript abundance could result in a wide range of phenotypic effects through an indirect influence on ribosome biosynthesis and, hence, protein synthesis. It is of interest to note that high levels of CycE expression occur in ovaries, supporting the notion that developmental exposure to ethanol in Drosophila could serve as a model for fetal alcohol spectrum disorder.
Studies on rodents have proposed that prenatal alcohol exposure can give rise to epigenetic modifications (Perkins et al. 2013; Brown and Feng 2017; Chater-Diehl et al. 2017; Tulisiak et al. 2017). Future studies in Drosophila can assess whether prenatal alcohol exposure results in histone modifications that drive altered gene expression and, if so, to what extent such modifications persist across generations.
Human orthologs can be superimposed on genes contained within the Drosophila genetic interaction network (Figure 6). More than half of these have been previously implicated with effects of developmental ethanol exposure in cell culture models (ERBB2, RAC1, ROBO1, SLIT1 (Tyler and Allan 2014); CDC42, CTNNB1, HSP90AB1, ROBO1 (Garic et al. 2014));, in chicken embryos (CTNNB1, CDC42, GSK3B, HSP90AB1, MYH10 (Berres et al. 2017)); in human embryonic cells (RHOB (Mandal et al., 2015)); in a GWA study for alcohol dependence in people (CTNNB1, SLIT1 (Edenberg et al. 2010)); and in gene expression microarray analysis of human frontal cortex (HSP90AB1 (Lewohl et al. 2000); CUL1 (Liu et al. 2016)).
In conclusion, exploration of the genetic underpinnings of developmental sensitivity to alcohol exposure in Drosophila has pinpointed CycE as a central hub gene and identified human orthologs of Drosophila candidate genes for future studies on human populations to uncover risk alleles for FASD.
Acknowledgments
This work was supported by NIH grant R01-AA016560 and U01-DA041613 to TFCM and RRHA.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6213629.
Communicating editor: D. Threadgill
Literature Cited
- Antonov A. V., Schmidt T., Wang Y., Mewes H. W., 2008. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 36: W347–W351. 10.1093/nar/gkn239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov A. V., 2011. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res. 39: W323–W327. 10.1093/nar/gkr372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. 10.1038/ng.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743. 10.1534/genetics.111.126995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta P., Gallant P., 2010. Myc function in Drosophila. Genes Cancer 1: 542–546. 10.1177/1947601910377490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S., 1967. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. USA 58: 1112–1119. 10.1073/pnas.58.3.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berres M. E., Garic A., Flentke G., Smith S. M., 2017. Transcriptome profiling identifies ribosome biogenesis as a target of alcohol teratogenicity and vulnerability during early embryogenesis. PLoS One 12: e0169351 10.1371/journal.pone.0169351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. N., Feng J., 2017. Drug addiction and DNA modifications. Adv. Exp. Med. Biol. 978: 105–125. 10.1007/978-3-319-53889-1_6 [DOI] [PubMed] [Google Scholar]
- Cao J., Albertson R., Riggs B., Field C. M., Sullivan W., 2008. Nuf, a Rab11 effector, maintains cytokinetic furrow integrity by promoting local actin polymerization. J. Cell Biol. 182: 301–313. 10.1083/jcb.200712036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M. A., Yamamoto A., Huang W., Lyman R. A., Meadors T. B., et al. , 2016. Genetic architecture of natural variation in visual senescence in Drosophila. Proc. Natl. Acad. Sci. USA 113: E6620–E6629. 10.1073/pnas.1613833113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavieres M. F., Smith S. M., 2000. Genetic and developmental modulation of cardiac deficits in prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 24: 102–109. 10.1111/j.1530-0277.2000.tb04559.x [DOI] [PubMed] [Google Scholar]
- Chater-Diehl E. J., Laufer B. I., Singh S. M., 2017. Changes to histone modifications following prenatal alcohol exposure: An emerging picture. Alcohol 60: 41–52. 10.1016/j.alcohol.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Debelak K. A., Smith S. M., 2000. Avian genetic background modulates the neural crest apoptosis induced by ethanol exposure. Alcohol. Clin. Exp. Res. 24: 307–314. 10.1111/j.1530-0277.2000.tb04612.x [DOI] [PubMed] [Google Scholar]
- Dembeck L. M., Huang W., Carbone M. A., Mackay T. F. C., 2015. Genetic basis of natural variation in body pigmentation in Drosophila melanogaster. Fly (Austin) 9: 75–81. 10.1080/19336934.2015.1102807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembeck L. M., Huang W., Magwire M. M., Lawrence F., Lyman R. F., et al. , 2015. Genetic architecture of abdominal pigmentation in Drosophila melanogaster. PLoS Genet. 11: e1005163 10.1371/journal.pgen.1005163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Downing C., Flink S., Florez-McClure M. L., Johnson T. E., Tabakoff B., et al. , 2012. Gene expression changes in C57BL/6J and DBA/2J mice following prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 36: 1519–1529. 10.1111/j.1530-0277.2012.01757.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg H. J., Koller D. L., Xuei X., Wetherill L., McClintick J. N., et al. , 2010. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 34: 840–852. 10.1111/j.1530-0277.2010.01156.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish E. W., Murdaugh L. B., Sulik K. K., Williams K. P., Parnell S. E., 2017. Genetic vulnerabilities to prenatal alcohol exposure: Limb defects in sonic hedgehog and GLI2 heterozygous mice. Birth Defects Res. 109: 860–865. 10.1002/bdr2.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fochler S., Morozova T. V., Davis M. R., Gearhart A. W., Huang W., et al. , 2017. Genetics of alcohol consumption in Drosophila melanogaster. Genes Brain Behav. 16: 675–685. 10.1111/gbb.12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P., 2009. Drosophila Myc. Adv. Cancer Res. 103: 111–144. 10.1016/S0065-230X(09)03005-X [DOI] [PubMed] [Google Scholar]
- Garic A., Berres M. E., Smith S. M., 2014. High-throughput transcriptome sequencing identifies candidate genetic modifiers of vulnerability to fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 38: 1874–1882. 10.1111/acer.12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlapow M. E., Huang W., Yarboro M. T., Peterson K. R., Mackay T. F. C., 2015. Quantitative genetics of food intake in Drosophila melanogaster. PLoS One 10: e0138129 10.1371/journal.pone.0138129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder D., Mandal C., Lee B. H., Lee J. S., Choi M. R., et al. , 2015. PCDHB14- and GABRB1-like nervous system developmental genes are altered during early neuronal differentiation of NCCIT cells treated with ethanol. Hum. Exp. Toxicol. 34: 1017–1027. 10.1177/0960327114566827 [DOI] [PubMed] [Google Scholar]
- Hoyme H. E., May P. A., Kalberg W. O., Kodituwakku P., Gossage J. P., et al. , 2005. A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 Institute of Medicine criteria. Pediatrics 115: 39–47. 10.1542/peds.2004-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., et al. , 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., et al. , 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24: 1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karak S., Jacobs J. S., Kittelmann M., Spalthoff C., Katana R., et al. , 2015. Diverse roles of axonemal dyneins in Drosophila auditory neuron function and mechanical amplification in hearing. Sci. Rep. 5: 17085 10.1038/srep17085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber M. L., Wright E., Singh S. M., 2011. Maternal voluntary drinking in C57BL/6J mice: advancing a model for fetal alcohol spectrum disorders. Behav. Brain Res. 223: 376–387. 10.1016/j.bbr.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Kleiber M. L., Laufer B. I., Wright E., Diehl E. J., Singh S. M., 2012. Long-term alterations to the brain transcriptome in a maternal voluntary consumption model of fetal alcohol spectrum disorders. Brain Res. 1458: 18–33. 10.1016/j.brainres.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Kojima T., Tsuji T., Saigo K., 2005. A concerted action of a paired-type homeobox gene, aristaless, and a homolog of Hox11/tlx homeobox gene, clawless, is essential for the distal tip development of the Drosophila leg. Dev. Biol. 279: 434–445. 10.1016/j.ydbio.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Kopp A., Duncan I., Godt D., Carroll S. B., 2000. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408: 553–559. 10.1038/35046017 [DOI] [PubMed] [Google Scholar]
- Lacin H., Rusch J., Yeh R. T., Fujioka M., Wilson B. A., et al. , 2014. Genome-wide identification of Drosophila Hb9 targets reveals a pivotal role in directing the transcriptome within eight neuronal lineages, including activation of nitric oxide synthase and Fd59a/Fox-D. Dev. Biol. 388: 117–133. 10.1016/j.ydbio.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl J. M., Wang L., Miles M. F., Zhang L., Dodd P. R., et al. , 2000. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol. Clin. Exp. Res. 24: 1873–1882. 10.1111/j.1530-0277.2000.tb01993.x [DOI] [PubMed] [Google Scholar]
- Liu J., Lewohl J. M., Harris R. A., Iyer V. R., Dodd P. R., et al. , 2006. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacol. 31: 1574–1582. 10.1038/sj.npp.1300947 [DOI] [PubMed] [Google Scholar]
- Logan-Garbisch T., Bortolazzo A., Luu P., Ford A., Do D., et al. , 2014. Developmental ethanol exposure leads to dysregulation of lipid metabolism and oxidative stress in Drosophila. G3 (Bethesda) 5: 49–59. 10.1534/g3.114.015040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal C., Park K. S., Jung K. H., Chai Y. G., 2015. Ethanol-related alterations in gene expression patterns in the developing murine hippocampus. Acta Biochim. Biophys. Sin. (Shanghai) 47: 581–587. 10.1093/abbs/gmv050 [DOI] [PubMed] [Google Scholar]
- Manning M. A., Eugene Hoyme H., 2007. Fetal alcohol spectrum disorders: a practical clinical approach to diagnosis. Neurosci. Biobehav. Rev. 31: 230–238. 10.1016/j.neubiorev.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Maples T., Rothenfluh A., 2011. A simple way to measure ethanol sensitivity in flies. J. Vis. Exp. 48: 2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt K., Brigman J. L., 2016. The impact of prenatal alcohol exposure on social, cognitive and affective behavioral domains: Insights from rodent models. Alcohol 51: 1–15. 10.1016/j.alcohol.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure K. D., French R. L., Heberlein U., 2011. A Drosophila model for fetal alcohol syndrome disorders: role for the insulin pathway. Dis. Model. Mech. 4: 335–346. 10.1242/dmm.006411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J., FlyBase Consortium , 2012. FlyBase 101–the basics of navigating FlyBase. Nucleic Acids Res. 40: D706–D714. 10.1093/nar/gkr1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memo L., Gnoato E., Caminiti S., Pichini S., Tarani L., 2013. Fetal alcohol spectrum disorders and fetal alcohol syndrome: the state of the art and new diagnostic tools. Early Hum. Dev. 89: S40–S43. 10.1016/S0378-3782(13)70013-6 [DOI] [PubMed] [Google Scholar]
- Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., et al. , 2010. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38: D204–D210. 10.1093/nar/gkp1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami R., Wakabayashi M., Sugimori S., Taniguchi K., Kokuryo A., et al. , 2012. The homeodomain protein defective proventriculus is essential for male accessory gland development to enhance fecundity in Drosophila. PLoS One 7: e32302 10.1371/journal.pone.0032302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova T. V., Huang W., Pray V. A., Whitham T., Anholt R. R. H., et al. , 2015. Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult Drosophila. BMC Genomics 16: 865 10.1186/s12864-015-2064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y., Fujiwara-Fukuta S., Yorimitsu T., Tanaka S., Minami R., et al. , 2011. Spatial and temporal requirement of defective proventriculus activity during Drosophila midgut development. Mech. Dev. 128: 258–267. 10.1016/j.mod.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Perkins A., Lehmann C., Lawrence R. C., Kelly S. J., 2013. Alcohol exposure during development: Impact on the epigenome. Int. J. Dev. Neurosci. 31: 391–397. 10.1016/j.ijdevneu.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent C., Glover D. M., Giet R., 2005. Drosophila Nek2 protein kinase knockdown leads to centrosome maturation defects while overexpression causes centrosome fragmentation and cytokinesis failure. Exp. Cell Res. 303: 1–13. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2016. R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Quinn L. M., Secombe J., Hime G. R., 2013. Myc in stem cell behaviour: insights from Drosophila. Adv. Exp. Med. Biol. 786: 269–285. 10.1007/978-94-007-6621-1_15 [DOI] [PubMed] [Google Scholar]
- Richardson H. E., O’Keefe L. V., Reed S. I., Saint R., 1993. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development 119: 673–690. [DOI] [PubMed] [Google Scholar]
- Richardson H., O’Keefe L. V., Marty T., Saint R., 1995. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development 121: 3371–3379. [DOI] [PubMed] [Google Scholar]
- Riggs B., Rothwell W., Mische S., Hickson G. R., Matheson J., et al. , 2003. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J. Cell Biol. 163: 143–154. 10.1083/jcb.200305115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogic S., Wong A., Pavlidis P., 2016. Meta-analysis of gene expression patterns in animal models of prenatal alcohol exposure suggests role for protein synthesis inhibition and chromatin remodeling. Alcohol. Clin. Exp. Res. 40: 717–727. 10.1111/acer.13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J. Y., Legent K., Janody F., Treisman J. E., 2010. The transcriptional co-factor Chip acts with LIM-homeodomain proteins to set the boundary of the eye field in Drosophila. Development 137: 273–281. 10.1242/dev.041244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Chakraborty G., Hui M., Masiello K., 2016. Ethanol-induced neurodegeneration and glial activation in the developing brain. Brain Sci. 6:pii E31 10.3390/brainsci6030031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S., Marrs J. A., 2017. Embryonic ethanol exposure affects early- and late-added cardiac precursors and produces long-lasting heart chamber defects in Zebrafish. Toxics 5: E35 10.3390/toxics5040035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Knoblich J. A., Richardson H., Lehner C. F., 1995. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 9: 1327–1339. 10.1101/gad.9.11.1327 [DOI] [PubMed] [Google Scholar]
- Schambra U. B., Goldsmith J., Nunley K., Liu Y., Harirforoosh S., et al. , 2015. Low and moderate prenatal ethanol exposures of mice during gastrulation or neurulation delays neurobehavioral development. Neurotoxicol. Teratol. 51: 1–11. 10.1016/j.ntt.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C., Huang D., Bjorklund M., Bischof J., Yin D., et al. , 2013. Systematic screening of a Drosophila ORF library in vivo uncovers Wnt/Wg pathway components. Dev. Cell 25: 207–219. 10.1016/j.devcel.2013.02.019 [DOI] [PubMed] [Google Scholar]
- Shcherbata H. R., Althauser C., Findley S. D., Ruohola-Baker H., 2004. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131: 3169–3181. 10.1242/dev.01172 [DOI] [PubMed] [Google Scholar]
- Smith S. M., Garic A., Flentke G. R., Berres M. E., 2014. Neural crest development in fetal alcohol syndrome. Birth Defects Res. C Embryo Today 102: 210–220. 10.1002/bdrc.21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B., Debelak K. A., Tessmer L. L., Cartwright M. M., Smith S. M., 2001. Genetic influences on craniofacial outcome in an avian model of prenatal alcohol exposure. Alcohol. Clin. Exp. Res. 25: 60–69. 10.1111/j.1530-0277.2001.tb02128.x [DOI] [PubMed] [Google Scholar]
- Tulisiak C. T., Harris R. A., Ponomarev I., 2017. DNA modifications in models of alcohol use disorders. Alcohol 60: 19–30. 10.1016/j.alcohol.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunstall N. E., Herr A., de Bruyne M., Warr C. G., 2012. A screen for genes expressed in the olfactory organs of Drosophila melanogaster identifies genes involved in olfactory behaviour. PLoS One 7: e35641 10.1371/journal.pone.0035641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler C. R., Allan A. M., 2014. Prenatal alcohol exposure alters expression of neurogenesis-related genes in an ex vivo cell culture model. Alcohol 48: 483–492. 10.1016/j.alcohol.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Zwarts L., Callaerts P., Norga K., Mackay T. F. C., et al. , 2008. Neurogenetic networks for startle-induced locomotion in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 105: 12393–12398. 10.1073/pnas.0804889105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DGRP lines are publicly available from the Bloomington stock center, IN. Raw phenotypic data for line means are presented in Table S1. Supplemental material is available at Figshare: https://doi.org/10.25387/g3.6213629.