Abstract
Purpose
Many hospital-acquired infections (HAIs) can be transmitted by pathogens contaminating hospital surfaces, not efficiently controlled by conventional sanitation, which can indeed contribute to the selection of MDR strains. Bacteriophages have been suggested as decontaminating agents, based on their selective ability to kill specific bacteria. However, there are no data on their stability in detergents and their potential use in routine sanitation. On the other hand, a probiotic-based sanitation system (Probiotic Cleaning Hygiene System, PCHS) was recently shown to stably reduce pathogens on treated surfaces. However, its action is not specific and slow, being based on competitive antagonism. This work aimed to assess the effectiveness of a combined use of phages and PCHS in removing HAI-associated pathogens from different hard surfaces.
Materials and methods
The decontamination ability of phages in PCHS was tested in vitro and in situ, against drug-susceptible or resistant Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa strains, and using bacterial densities similar to those detected on hospital surfaces.
Results
Phages targeted efficiently all tested bacteria, maintaining their full activity when added to the PCHS detergent. Notably, the combined use of phages and PCHS not only resulted in a rapid reduction (up to >90%) of the targeted pathogens, but also, due to the stabilizing effect of probiotics, the pathogens were maintained at low levels (>99%) at later times too, when instead the effect of phages tends to diminish.
Conclusion
These results suggest that a combined biological system might be successfully used in hospital sanitation protocols, potentially leading to effective and safe elimination of MDR pathogens from the hospital environment.
Keywords: drug-resistant bacteria, hospital infections, biological decontamination, bacteriophages, probiotics
Plain language summary
The so-called hospital-acquired infections are often transmitted by microbes contaminating hospital surfaces, which are also often resistant to drugs, consequently causing infections very hard to treat, responsible for millions of deaths in the western world. Unfortunately, conventional chemicals-based cleaning is not effective in eliminating in a stable way such pathogenic microbes, indeed promoting their resistance to disinfectants and drugs. In an attempt to find a method capable of fighting such pathogens, we recently studied a biological approach based on the use of beneficial bacteria, showing that they can abate pathogens without inducing drug resistance. However, probiotic action is neither rapid nor specific. By contrast, bacteriophages are able to kill specific bacteria very rapidly, but their action is limited in time. Consequently, based on the properties of probiotics and bacteriophages, we wanted to test their combined use as a potential system for stably eliminating bacteria mainly responsible for hospital infections, with particular attention to drug-resistant ones. Our results, obtained using an eco-friendly cleanser added with bacteriophages and probiotics, showed that this biological approach is effective in stably eliminating surface pathogens, as it combines the rapid and specific action of bacteriophages with the stabilizing and general action of probiotics. This approach opens new perspectives in the management of infection control in the hospital environment.
Introduction
Healthcare-associated or hospital-acquired infections (HAIs) represent one of the major concerns in the western world, impairing the clinical outcome of up to 15% of all hospitalized patients.1 Every year in the European community about 3.2 million patients acquire an HAI, and 37,000 die as a direct consequence of HAI, also because of the growing presence of multidrug-resistant (MDR) pathogens.1,2
Based on several evidences, the health care environment can significantly contribute to HAI transmission,3–8 as hospital surfaces represent the reservoir of pathogens spread by hospital inpatients and personnel.6 Persistently contaminated surfaces and objects, in fact, continually come into contact with hospitalized subjects, threatening patients’ health just because of hospitalization.1,2 Several studies have shown that hospital surfaces are indeed persistently contaminated by several, often drug-resistant, pathogens,3,5,7–9 most frequently including Staphylococcus spp. (including methicillin-resistant Staphylococcus aureus, MRSA), Enterobacteriaceae (Escherichia coli, Klebsiella pneumoniae), and Pseudomonas spp.3,10–12
S. aureus is a leading cause of hospital-acquired infections in developed countries.13 It is transmissible between patients, via hospital staff, and by the contaminated environment. Currently, S. aureus is the species most commonly associated to blood stream, lung, soft tissue, and skin infections.14 In addition, S. aureus has evolved resistance to multiple antibiotics in the recent decades, and the MRSA group is also resistant to erythromycin, levofloxacin, tetracycline, clindamycin, gentamicin, trimethoprim, and doxycycline, while being usually susceptible to vancomycin.15 However, vancomycin-resistant MRSA might become predominant in the future, leaving clinicians without any treatment option.
Among Enterobacteriaceae, E. coli represents a major cause of several HAIs, especially because of the rising antibiotic resistance in particularly virulent E. coli types, such as Shiga toxin producing E. coli and enteropathogenic E. coli strains.16
Similarly, another Gram-negative bacterium, the opportunistic pathogen Pseudomonas aeruginosa, represents a leading cause of nosocomial infections, mainly in subjects with compromised immune defence. Due to its mechanisms for adaptation, survival, and resistance to a wide range of antibiotics, infections sustained by P. aeruginosa represent an increasing public health threat.
A characteristic shared by all mentioned HAI-associated bacteria is the high prevalence of multidrug resistance. The proportion of Enterobacteriaceae producing extended-spectrum β-lactamases (which confer resistance to many β-lactam antibiotics) and carbapenemases is increasing around the world, and infections sustained by these organisms are often associated with high mortality.17,18 Not surprisingly, all the three mentioned groups have been included in the global priority list of antibiotic-resistant bacteria by the World Health Organization (WHO), with Enterobacteriaceae and P. aeruginosa in the “critical” priority group and S. aureus in the “high” priority group.19
Despite the efforts to prevent infections by such pathogens, their transmission still occurs, suggesting that more effective strategies are needed to eliminate the risk of contracting pathogens from hospital environments.
So far, removal of pathogens from hospital surfaces has been addressed by conventional chemicals-based sanitation, which, however, show important limitations, as it has a temporary effect, a high environmental impact, it is not targeted toward specific pathogens, and most importantly can contribute to the selection of both disinfectant-resistant and antibiotic-resistant pathogens.20,21 This last aspect, in particular, represents a highly undesirable side effect of chemical cleaning, as MDR pathogens have been constantly and rapidly growing in the recent decades and a high proportion of HAIs is currently sustained by them.22,23 Actually, the rising antibiotic resistance of bacterial pathogens appears as one of the most important challenges facing modern medicine, and antibiotic resistance has become so widespread that WHO reports that it is now “one of the biggest threats to global health, food security, and development.”24
In addition, it has been shown that the risk to acquire an infection sustained by a specific pathogen increases for patients occupying the rooms where an infected/colonized patient with such pathogen was previously present in the same hospital room.25–27 Thus, it would be important to develop sanitation procedures that are able to fight specific MDR pathogens and outbreaks associated to the spread of specific pathogens, as conventional disinfectants are not capable of doing so.
Based on these observations, an ideal sanitation approach should be eco-sustainable, able to remove specific HAI-associated pathogens, including those that are antibiotic-resistant, and devoid of undesirable side effect such as drug resistance selection and/or induction.
In the search for effective methods, we recently studied a probiotic-based sanitation system consisting of an eco-friendly cleaning solution added with spores of probiotic bacteria belonging to the Bacillus genus (Probiotic Cleaning Hygiene System, PCHS), showing that it is efficient in stably abating pathogens on hospital surfaces, including drug-resistant strains,28–30 being also safe for hospital inpatients.31 However, being based on competitive antagonism,28 PCHS action is not addressed toward specific microbial targets, and is quite slow, as several weeks are needed to achieve maximum inhibition of pathogens growth on treated surfaces.
By contrast, bacteriophages are characterized as having a very rapid action against specific bacteria, and have consequently been suggested and tested as potential decontaminating agents. Phage application has been proved effective against foodborne bacteria, for treatment of food or food processing surfaces,32–34 as well as against various bacterial targets, including drug-resistant S. aureus and E. coli strains.16,32,35,36 Based on these data, US Food and Drug Administration (FDA) approved the use of specific phages as antimicrobial agents against food contamination by Listeria monocytogenes.37
However, results reported so far showed the need for prolonged contact between phages and target bacteria in aqueous solution,32 which is scarcely compatible with routine sanitation protocols and inpatients presence. Also, phage activity was tested using high bacterial densities, around 108 colony forming units (CFU) per square meter,36 that favor the encounter between phages and target bacteria, facilitating phage infection of bacterial targets, but are not relevant to health care settings surfaces, where the average level of contamination is consistently lower (between 103 and 105 CFU/m2) and dispersed on huge surfaces. This implies that, to be predictive for routine surface sanitation, in vitro tests might be performed using bacterial amounts comparable to those found on hospital surfaces, and limiting as far as possible the time of contact in aqueous solution between phages and target bacteria. In addition, phages are not particularly resistant in a dry environment; thus, the decontamination obtained by phages is unlikely to result in a stable abatement of the targeted pathogens, instead needing repeated treatments to guarantee a low level of the targeted pathogens.
Based on phages and PCHS characteristics, we wanted to assess their potential to be used as a combined product, testing their activity on hard surfaces, against the most common HAI-associated, even drug-resistant, nosocomial pathogens, namely S. aureus, E. coli, and P. aeruginosa.
The results showed that the combined use of phages and PCHS resulted in a strong and stable abatement of the targeted species, suggesting that biological sanitization might be applied in routine cleaning protocols, being potentially able to control a high number of HAI-associated pathogens.
Materials and methods
Bacterial species
The bacterial strains used in in vitro experiments included strains obtained from American Type Culture Collection (ATCC), and wild-type strains isolated from hospital surfaces. ATCC strains included S. aureus (ATCC 25923), E. coli (ATCC 25922), and P. aeruginosa (ATCC BAA-47). Hospital isolates included three strains selected for their drug resistance characteristics: S. aureus (SA2-R73), E. coli (EC-R60), and P. aeruginosa (PA-V6). Hospital isolates were collected by direct sampling of hospital surfaces with 55 mm diameter contact Rodac plates (24 cm2 surface), containing the following selective media: Baird–Parker agar (for Staphylococcus spp., cat. n. 146189), MacConkey agar (for Enterobacteriaceae spp., cat. n. 146427), and cetrimide agar (for Pseudomonas spp., cat. n. 146768) (all bacterial media were from Merck Millipore, Billerica, MA, USA). Grown colonies with morphological S. aureus, E. coli, and P. aeruginosa features were further streaked on the corresponding selective medium and incubated at 37°C for 24 hours, to isolate single pure colonies. Each individual colony was then characterized by Gram-staining, and identified by appropriate biochemical tests (API-Staph, cat. n. 20500, and API-20E, cat. n. 20100; Biomerieux, Florence, Italy).
After identification, all isolates were expanded overnight in tryptic soy broth (TSB, cat. n. 146599; Merck Millipore) at 37°C, frozen in 50% sterile glycerol, and kept at −80°C until use. Each isolate was characterized for antibiotic resistance by conventional disc-diffusion Kirby–Bauer antibiograms, using Mueller–Hinton agar plates (cat. n. 105437; Merck Millipore), testing the following antibiotics: penicillin G (cat. n. CT0043B; Oxoid, Altrincham, UK), ampicillin (Oxoid; cat. n. CT0003B; Oxoid), vancomycin (cat. n. CT0058B; Oxoid), oxacillin (cat. n. CT0040B; Oxoid), ofloxacin (cat. n. CT0446B; Oxoid), cefotaxime (cat. n. CT066B; Oxoid), cefoxitin (cat. n. CT0119B; Oxoid), gentamicin (cat. n. 9026; Liofilchem, Italy), imipenem (cat. n. CT0455B; Oxoid), aztreonam (cat. n. 9008; Liofilchem, Liofilchem, Teramo, Italy), meropenem (cat. n. 9068; Liofilchem), and colistin (cat. n. CT0017B; Oxoid). Zone inhibition diameters were interpreted according to the European Committee on Antimicrobial Susceptibility Testing breakpoint tables for interpretation of minimum inhibitory concentration (MIC) and inhibition zone diameters38 and to the Clinical and Laboratory Standards Institute manual (26th edition).39 In addition, MICs of resistant strains were also measured, accordingly to European Food Safety Authority guidelines, by using antibiotic stripes containing serial dilutions of each antibiotic (cat. n. 92003, 92033, 92006, 92066, 92141, 92009, 92054, 92085, 92099, 92015, 92102, 92057; Liofilchem).
The concentrated PCHS detergent included, as previously described,28 107/mL spores of three species of probiotics belonging to the Bacillus genus, namely Bacillus subtilis, Bacillus pumilus, and Bacillus megaterium.
Bacteriophages
All bacteriophages used in this study were obtained from Eliava Institute (Staphylococcal phage and Pyophage; GA, USA). Phage preparations contained a mixture of selected lytic phages directed against Staphylococcus spp. (staphylococcal phage and Pyophage), as well as phages against Streptococcus spp., Proteus spp., E. coli and P. aeruginosa (Pyophage), at a concentration corresponding to 105–106 total plaque forming units (PFU)/mL. Phage mixtures were stored at 4°C until use, as indicated by manufacturer instructions. Each individual phage component was titrated by PFU counting on the correspondent ATCC bacterial target. Briefly, phage stocks were serially diluted in TSB; then, 100 µL of diluted phages were mixed with 100 µL of bacterial suspension in logarithmic growth phase (OD600nm =0.4; spectrophotometer DU-640B; Beckman Coulter, Brea, CA, USA). The bacteria and phages mixture was then added to 3 mL of soft agar, finally poured onto tryptic soy agar (TSA, cat. n. 146431; Merck Millipore) plates, and allowed to solidify for 15 minutes. Samples were performed in triplicate. After 24 hours incubation at 37°C, PFU were counted at the appropriate dilution.
Individual phage preparations specifically targeting each single bacterial species were obtained from the mixtures by the lysis plaque elution method. Briefly, phage plaques obtained on the plates were collected, disrupted by pulse-vortexing in 1 mL of TSB, and added to 5 mL of the appropriate bacterial culture at OD600nm =0.4. The suspension was incubated at 37°C under mild agitation until the solution was clear. The solution was then filtered through a 0.22 µm pore size membrane filter, added with 15% sterile glycerol, and stored at −80°C until use.
Each individual phage stock was titrated as already described; the final titer of each individual phage preparation was 1010 PFU/mL.
Host range analysis
The host range of each single phage stock was determined by spot testing, performed against all the bacterial strains used in the study. Briefly, overnight bacterial cultures in TSB were subcultured by 1:10 dilution and grown at 37°C under agitation until the suspension reached OD600nm =0.4. Aliquots (100 µL) of bacterial subculture were added to 3 mL of soft agar, overlaid on TSA plates, and allowed to solidify at room temperature for 15 minutes. Phage stocks were serially diluted in phosphate buffered saline (PBS) (with 10-fold increments) and 10 µL aliquots of phage dilutions were added to bacterial lawns, checking their lytic activity after 24 hours of incubation at 37°C. Spot tests were performed in triplicate.
Decontamination tests
The ability of phages to lyse S. aureus, E. coli, and P. aeruginosa on different kinds of hard nonporous surfaces was assessed by in vitro decontamination assays.40 Briefly, target bacteria were grown in TSB until reaching the logarithmic growth phase (checked by spectrophotometric reading, OD600nm =0.4), and then diluted to obtain a final concentration of 4×106 CFU/mL. To mimic the bacterial load detectable on hospital surfaces, 10 µL of suspension were spread on a 24 cm2 surface, obtaining a final density of 100 CFU/24 cm2 (ie, 4×104 CFU/m2), which represents the average value of microbial contamination found on different types of hospital surfaces as detected in previous studies.28,29 Tested surfaces included plastic, glass, and ceramic surfaces (respectively represented by irradiated sterile plastic plates, or glass plates, and ceramic tiles sterilized by autoclave).
Seeded bacteria were allowed to dry for 15 minutes at room temperature; then, 50 µL of the concentrated individual phage solution diluted in PBS at a multiplicity of infection (MOI) of 10, 100, and 1000 were spread on the surface and allowed to dry in a maximum drying time of 10 minutes. Mock treatment with phage buffer alone was used as a control. Denatured alcohol was used as a positive control. Each sample was performed in triplicate.
After 1, 3, 6, and 24 hours, surfaces were directly sampled by contact Rodac plates containing the appropriate selective medium, to collect residual viable bacteria. Each plate, containing samples taken at the different time points, was incubated for 24 hours at 37°C and bacterial load was evaluated by enumerating plate CFU.
The same assays were performed by diluting phage preparations in PCHS eco-sustainable detergent (PCHS; Copma, Ferrara, Italy), already used for routine hospital cleaning.28 The detergent was diluted 1:100 (v/v) in bi-distilled sterile water to obtain the work dilution as indicated by the manufacturer, and then used to suspend and dilute phage stocks. Phage stability was measured after 1, 2, 3, and 7 days at room temperature, by PFU titration on the specific bacterial targets, after removing the bacterial component by centrifugation. Following titration, the decontamination potential of the PCHS added with phages was tested in vitro by the already described decontamination assays, performed using 100 CFU/24 cm2 of target bacteria and phage at 1000 MOI. In parallel, decontamination assays were also performed in situ, using the ceramic sink of a bathroom specifically isolated and artificially contaminated with 105 CFU/m2 of S. aureus (ATCC strain), and treated with PCHS containing phages at 108 PFU/m2 (MOI 1000).
Statistical analysis
Statistical significance was measured by the unpaired one-tailed Student’s t-test (STAT View software; SAS Institute Inc., Cary, NC, USA). Values of p<0.05 were considered statistically significant.
Results
Phage susceptibility of tested bacteria
S. aureus, E. coli, and P. aeruginosa spp., which are also frequently associated to HAI onset, are among the most common pathogens detected as persistent contaminants on hospital surfaces. Based on this observation, we wanted to assess phages effectiveness against such pathogens, using both ATCC reference strains and wild-type MDR hospital isolates belonging to the same target bacterial species. MDR hospital strains were isolated from hospital surfaces and biochemically identified. Prior to use, all bacterial isolates were characterized for antibiotic susceptibility. Drug susceptibility of bacterial targets used in the decontamination assays is summarized in Table 1. For MDR hospital isolates, the MIC is also provided.
Table 1.
Bacterial strains used in decontamination assays
| Bacterial strains | Drug resistance
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | ATM | CTX | FOX | COL | CN | IPM | MEM | OFX | OX | P | VA | |
| Staphylococcus aureus | S | R | S | S | R | S | S | S | S | S | S | S |
| ATCC 25923 | ||||||||||||
| Escherichia coli | I | S | S | S | S | S | S | S | S | R | R | R |
| ATCC 25922 | ||||||||||||
| Pseudomonas aeruginosa | R | S | S | R | S | S | S | S | S | R | R | R |
| ATCC BAA-47 | ||||||||||||
| S. aureus | R | R | R | R | R | I | R | R | R | R | R | S |
| SA2-R73 | (128) | (>256) | (>256) | (>256) | (>256) | (0.25) | (>256) | (48) | (8) | >256 | (>256) | (3) |
| E. coli | R | S | S | R | R | S | S | S | S | R | R | R |
| EC-R60 | (>256) | (0.47) | (0.64) | (96) | (128) | (2) | (0.38) | (0.64) | (0.19) | (>256) | (>256) | (>256) |
| P. aeruginosa | R | S | R | R | S | R | S | S | S | R | R | R |
| PA-V6 | (>256) | (24) | (48) | (>256) | (8) | (12) | (4) | (0.75) | (2) | (>256) | (>256) | (>256) |
Note: MICs (µg/mL) of hospital isolates are also reported in parentheses.
Abbreviations: AMP, ampicillin 10 µg; ATM, aztreonam 30 µg; CTX, cefotaxime 30 µg; FOX, cefoxitin 30 µg; COL, colistin 10 µg; CN, gentamicin 10 µg; IPM, imipenem 10 µg; MEM, meropenem 10 µg; OFX, ofloxacin 5 µg; OX, oxacillin 1 µg; P, penicillin 10 IU; VA, vancomycin 30 µg; MIC, minimal inhibitory concentration; ATCC, American Type Culture Collection; S, susceptible; I, intermediate; R, resistant.
All bacterial targets, including wild-type MDR isolates, were analyzed for phage susceptibility, by spot testing on soft agar.
Each individual phage stock was active in lysing ATCC strains and, with a slightly inferior efficiency, also MDR isolates (Figure 1). Based on these results, decontamination assays phages were tested at 10, 100, and 1000 MOI against ATCC strains, and at 1000 MOI against MDR isolates.
Figure 1.
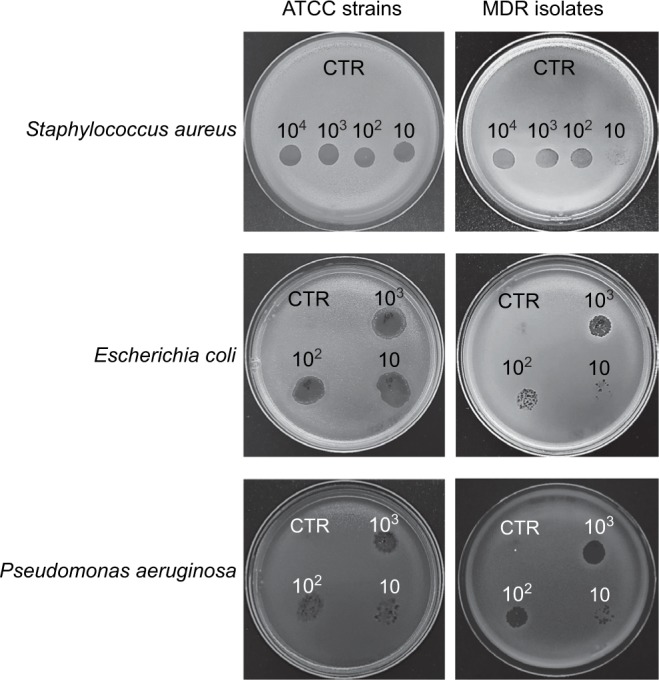
Phage activity against S. aureus, E. coli, and P. aeruginosa (ATCC or MDR strains). Bacteriophage activity was verified by spot tests. Briefly, after suspension in soft agar, bacterial cultures were overlaid on TSA plates; serially diluted phage stocks were added to bacterial lawns, checking their lytic activity after 24 hours of incubation at 37°C. Results are representative of triplicate samples.
Abbreviations: ATCC, American Type Culture Collection; MDR, multidrug-resistant; TSA, tryptic soy agar; CTR, control.
Phage decontaminating potential on hard surfaces
Decontamination assays were carried out first on the ATCC bacterial strains. To mimic contamination conditions similar to those detected in clinical settings, we took as a reference the bacterial load detected on hospital surfaces in previous studies, performed in European hospitals.28,29 Bacterial cells were spread on surfaces at a density of 102 CFU/24 cm2, corresponding to 4×104 CFU/m2. Tested hard nonporous surfaces included sterile plastic, glass, and ceramic. After spreading, bacteria were left to dry for 15 minutes; then, phage preparations, diluted in PBS, were applied uniformly in a 50 µL volume, sufficient to cover the 24 cm2 contaminated area, followed by drying for not more than 10 minutes at room temperature. Mock treatments were performed by using PBS alone, whereas positive control treatments were performed by using a chemical disinfectant (denatured ethanol).
After 1, 3, 6, and 24 hours of incubation at room temperature, surfaces were sampled by applying contact Rodac plates, containing the appropriate selective medium. Residual bacterial CFU on artificially contaminated surfaces were then measured by enumerating grown colonies after 24 hours incubation at 37°C.
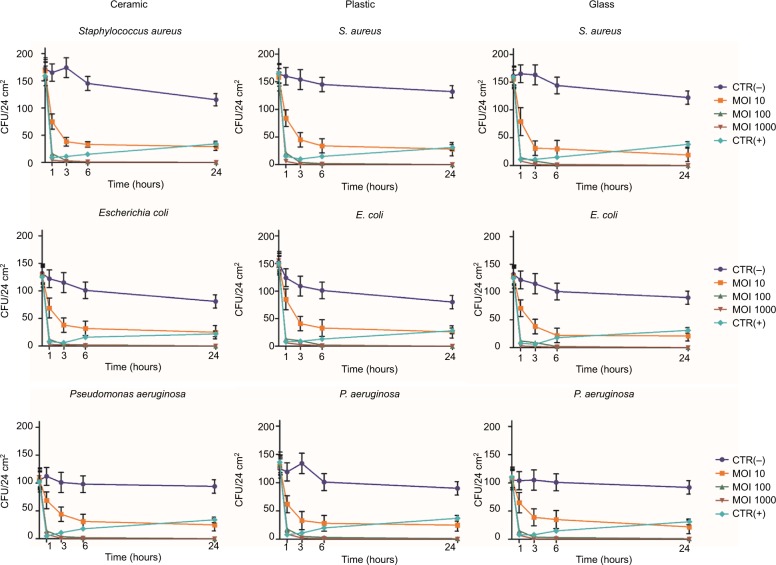
The results evidenced that phages can efficiently reduce viable bacterial cells on contaminated surfaces, even when the bacterial density is relatively low, with a reduction of up to 90±8% compared to mock-treated surfaces, independently of the surface type (ceramic, plastic, glass) and bacterial species (S. aureus, E. coli, P. aeruginosa) (Figure 2). A significant reduction, compared to controls, was detected at 1 hour post treatment at 10 MOI (−40±15%, p<0.05). Phage efficiency increased with increasing MOI, with statistically significant differences between 10 and higher MOIs (100–1000) at all times tested (p<0.01), whereas no significant difference was observed between 100 and 1000 MOI.
Figure 2.
Reduction of bacterial load on different types of hard surfaces treated with specific phages. One hundred CFU of each ATCC bacterial strain (S. aureus, E. coli, and P. aeruginosa) were spread on the indicated sterile surfaces, allowed to dry, and subsequently treated with the specific phages, suspended in PBS, at the indicated MOI. After 1, 3, 6, and 24 hours at room temperature, residual viable cells were measured by Rodac sampling with specific selective media and subsequent CFU count after 24 hours of incubation at 37°C. Negative CTR(−) samples were treated with PBS alone. Positive CTR(+) samples were treated with denatured alcohol. Results represent the mean ± SD of triplicate samples in two independent assays per bacterial target.
Abbreviations: ATCC, American Type Culture Collection; CFU, colony forming units; PBS, phosphate-buffered saline; MOI, multiplicity of infection; CTR, control.
Phage activity also increased with time, as almost no survivors were detected after 6 hours, when using 1000 MOI, and the reduction was maintained in the subsequent 24 hours. By contrast, disinfectant-treated control surfaces showed an evident drop of bacterial cell number within the first 3–6 hours, followed by new detection of viable bacteria after 24 hours, suggesting a bacteriostatic effect, rather than true bacterial killing, had occurred on the surfaces.
Phage effectiveness against MDR isolates spread on hard surfaces
Based on the results obtained on ATCC strains, phage activity was assessed using wild-type drug-resistant S. aureus, E. coli, and P. aeruginosa isolates, collected from hospital surfaces.
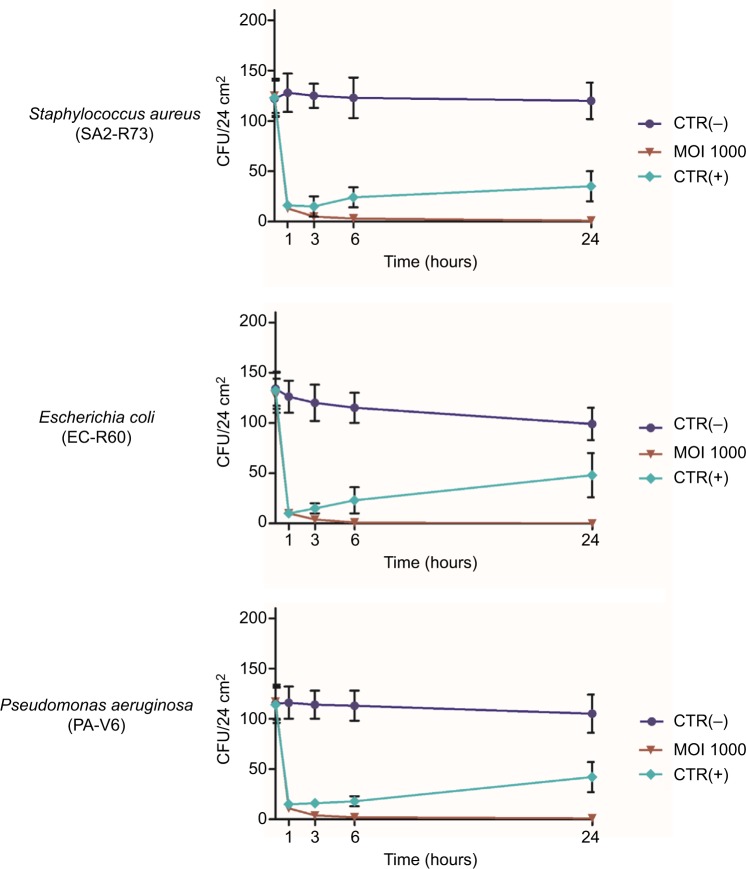
The assays were performed as done for ATCC strains, using an MOI of 1000 and testing all different types of surfaces (ceramic, plastic, glass). Results showed that phages significantly reduced also MDR bacteria on treated surfaces (Figure 3), with no significant differences in the percentage of reduction of drug-resistant strains compared to ATCC strains. In addition, no differences were observed among the different types of hard surfaces used.
Figure 3.
Reduction of MDR hospital isolates on hard surfaces treated with specific phages. One hundred CFU of each bacterial isolate (wild-type S. aureus, E. coli, and P. aeruginosa strains) were spread on ceramic, plastic, or glass surfaces, allowed to dry, and subsequently treated with the specific phages, suspended in PBS, at 1000 MOI. After 1, 3, 6, and 24 hours at room temperature, residual viable cells were measured by Rodac sampling with specific selective media, and subsequent CFU count after 24 hours of incubation at 37°C. Negative CTR(−) samples were treated with PBS alone. Positive CTR(+) samples were treated with denatured alcohol. Results represent the mean of triplicate samples in two independent experiments, for each surface type. As no significant differences were observed between surface types, graphed values represent the mean ± SD of all the measured samples (18 total samples).
Abbreviations: MDR, multidrug-resistant; CFU, colony forming units; PBS, phosphate-buffered saline; MOI, multiplicity of infection; CTR, control.
Phage stability in probiotic detergent
As one objective of this study was to determine whether phages could be used as decontaminants during routine sanitation procedures performed in hospital cleaning, we tested phage stability in the eco-sustainable detergent (PCHS), containing spores of probiotic Bacilli and already used for routine cleaning of hospital surfaces in several Italian hospitals (Copma). Prior to use, concentrated PCHS detergent (pH =8.4) was diluted 1:100 in sterile water, as indicated by the manufacturer, and used to suspend phages at 107 PFU/mL. Phage stability was measured after 1, 2, 3, and 7 days of incubation at room temperature, by PFU titration on the specific bacterial targets, after removing the Bacilli component by centrifugation. The results showed that phages retained 100% activity even after 7 days after dilution (Figure 4). Indeed, both the number and diameter of lysis plaques obtained with phages in PCHS were larger on the same tested bacterial strains, compared to those obtained with phages in PBS, although the differences were not statistically significant (Table 2).
Figure 4.
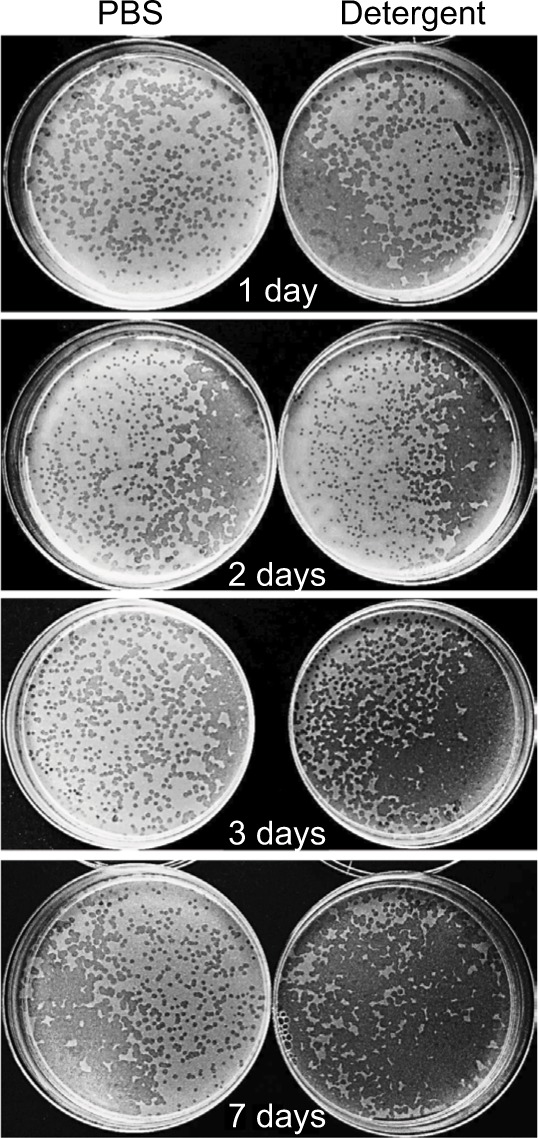
Phage stability in PCHS detergent. Phage stocks were suspended in PBS or in PCHS detergent diluted in water as indicated by the manufacturer, and kept at room temperature in closed plastic tubes for 1, 2, 3, or 7 days. After the indicated times, aliquots were collected and titrated by PFU counting on the corresponding ATCC bacterial target. Samples were performed in duplicate. Pictures refer to anti-E. coli phages. Superimposable results were obtained with the phages directed against S. aureus and P. aeruginosa.
Abbreviations: ATCC, American Type Culture Collection; PCHS, Probiotic Cleaning Hygiene System; PFU, plaque forming units; PBS, phosphate-buffered saline.
Table 2.
PFU titration of phage preparation in detergent and PBS
| Bacteriophage type | Bacteriophage titera
|
||
|---|---|---|---|
| PBS | PCHS | p | |
| Staphylococcus aureus | 7.7±1.9 | 8.5±1.3 | ns |
| Escherichia coli | 7.6±2.0 | 8.4±1.8 | ns |
| Pseudomonas aeruginosa | 7.8±1.8 | 8.6±2.0 | ns |
Notes:
Phages were titrated by soft agar method after 7 days in PBS or PCHS solution. Results are expressed as mean PFU ± SD ×106/mL in triplicate experiments.
Abbreviations: PBS, phosphate-buffered saline; PCHS, Probiotic Cleaning Hygiene System; PFU, plaque forming units; ns, not significant.
Phage/probiotic decontaminating effectiveness “in situ”
Since previous results showed that probiotic-based PCHS detergents were able to gradually and stably abate pathogens on surfaces,28 based on results showing phage stability in PCHS detergent, we wanted to assess the effectiveness of a combined phage–probiotic detergent in reducing bacterial load on surfaces. To this aim, we performed decontamination assays in situ, using S. aureus as the target pathogen. Briefly, a 102 CFU/mL culture of S. aureus was uniformly spread on the ceramic sink of an isolated bathroom, allowed to dry for 24 hours, and then treated by spraying PCHS detergent alone, or added with 105 PFU/mL of anti-staphylococcal phages, corresponding to 1000 MOI.
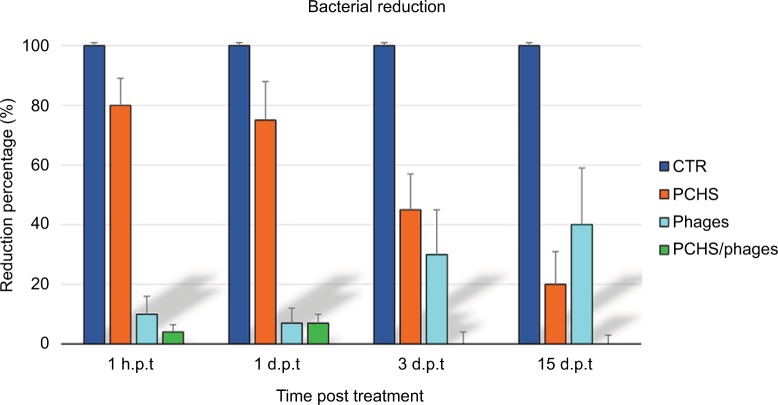
The detergent solution sprayed on the tested surfaces was kept low enough to dry completely in <10 minutes, mimicking routine surface cleaning. After treatment, surface contamination was assayed by application of Baird–Parker Rodac plates after 1 hour, and 1, 3, and 15 days. Both S. aureus and PCHS-Bacilli can grow on Baird–Parker agar, being, however, clearly distinguishable as S. aureus originates black round colonies surrounded by a clear zone, whereas Bacilli give rise to irregular gray-brown colonies. Collected results, summarized in Figure 5, confirmed that phage treatment alone significantly reduced bacterial CFU at early times post treatment (90±5% at 1 and 24 hours after treatment) (p<0.01), as already observed in in vitro assays. However, the reduc tion was not maintained at later times (15 days), likely due to the loss of intact and therefore infectious phages on the surface. By contrast, probiotics alone, began to be active after 3 days, as expected, with a maximum reduction of 75±6% at 15 days after application. Interestingly, the combined use of phages and probiotics resulted not only in a rapid reduction of the target bacteria (94±4% within 1 hour), but also in the persistence of CFU reduction even after 15 days (99±1%). The graph in Figure 6 represents the CFU amount detected in in situ assays following the application of phage, probiotic, or phage–probiotic-based compounds.
Figure 5.
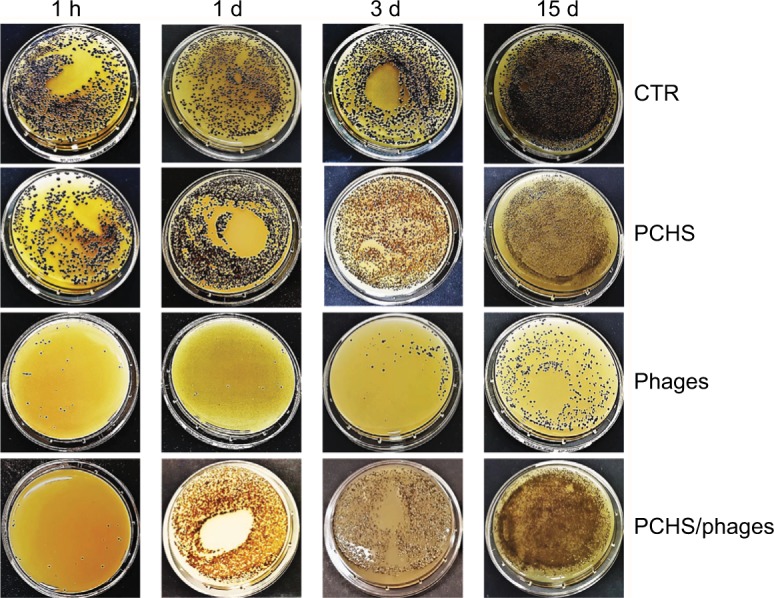
Reduction of Staphylococcus aureus contamination in situ, by a combined phage–probiotic detergent. About 100 CFU of S. aureus (ATCC strain) per 24 cm2 were uniformly spread on the surface of a ceramic sink. After 24 hours, the artificially contaminated surface was treated with water (CTR), probiotic detergent alone (PCHS), anti-staphylococcal phages in PBS alone (phages), or probiotic detergent including anti-staphylococcal phages (PCHS + phages). Phages were used at 1000 MOI. After 1 hour, and 1, 3, and 15 days, surfaces were sampled by Baird–Parker Rodac plates, and residual S. aureus viable cells were counted by enumerating black round colonies after 24 hours of incubation at 37°C. PCHS-Bacilli gave rise to gray-brown irregular colonies on Baird–Parker medium, easily distinguishable from the S. aureus ones. Results are representative of duplicate samples in three independent experiments.
Abbreviations: h, hours; d, days; ATCC, American Type Culture Collection; PCHS, Probiotic Cleaning Hygiene System; CFU, colony forming units; PBS, phosphate-buffered saline; MOI, multiplicity of infection; CTR, control.
Figure 6.
Average reduction of Staphylococcus aureus contamination in situ, by a combined phage–probiotic detergent. Surfaces were artificially contaminated with S. aureus and subsequently treated as described in Figure 5. Results are expressed as mean value ± SD of duplicate samples from three independent experiments. CTR, water; PCHS, probiotic detergent alone; phages, anti-staphylococcal phages in PBS alone; PCHS + phages, probiotic detergent including anti-staphylococcal phages.
Abbreviations: PCHS, Probiotic Cleaning Hygiene System; PBS, phosphate-buffered saline; CTR, control; h.p.t, hours post treatment; d.p.t, days post treatment.
Discussion
Phages have been repeatedly suggested as potential decontaminating agents against foodborne pathogens on food and food processing surfaces,32–34 leading to approval by the FDA of the use of phages for decontamination from the food pathogen L. monocytogenes.37 More recently, the emergence of increasing antibiotic-resistant bacterial pathogens has directed attention on methods capable of controlling their spread and growth, both in infected subjects and surfaces of health care settings. Toward this aim, phages have been recently tested against drug-resistant strains of E. coli and S. aureus, showing good activity on surfaces and fomites,32,35,36 and suggesting their potential use as disinfectants substitutes or disinfectants supplements.
Although promising, most reported phage-based studies appear barely applicable in routine sanitation, as prolonged contact between phages and target bacteria in aqueous solution is generally needed,32 consequently needing that surfaces remain wet for a long time, that is not compatible with inpatients presence. In addition, in vitro phage activity was tested on highly contaminated surfaces, using bacterial densities (around 108 CFU/m2), which facilitate the encounter between phages and target bacteria,36 but are not consistent with what is found on health care settings surfaces, where the mean level of contamination is 3–5 logs lower (around 103–105 CFU/m2). This implies that, to be predictive, in vitro tests should be performed using bacterial amounts comparable to those detectable on hospital surfaces. Interestingly, however, a recent paper reported a decrease of Acinetobacter baumannii infections upon using the application by aerosol of specific anti-Acinetobacter phages in addition to chlorine-based disinfection for terminal sanitation of intensive care unit (ICU) rooms,41 suggesting they might be used effectively to reduce specific pathogens in hospital rooms, although the modality used for phage application was only compatible with sporadic use, such as during terminal sanitation, when the room is empty. Also, a high-phage concentration was used to treat a small space, and no information about the relative abundance of the bacterial target and the specific bacteriophages was available, rendering it difficult to draw a conclusion about the possible general use of phages in routine sanitation. On the other hand, a probiotic-based sanitation procedure has been shown to stably abate surface pathogens,28–30 potentially controlling a high number of HAI-associated pathogens. The system, named PCHS, consists in a mild eco-friendly chemical detergent added with spores of Bacillus probiotics. Due to the action, essentially based on competitive antagonism mechanisms, PCHS effects are slow and not specific.
Based on the promising and complementary characteristics of bacteriophages and probiotics, the aim of this study was to analyze the stability of phages in the PCHS detergent, to ascertain the feasibility of a combined phage/probiotic sanitation in routine protocols used for hospital environments.
To this purpose, we used some of the HAI-associated pathogens most frequently detected as persistent contaminants on hospital surfaces, namely S. aureus, E. coli, and P. aeruginosa, as target bacteria. Tested hard surfaces included ceramic, plastic, and glass, as these surface typologies are often present in hospital rooms and environment. Furthermore, as hospital pathogens are often MDR, because of the selective pressure exerted by the use of antibiotics, we also examined phage ability to eliminate drug-resistant strains of S. aureus, E. coli, and P. aeruginosa. Such strains were isolated from hospital surfaces, thus representing more closely what can be actually found in hospital environments.
To mimic bacterial densities comparable to those found on hospital surfaces, target bacteria were seeded at 100 CFU/24 cm2, corresponding to 4×104 CFU/m2, representing a realistic contamination value, based on previous studies.29 In addition, to imitate what is done during sanitation procedures, we kept the contact time between phages and bacteria in water solution as low as far as possible, using phage volumes drying in a maximum time of 10 minutes.
The results showed that phages can be active in decontaminating all the types of hard surfaces tested, without any detectable difference between surface types and bacterial strains, indicating that phages in vitro are active in removing pathogen levels similar to those detected on field on hospital surfaces. In addition, phages proved to be comparably active against MDR hospital isolates, highlighting their ability to eliminate drug-resistant pathogens, not only in infected subjects, where bacterial concentration and growth rate are high, but also on inanimate surfaces, where the bacterial density and growth rate are sensibly lower.
Notably, phages not only retained their full activity when suspended in PCHS detergent at work dilution, but indeed also exhibited stronger antibacterial activity compared to that observed when phages are suspended in PBS. This suggests that the cleaning chemicals contained in the eco-friendly probiotic detergent might somehow stabilize phages at room temperature, favor the contact between phage and bacterial targets, or facilitate the entrance/action of phages in the bacterial cell.
Consistently with the results obtained in vitro, as well as with previous observations conducted with probiotic-detergent on hospital surfaces, in situ assays showed that the combined probiotic–phage application resulted in remarkable decontaminating activity, compared to the individual probiotic and phage components alone. In fact, viable bacterial targets dropped very rapidly (within 1 hour), in contrast to what was observed with probiotics alone, likely as a consequence of the killing ability of phages. By contrast, contrarily to what was observed with phages alone, bacterial contamination was maintained stably low for a prolonged time interval (till 2 weeks, representing the assay time end), likely due to the stabilizing action of the probiotic component of the detergent, that, acting by competitive inhibition of target bacteria, is able to overcome the problem associated to loss of infectivity of phages on treated surfaces.
Conclusion
Our data indicate that a phage/probiotic detergent might be suitable for use as routine sanitizing agent. This, especially in consideration of the high proportion of antibiotic-resistant isolates on hospital surfaces, opens new perspectives for the development of innovative products and systems aimed at the prevention of HAIs transmitted by contaminated surfaces in the hospital environment.
Data availability
The datasets supporting the conclusions of this article are included within the article and its additional files.
Acknowledgments
We thank Dr Mzia Kutateladze for critical revision of the manuscript. We thank Iva Pivanti and Maddalena Coccagna for the excellent technical support. We acknowledge the assistance offered by Linda Sartor for revising the English manuscript.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Allegranzi B, Bagheri Nejad S, Combescure C, et al. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet. 2011;377(9761):228–241. doi: 10.1016/S0140-6736(10)61458-4. [DOI] [PubMed] [Google Scholar]
- 2.Suetens C, Hopkins S, Kolman J, Diaz Högberg L. Point Prevalence Survey of Healthcare Associated Infections And Antimicrobial Use in European Acute Care Hospitals. Stockholm, Sweden: European Centre for Disease Prevention and Control; 2013. [Google Scholar]
- 3.Boyce JM. Environmental contamination makes an important contribution to hospital infection. J Hosp Infect. 2007;65(Suppl 2):50–54. doi: 10.1016/S0195-6701(07)60015-2. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell BG, Dancer SJ, Anderson M, Dehn E. Risk of organism acquisition from prior room occupants: a systematic review and meta-analysis. J Hosp Infect. 2015;91(3):211–217. doi: 10.1016/j.jhin.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Hota B. Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin Infect Dis. 2004;39(8):1182–1189. doi: 10.1086/424667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dancer SJ. Controlling hospital-acquired infection: focus on the role of the environment and new technologies for decontamination. Clin Microbiol Rev. 2014;27(4):665–690. doi: 10.1128/CMR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otter JA, Yezli S, French GL. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect Control Hosp Epidemiol. 2011;32(7):687–699. doi: 10.1086/660363. [DOI] [PubMed] [Google Scholar]
- 8.Dancer SJ. The role of environmental cleaning in the control of hospital-acquired infection. J Hosp Infect. 2009;73(4):378–385. doi: 10.1016/j.jhin.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Weber DJ, Anderson D, Rutala WA. The role of the surface environment in healthcare-associated infections. Curr Opin Infect Dis. 2013;26(4):338–344. doi: 10.1097/QCO.0b013e3283630f04. [DOI] [PubMed] [Google Scholar]
- 10.Weber DJ, Rutala WA. Role of environmental contamination in the transmission of vancomycin-resistant enterococci. Infect Control Hosp Epidemiol. 1997;18(5):306–309. doi: 10.1086/647616. [DOI] [PubMed] [Google Scholar]
- 11.Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38(5 Suppl 1):S25–S33. doi: 10.1016/j.ajic.2010.04.196. [DOI] [PubMed] [Google Scholar]
- 12.Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119(9):2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diekema DJ, Pfaller MA, Schmitz FJ, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 15.McDougal LK, Fosheim GE, Nicholson A, et al. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob Agents Chemother. 2010;54(9):3804–3811. doi: 10.1128/AAC.00351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamal M, Hussain T, Rajanna Das C, Andleeb S. Isolation and characterization of a myoviridae MJ1 bacteriophage against multi-drug resistant Escherichia coli 3. Jundishapur J Microbiol. 2015;8(11):e25917. doi: 10.5812/jjm.25917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaden JT, Park LP, Maskarinec SA, Ruffin F, Fowler VG, Jr, van Duin D. Results from a 13-year prospective cohort study show increased mortality associated with bloodstream infections caused by pseudomonas aeruginosa compared to other bacteria. Antimicrob Agents Chemother. 2017;61(6):e02671–16. doi: 10.1128/AAC.02671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palacios-Baena ZR, Gutierrez-Gutierrez B, De Cueto M, et al. Development and validation of the INCREMENT-ESBL predictive score for mortality in patients with bloodstream infections due to extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother. 2017;72(3):906–913. doi: 10.1093/jac/dkw513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017. [Accessed April 4, 2018]. Available from: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 20.Bock LJ, Wand ME, Sutton JM. Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J Hosp Infect. 2016;93(1):42–48. doi: 10.1016/j.jhin.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Wand ME, Bock LJ, Bonney LC, Sutton JM. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother. 2017;61(1):e01162–16. doi: 10.1128/AAC.01162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornejo-Juarez P, Vilar-Compte D, Perez-Jimenez C, Namendys-Silva SA, Sandoval-Hernandez S, Volkow-Fernandez P. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int J Infect Dis. 2015;31:31–34. doi: 10.1016/j.ijid.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Caini S, Hajdu A, Kurcz A, Borocz K. Hospital-acquired infections due to multidrug-resistant organisms in Hungary, 2005–2010. Euro Surveill. 2013;18(2):20352. [PubMed] [Google Scholar]
- 24.WHO Antimicrobial Resistance: Global Report on Surveillance, 2014. 2014. [Accessed April 04, 2018]. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en/
- 25.Huang SS, Datta R, Platt R. Risk of acquiring antibiotic-resistant bacteria from prior room occupants. Arch Intern Med. 2006;166(18):1945–1951. doi: 10.1001/archinte.166.18.1945. [DOI] [PubMed] [Google Scholar]
- 26.Drees M, Snydman DR, Schmid CH, et al. Prior environmental contamination increases the risk of acquisition of vancomycin-resistant enterococci. Clin Infect Dis. 2008;46(5):678–685. doi: 10.1086/527394. [DOI] [PubMed] [Google Scholar]
- 27.Nseir S, Blazejewski C, Lubret R, Wallet F, Courcol R, Durocher A. Risk of acquiring multidrug-resistant Gram-negative bacilli from prior room occupants in the intensive care unit. Clin Microbiol Infect. 2011;17(8):1201–1208. doi: 10.1111/j.1469-0691.2010.03420.x. [DOI] [PubMed] [Google Scholar]
- 28.Caselli E, D’Accolti M, Vandini A, et al. Impact of a probiotic-based cleaning intervention on the microbiota ecosystem of the hospital surfaces: focus on the resistome remodulation. PLoS One. 2016;11(2):e0148857. doi: 10.1371/journal.pone.0148857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandini A, Temmerman R, Frabetti A, et al. Hard surface biocontrol in hospitals using microbial-based cleaning products. PLoS One. 2014;9(9):e108598. doi: 10.1371/journal.pone.0108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caselli E. Hygiene: microbial strategies to reduce pathogens and drug resistance in clinical settings. Microb Biotechnol. 2017;10(5):1079–1083. doi: 10.1111/1751-7915.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caselli E, Antonioli P, Mazzacane S. Safety of probiotics used for hospital environmental sanitation. J Hosp Infect. 2016;94(2):193–194. doi: 10.1016/j.jhin.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Abuladze T, Li M, Menetrez MY, Dean T, Senecal A, Sulakvelidze A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157:H7. Appl Environ Microbiol. 2008;74(20):6230–6238. doi: 10.1128/AEM.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greer GG. Bacteriophage control of foodborne bacteriat. J Food Prot. 2005;68(5):1102–1111. doi: 10.4315/0362-028x-68.5.1102. [DOI] [PubMed] [Google Scholar]
- 34.Tomat D, Quiberoni A, Mercanti D, Balague C. Hard surfaces decontamination of enteropathogenic and Shiga toxin-producing Escherichia coli using bacteriophages. Food Res Int. 2014;57:123–129. [Google Scholar]
- 35.Sulakvelidze A. Phage therapy: an attractive option for dealing with antibiotic-resistant bacterial infections. Drug Discov Today. 2005;10(12):807–809. doi: 10.1016/S1359-6446(05)03441-0. [DOI] [PubMed] [Google Scholar]
- 36.Jensen KC, Hair BB, Wienclaw TM, et al. Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS One. 2015;10(7):e0131714. doi: 10.1371/journal.pone.0131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regulations C-CoF Title 21: Food and drugs Chapter 1: Food and Drug Administration. Department of Health and Human Services Subchaper B: Food for human consumption. PART 172: Food additives permitted for direct addition to food for human consumption. Subpart H: Other Specific Usage Additives. Sec. 172.785 Listeria-specific bacteriophage preparation. 2017. [Accessed April 04, 2018]. Available from: https://www.gpo.gov/fdsys/pkg/CFR-2011-title21-vol3/pdf/CFR-2011-title21-vol3-chapI.pdf.
- 38.EUCAST The European Committee on Antimicrobial Susceptibility Testing – EUCAST. Oct 20, 2017. [Accessed July 12, 2018]. Available from: http://www.eucast.org/
- 39.CLSI Performance standards for antimicrobial susceptibility testing. 28th edition. 2018. [Accessed July 12, 2018]. Available from: https://clsi.org/standards/products/microbiology/documents/m100/
- 40.Caselli E. Testing antibacterial activity of bacteriophages on hard surfaces. [Accessed July 12, 2018]. http://www.protocols.io.2018; Available from: dx.org/10.17504/protocols.io.nwmdfc6.
- 41.Ho YH, Tseng CC, Wang LS, et al. Application of bacteriophage-containing aerosol against nosocomial transmission of carbapenem-resistant Acinetobacter baumannii in an intensive care unit. PLoS One. 2016;11(12):e0168380. doi: 10.1371/journal.pone.0168380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.