Abstract
Short tandem repeat (STR) mutations may comprise more than half of the mutations in eukaryotic coding DNA, yet STR variation is rarely examined as a contributor to complex traits. We assessed this contribution across a collection of 96 strains of Arabidopsis thaliana, genotyping 2046 STR loci each, using highly parallel STR sequencing with molecular inversion probes. We found that 95% of examined STRs are polymorphic, with a median of six alleles per STR across these strains. STR expansions (large copy number increases) are found in most strains, several of which have evident functional effects. These include three of six intronic STR expansions we found to be associated with intron retention. Coding STRs were depleted of variation relative to noncoding STRs, and we detected a total of 56 coding STRs (11%) showing low variation consistent with the action of purifying selection. In contrast, some STRs show hypervariable patterns consistent with diversifying selection. Finally, we detected 133 novel STR-phenotype associations under stringent criteria, most of which could not be detected with SNPs alone, and validated some with follow-up experiments. Our results support the conclusion that STRs constitute a large, unascertained reservoir of functionally relevant genomic variation.
Rates of mutation vary by several orders of magnitude across different elements in genomes (Acuna-Hidalgo et al. 2016), from ∼10−8 to 10−9 for substitutions to 10−3 to 10−4 for short tandem repeat (STR) mutations (Sun et al. 2012; Willems et al. 2016; Gymrek et al. 2017). STR mutations generally occur through the addition or subtraction of repeat units. Given the prevalence of STR loci in eukaryotic genomes, we would expect more de novo STR mutations than de novo single nucleotide substitutions in the human genome per generation (Willems et al. 2016). Thus, while the overall mutation rate is under the strong control of natural selection (Lynch 2010), some loci experience a heavier mutational burden than others in the form of recurrent mutation (Harpak et al. 2016). The existence of such highly mutable loci violates simplifying assumptions of the infinite sites model of population genetics (Haasl and Payseur 2010), namely, that no locus mutates more than once in a population, as well as quantitative genetic models assuming contributions from many independent loci (Yang et al. 2010).
In spite of the large effects that STRs can have on complex traits and diseases in model organisms and humans (Fondon et al. 2008; Hannan 2010; Press et al. 2014), their variation is rarely considered in genotype–phenotype association studies, because technical obstacles hinder their ascertainment. Most prominently, dozens of human neurodevelopmental disorders are thought to be caused by STR expansions, i.e., large increases in copy number at specific STR loci (Hannan 2018). However, STR genotyping methods of sufficient accuracy, throughput, and cost-effectiveness to ascertain STR alleles at high throughput have recently become available (Highnam et al. 2013; Carlson et al. 2015; Willems et al. 2017). Studies leveraging these methods suggest considerable contributions of STRs to heritable phenotypic variation (Carlson et al. 2015; Gymrek et al. 2015).
From an evolutionary perspective, the high mutation rate and strong phenotypic effects of STRs have been speculated to provide accessible evolutionary paths for rapid adaptation (Moxon et al. 1994; King et al. 1997; Kashi and King 2006; Gemayel et al. 2010; King 2012). Variation of STRs in various nonhuman species is associated with traits under strong selection such as morphogenesis and reproductive phenology, e.g., in dogs (Laidlaw et al. 2007) and in the flowering plant Arabidopsis thaliana (Undurraga et al. 2012; Rival et al. 2014; Press and Queitsch 2017). Over longer time scales, the presence of highly mutable STRs within coding regions is thought to be maintained by selection (Yu et al. 2005; Mularoni et al. 2010; Sawaya et al. 2012). In plants specifically, microsatellites tend to be associated with otherwise nonrepetitive DNA and covary in number with the amount of transcribed DNA rather than total genome size (Morgante et al. 2002). These observations argue for important roles of STRs as a reservoir of functional genetic variation.
In the present study, we apply massively parallel STR genotyping to a diverse panel of well-characterized A. thaliana strains. We use these data to generate and test hypotheses about the functional effects of STR variation, combining observations of gene disruption by STR expansion, inferences about STR conservation, and analyses of phenotypic association. Based on our results, we argue that STRs must be included in any comprehensive account of phenotypically relevant genomic variation.
Results
STR genotyping reveals complex allele frequency spectra
We targeted 2046 STR loci for genotyping with molecular inversion probes (MIPs) (Carlson et al. 2015) across a core collection of 96 inbred A. thaliana strains (Methods). These inbred strains, maintained by single-seed descent, are considered to be effectively homozygous (Koornneef and Meinke 2010). Targeted STR loci were all less than 200 bp in length and had nucleotide purity of at least 89%, encompassing nearly all gene-associated STRs (including STRs in coding regions, introns, and untranslated regions [UTRs]), as well as ∼40% of intergenic STRs (Fig. 1A). We focused on genic STRs as those most likely to have phenotypic relevance, but also include intergenic STRs as a reference. For additional details regarding STR annotation, STR selection for targeting, and MIP design, see Methods and Supplemental Text. We used comparisons with the Col-0 reference genome, PCR analysis of selected STRs, and dideoxy sequencing to estimate that MIP STR genotype calls were ∼95% accurate, and inaccurate calls were generally only one to two units away from the correct copy number (Supplemental Text; Supplemental Figs. S1–S4; Supplemental Table S1). In this and previous work (Carlson et al. 2015), we did not observe STR heterozygosity within these strains, although we cannot formally exclude trace levels of heterozygosity (although duplicated STRs of differing copy number were observed in the previous study). For additional details regarding MIP assay performance, see Supplemental Text.
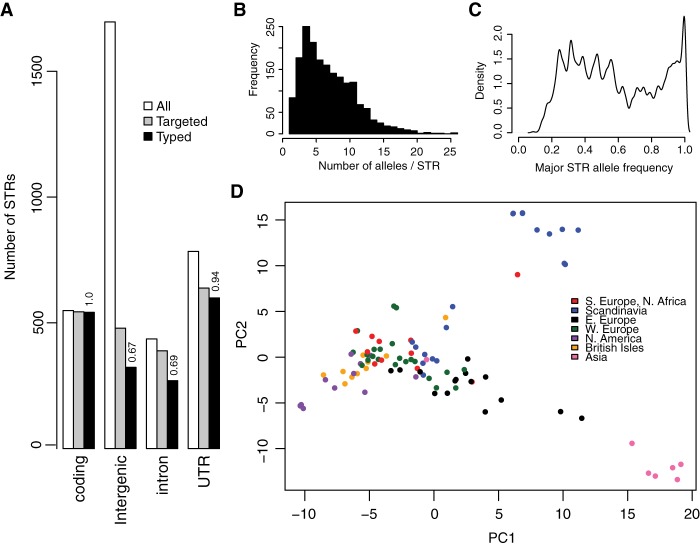
Figure 1.
STRs in A. thaliana show a complex allele frequency distribution and geographic differentiation. (A) Distribution and ascertainment of STR loci. (All) All STRs matching the definition of STRs for this study, e.g., ≤200 bp length in TAIR10, ≥89% purity in TAIR10, 2–10 bp nucleotide motif. (Targeted) The 2046 STRs targeted for MIP capture. (Typed) STRs successfully genotyped in the Col-0 genome in a MIPSTR assay. Numbers above the bars indicate the proportion of targeted STRs in the relevant category that were successfully genotyped. (B) The distribution of allele counts across all genotyped STRs. (C) The distribution of major allele frequencies (frequency of the most frequent allele at each locus) across genotyped STRs. y-Axis is arbitrary units indicating density of loci showing the relevant frequency signature. (D) Principal component analysis (PCA) reveals substantial geographic structure according to STR variation. PC1 and PC2 correspond, respectively, to 5.2% and 4.0% of total STR allele variance.
Across genotyped loci, we observed that 95% of STRs were polymorphic. Most STRs were highly multiallelic across strains (mean = 6.4 alleles, median = 6 alleles) (Fig. 1B), and this variation was mostly unascertained by the 1001 Genomes resource for A. thaliana (Supplemental Fig. S3A; The 1001 Genomes Consortium 2016). Coding STRs were only slightly less polymorphic than noncoding STRs (mean = 4.5 alleles, median = 4 alleles; 2.6 ± 0.25 SEM fewer alleles on average than intergenic STRs), although it is unknown whether this difference is due to purifying selection or variation in mutation rates. Highlighting the massive variation segregating at STR loci, 45% of STRs had a major allele with frequency less than 0.5. This complicates the familiar concepts of major and minor alleles, which have provided a common framework for detecting genotype associations (Fig. 1C). Specifically, the Col-0 reference strain carries the major STR allele at only 48% of STR loci. Moreover, rarefaction analysis implied that more STR alleles at these loci are expected with further sampling of A. thaliana strains (Supplemental Fig. S4C).
Principal component analysis of STR variation revealed genetic structure corresponding to Eurasian geography (Fig. 1D; Supplemental Fig. S5), consistent with previous observations that genetic population structure is correlated with the geographic distribution of A. thaliana (Nordborg et al. 2005; The 1001 Genomes Consortium 2016). By corroborating previous observations from a much larger set of genome-wide single nucleotide polymorphism markers, this result demonstrates that a comparatively small panel of STRs suffices to capture detailed population structure in A. thaliana strains (Fig. 1D; Supplemental Fig. S5).
Novel STR expansions are associated with splicing disruptions
We next examined the frequency and functional consequences of STR expansions in A. thaliana. STR expansions are high-copy-number variants of comparatively short STRs that are widely recognized as contributors to human diseases (Usdin 2008) and other phenotypes (Sureshkumar et al. 2009). Although large (>150 bp) expansions are difficult to infer due to limitations of MIP technology, we detected modest STR expansions using a simple heuristic that compares the longest allele to the median allele observed at each locus (Fig. 2A; Methods). We identified expansions in 64 of 96 A. thaliana strains, each carrying at least one expanded STR allele from one of 28 expansion-prone STRs (nine coding, six intronic, eight UTR, five intergenic) (Supplemental Table S2). Most expansions were found in multiple strains (Fig. 2B), although expansion frequencies were likely underestimated due to a higher rate of missing data at these loci. We ascertain expansions of up to about 50 copies, whereas coding STR expansions associated with human disease can be as small as 20 copies, suggesting our expansions can be functionally relevant (Usdin 2008; Hannan 2018).
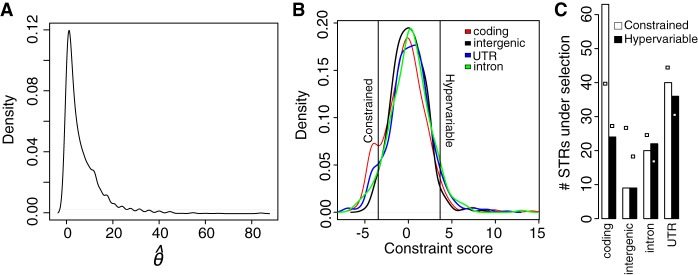
Figure 2.

Inferring and assessing the functional effects of modest STR expansions. (A) The distribution of expansion scores across STRs, where the expansion score is computed as [max(STR length) – median(STR length)]/median STR length. We called any STRs with a score >2 a modest expansion (indicated). (B) Distribution of allele frequencies of the 28 expanded STR alleles. (C,D) Distribution of STR copy number of the intronic STR (motif CAA) in the NTM1 gene and the 3′ UTR STR (motif AT) in the MEE36 gene. (E) RT-PCR demonstrates intron retention in NTM1 mRNA in the Mr-0 strain, which carries the STR expansion, yielding an aberrant 437-bp product. (F) MEE36 transcript abundances measured by qRT-PCR and normalized relative to UBC21 transcript levels. For each strain, two independent biological replicates are shown as points. Transcript levels are expressed relative to Col-0 levels (set to 1). (*) STR genotype corrected by follow-up dideoxy sequencing. Strains and order are the same between E and F.
We assayed the effects of STR expansions on expression of associated genes using qRT-PCR. The most dramatic expansions (with large relative copy number increase) affected an intronic STR in the NTM1 gene (five other expansions also resided in introns) (Fig. 2C) and a STR in the 3′ UTR of the MEE36 gene (Fig. 2D). These genes, respectively, have roles in cell proliferation and embryonic development. We next considered intronic STR disruptions, which may cause obvious splicing defects, by assaying the splicing of all six expanded intronic STRs. In three cases, the expanded allele was associated with partial or full retention of its intron, which we confirmed by dideoxy sequencing of cDNA (Fig. 2E; Supplemental Figs. S6, S7A; Supplemental Text). One of these retention events occurred in the major NTM1 splice form in the Mr-0 strain (Fig. 2E). This NTM1 intron retention is predicted to lead to a nonsense mutation truncating most of the NTM1 protein (Supplemental Fig. S7B). For the other two intron retentions, more complex and STR allele-specific mRNA species were formed (Supplemental Fig. S6; Supplemental Text). The MEE36 STR expansion alleles were associated with dramatically reduced MEE36 transcript levels (Fig. 2F), possibly due to the STR expansion altering transcript processing (Jackson 1993). Although due to numerous other polymorphisms between strains we cannot confidently ascribe causality to any specific polymorphism, these examples emphasize the potential for previously unascertained STR variation to modify gene function. Moreover, we show that the distribution of allele sizes itself can be informative, enabling predictions about functional effects of specific STR alleles based on copy number outliers.
Signatures of functional constraint on STR variation
Using the observed STR allele frequency distributions, we next attempted to infer selective processes acting on STRs. Although previous models for evaluating functional constraint on STRs are few (Haasl and Payseur 2013), there is consensus that selection shapes STR variation to at least some degree (Huntley and Clark 2007; Mularoni et al. 2010; King 2012; Haasl and Payseur 2013). Naïvely, we would expect that coding STRs should show increased constraint (lower variation). Consistent with this expectation, we observed that most invariant STRs are coding (53 of 84 invariant STRs genotyped across at least 70 strains; odds ratio = 4.5, P = 5 × 10−11, Fisher's exact test). However, methods of inferring selection by allele counting are confounded by population structure and mutation rate, which vary widely across STRs in this (Fig. 3A) and other studies (Schlötterer et al. 2004; Gymrek et al. 2017). Mutation rate specifically may be expected to differ between coding and other regions, given constraints on motif size and the generally lower purity of coding STRs compared to other regions (Lawson and Zhang 2006; Pramod et al. 2014).
Figure 3.
Detecting functionally constrained STRs. (A) The distribution of (Watterson's estimator, or estimated population mutation rate) (Haasl and Payseur 2010) across all genotyped STR loci. (B) Distribution of “selection scores” across all STRs, separated by locus category. Vertical lines indicate 2.5% and 97.5% quantiles of the distribution of intergenic STRs, which are used as thresholds for putative constraint and hypervariability, respectively. (C) STRs under selection, e.g., constrained or hypervariable STRs, separated by locus category. White boxes indicate the expected numbers for each bar, based on number of STRs in each locus category and number of STRs under different types of selection.
To therefore account for mutation rate and population structure, we used support vector regression (SVR) to model STR variability across these 96 strains, using well-established correlates of STR variability (e.g., STR unit number and STR purity) (Methods) (Legendre et al. 2007; Eckert and Hile 2009; Gymrek et al. 2017). Selection was defined as deviation from expected variation of a neutral STR among strains. We trained SVRs on the set of intergenic STRs, which should experience minimal selection relative to STRs associated with genes (Supplemental Figs. S8–S10). We used bootstrap aggregation of SVR models to compute a putative constraint score for each STR by comparing its observed variability to the expected distribution from bootstrapped SVR models (Fig. 3B; Supplemental Text; Supplemental File S1).
According to constraint scores, 132 STRs were less variable than expected under neutrality, suggesting purifying selection on these loci (Fig. 3C). Among these, coding STRs were overrepresented relative to their prevalence (OR = 2.4, P = 3.7 × 10−6, Fisher's exact test) (Fig. 3C), in accordance with our naïve analysis of invariant STRs above. Examples of constrained coding STRs included STRs encoding homologous polylysines adjoining the histone core in three different histone H2B proteins; notably, core histones are among the most conserved proteins across eukaryotes. Generally, coding STRs showing purifying selection encoded roughly half as many polyserines and twice as many acidic homopolymers as expected from proteome-wide averages (Supplemental Table S3; Karlin et al. 2002). The interpretation of this pattern is unclear, but it may be related to some structural role of such different classes of homopolymers in proteins. Although many more coding STRs are probably functionally constrained, our power to detect such constraints is limited by the size of the data set, as well as the potential for purifying selection acting on some STRs in the background set. We also observed high conservation of some STRs in noncoding regions (most commonly, intronic or UTR STRs), although this is less interpretable given the ambiguous relationship between sequence conservation and regulatory function in A. thaliana (Alexandre et al. 2017). Intergenic STRs specifically, as opposed to intronic and UTR STRs, showed low prevalence of constraint (∼2%), as expected for regions not associated with gene function (Fig. 3C). The most constrained intronic STR, in the BIN4 gene, which is required for endoreduplication and normal development (Breuer et al. 2007), shows a restricted allele frequency spectrum compared to similar STRs (Fig. 4A).
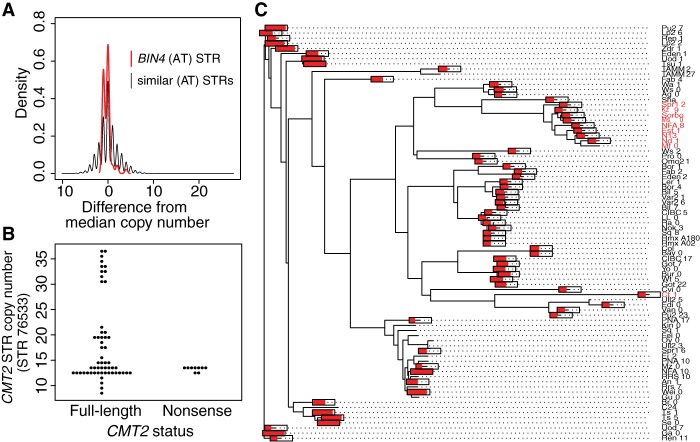
Figure 4.
Noncoding STRs showing non-neutral variation. (A) BIN4 intron STR is constrained relative to similar STRs. Allele frequency spectra are normalized by subtracting the median copy number (9 for the BIN4 STR). All pure STRs with TA/AT motifs and a median copy number between 7 and 12 are included in the “similar STRs” distribution. (B) Lack of association between near-expansion CMT2 STR alleles and previously described nonsense mutations. (C) Neighbor-joining tree of a 10-kb region of A. thaliana Chromosome 4 encompassing the CMT2 gene across 81 strains with available data, using Kimura's two-parameter distance model in APE (Paradis et al. 2004). Text labels in red indicate an adaptive nonsense mutation early in the first exon of CMT2, as noted previously (Shen et al. 2014). Red bars drawn on tips of the tree indicate the length of the CMT2 intronic STR (as a proportion of its maximum length, 36.5 units) in each of the 81 strains or tips. The bars are omitted for tips with missing STR data.
Hypervariable coding STRs (showing more alleles than expected) were too few for statistical arguments, but nonetheless showed several notable patterns (Supplemental Text; Supplemental Fig. S11). One noncoding hypervariable STR lies in an intron of the Chromomethylase 2 (CMT2) gene, which is under positive selection in A. thaliana (Shen et al. 2014). Specifically, CMT2 nonsense mutations in some populations are associated with temperature seasonality. We considered whether the extreme CMT2 STR alleles might be associated with these nonsense mutations. Instead, these extreme alleles exclusively occurred in strains with full-length CMT2 (Fig. 4B). Strains with the common CMT2 nonsense mutation form a tight clade in the CMT2 sequence tree, whereas the CMT2 STR length fluctuates rapidly throughout the tree and appears to converge on longer alleles independently in different clades (Fig. 4C). These convergent changes are consistent with a model in which the CMT2 STR is a recurrent target of positive selection.
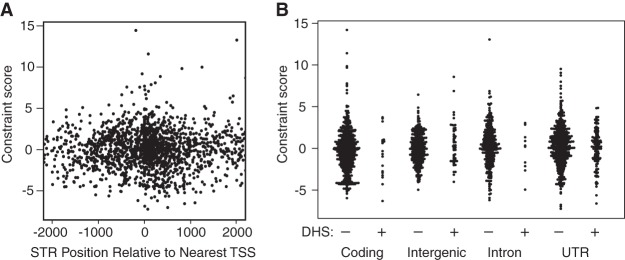
We further assessed whether STR conservation can be attributed to cis-regulatory function. Considering all STRs regardless of other annotations, we examined whether STRs near transcription start sites (TSSs) showed signatures of functional constraint (Fig. 5A). We found little evidence for reduced STR variation near TSSs, suggesting that cis-regulatory effects do not generally constrain STR variation in A. thaliana. Moreover, we found no relationship between constraint scores and location of STRs in accessible chromatin sites marking regulatory DNA (Fig. 5B).
Figure 5.
Relationship of STR constraint to putative gene regulatory elements. (A) Constraint score from Figure 3B plotted with respect to nearest TSS. (B) Constraint score from Figure 3B plotted with respect to STR annotations and presence of (putatively regulatory) DNase I hypersensitive sites (DHS).
We also investigated whether overlap of STRs with transposable elements affected conservation but observed no notable effect of this annotation on conservation scores (Supplemental Fig. S12).
STRs yield numerous novel genotype–phenotype associations
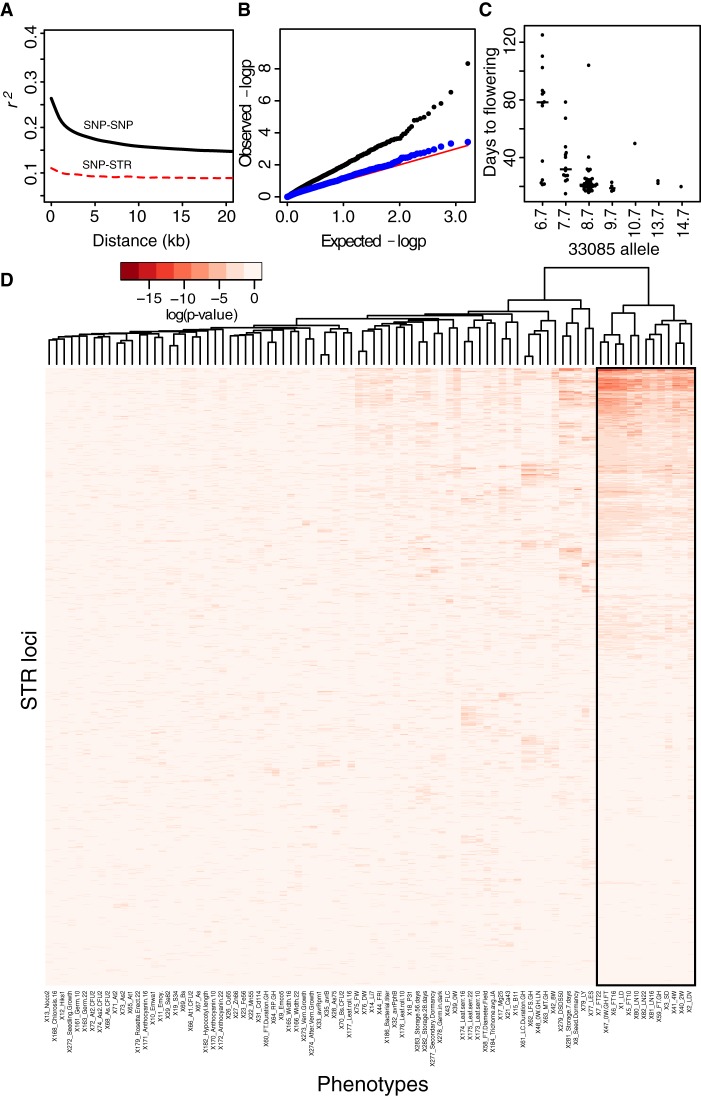
We next addressed the question of whether STR genotypes contribute new information for explaining phenotypic variation. One basic expectation is that linkage disequilibrium (LD) with other markers is substantially weaker for STRs than for SNPs, due to elevated mutation rates (Willems et al. 2014). Indeed, we found greatly reduced LD between STR and SNPs in A. thaliana, compared to SNP-SNP LD, as opposed to pairs of SNPs (Fig. 6A). Moreover, the observed LD around STR loci declined with increasing STR allele number (Supplemental Fig. S13), consistent with an expected higher mutation rate at multiallelic loci and numerical constraints on LD with large numbers of alleles (Haasl and Payseur 2010; Sawaya et al. 2015). This result suggests that STR-phenotype associations need to be directly tested rather than relying on linkage to SNPs. A. thaliana offers extensive high-quality phenotype data for our inbred strains, which have been previously used for SNP-based association studies (Atwell et al. 2010). We tested each polymorphic STR for associations across the 96 strains with each of 105 published phenotypes. For the subset of 32 strains for which RNA-seq data were available (Kawakatsu et al. 2016), we also tested for associations between STR genotypes and expression of genes (i.e., eQTLs) (Supplemental Text), using a range of distances from genes. Although our power to detect eQTLs was limited by sample size, we detected 12 significant associations. The strongest association was between a STR residing in long noncoding RNA gene AT4G07030 and expression of the nearby stress-responsive gene AtCPL1 (Supplemental Fig. S14; Supplemental Table S4).
Figure 6.
Diverse associations of STRs with quantitative phenotypes. (A) Multiallelic LD (Zaykin et al. 2008) estimates for STR and SNP loci. Lowess lines for each category are plotted. All values of r2 < 0.05 are omitted from lowess calculation for visualization purposes. (B) Quantile–quantile plot of P-values from tests of association between STRs and germination rate after 28 d of storage. (C) An example association between an STR (33085) and a phenotype (flowering time in long days after 4 wk vernalization) in A. thaliana strains. Median of each distribution is indicated by a bar proportional in width to the number of observations. (D) Heatmap showing pairwise associations between STRs and phenotypes, summarized by the P-value from a linear mixed model, fitting STR allele as a fixed effect and kinship as a random effect. Both rows and columns are clustered, although the row dendrogram was omitted for clarity. STRs with genotype information in fewer than 25 strains are not displayed. Flowering time phenotypes are boxed in black.
We next focused on organismal phenotypes, using a linear mixed-model framework to test STR loci for associations while correcting for population structure. Certain STRs showed associations with multiple phenotypes, and flowering time phenotypes were particularly correlated with one another (Fig. 6C,D). Similar to these patterns, SNPs have also shown associations with multiple phenotypes, and the various flowering time phenotypes are among the strongest associations previously detected in the same strains (Atwell et al. 2010). As in previous association studies using STRs (Gymrek et al. 2015), some inflation was apparent in test P-values compared to expectations (i.e., the test P-value distribution skews anticonservative), although the same tests using permuted STR genotypes showed negligible inflation (Fig. 6B; Supplemental Fig. S15). Negligible inflation with permuted genotypes has been used previously to exclude confounding from population structure (Gymrek et al. 2015), which we will also presume here (Supplemental Text). We found 133 associations between 61 STRs and 25 phenotypes at stringent genome-wide significance levels (Methods; Supplemental Table S5). Given the low LD observed between STRs and other variants, STR variants may themselves be causal, rather than merely tagging nearby causal variants. Our analysis found plausible candidate genes, such as COL9, which acts in flowering time pathways and contains a flowering time-associated STR, and RABA4B, which acts in the salicylic acid defense response (Antignani et al. 2015) and contains a STR associated with lesion formation. Many of these traits, such as reproductive phenology and disease resistance, are traits under strong selection in A. thaliana (Tian et al. 2002; Caicedo et al. 2004).
We evaluated whether these associations might have been found using SNP-based analyses. We found that STR effects on phenotype are largely not accounted for by nearby SNP variation. Considering the strongest association for each STR, only 18 of the 61 STRs were near potentially confounding SNP variants, and most such associations (14/18) were robust to adjustment for nearby SNP genotypes (Supplemental Table S6; Supplemental Text; Atwell et al. 2010). One notable exception was a STR closely linked to a well-known deletion of the RPS5 gene in a hypervariable region of Chromosome 1 (Tian et al. 2002) that causes resistance to bacterial infection (Karasov et al. 2014). RPS5 status is under balancing selection in A. thaliana, possibly due to a frequency-dependent model of pathogen resistance (Tian et al. 2002). In this case, the association and the linkage are apparently strong enough (the STR is ∼4 kb upstream of the deletion) that this STR tags RPS5’s effect on infection.
To assess the STR contribution to the variance of a specific trait, we performed a naïve variance decomposition of the long-day flowering phenotype into SNP and STR components, as represented by the loci showing associations with this trait. Our results suggested that STRs potentially contribute as much or more variance than SNPs to this phenotype (Supplemental Text; Supplemental Table S7). Estimated effect sizes for STR variants on this phenotype were similar to those of large-effect SNP variants (Supplemental Fig. S16; Atwell et al. 2010). However, this analysis involves a large number of parameters and should thus be treated as preliminary, especially as none of the loci reached nominal significance when modeled together (Supplemental Text).
Finally, we used mutant analysis to evaluate the two strongest flowering time associations. These included a coding STR in AGL65 and an intronic STR in the uncharacterized gene AT4G01390; neither locus had been associated with flowering time phenotypes. We found that disruptions of both STR-associated genes conferred modest early flowering effects (by ∼2 d and about one rosette leaf, P < 0.05 for each in linear mixed models) (Supplemental Fig. S17), supporting the robustness of our STR-phenotype associations. Taken together, our study suggests that STRs contribute substantially to phenotypic variation.
Discussion
Our results imply that STRs contribute substantially to trait heritability in A. thaliana. There is little support for the hypothesis that STRs are “junk DNA”: STRs are apparently constrained by functional requirements, STR variation can disrupt gene function, and STR variation is associated with phenotypic variation. Considering that STR variation is represented poorly by nearby SNPs, the failure to directly ascertain STRs will mask most phenotypic consequences of such STR effects. This finding contradicts the assumption that STR-phenotype associations should be captured through linkage to common single nucleotide polymorphisms (SNPs) (Payseur et al. 2008). Our finding is consistent with human STR data (Payseur et al. 2008; Willems et al. 2014) and simulation studies (Sawaya et al. 2015) indicating limited LD between STRs and SNPs, making it unlikely that SNP markers can generally tag STR genotypes. If STR variation contributes to the phenotypic variance, as we argue here, the scope of any genome-wide association study relying on SNP genotypes alone will be limited.
Compared to other classes of markers, STRs may exert an outsized influence on the phenotypic variance due to de novo mutations (Vm) (Willems et al. 2016). Estimates of Vm from model organisms are on the order of 1% but may vary substantially from trait to trait (Lynch 1988). STRs are good candidates for a substantial proportion of this variance, given their high mutation rate, residence in functional regions, and functional constraint demonstrated here (although we make no attempt to quantify Vm from STRs in this study). In previous work (Rival et al. 2014), we showed apparent copy number conservation of a STR in spite of a high mutation rate. In this case, deviation from the conserved copy number produced aberrant phenotypes. Our observation that constrained STRs are common suggests that STRs are a likely source of deleterious de novo mutations that are subsequently removed by selection. This finding likely generalizes beyond A. thaliana to humans, as the measured rates of de novo substitutions and indels are similar between the two species (Ossowski et al. 2010; Acuna-Hidalgo et al. 2016). Moreover, even if STR variants are not causal, they may tag certain hypervariable regions more effectively than SNPs. For example, our observation that the RPS5 locus is tagged by a STR leads us to speculate that STR variation holds information about genomic regions with complex mutational histories.
The extent to which STRs affect phenotype is only partially captured in this study. Specifically, we assayed two STRs shown to influence phenotypic variation in prior transgenic A. thaliana studies (Sureshkumar et al. 2009; Undurraga et al. 2012), but these STR loci did not show strong signatures of phenotype association or of selection. This lack of ascertainment suggests that many more functionally important STRs exist in A. thaliana than we can detect with the analyses presented here. For example, the polyQ-encoding STR in the ELF3 gene causes dramatic variation in developmental phenotypes (Undurraga et al. 2012), yet we find no statistical associations between this locus and phenotype across our 96 strains. In this case, the lack of phenotype association is expected, as ELF3 STR alleles interact epistatically with several other loci (Press and Queitsch 2017), and thus would require increased power or more sophisticated analyses to detect associations. Indeed, we have argued that STRs are more likely than less mutable classes of genomic variation to exhibit epistasis (Press et al. 2014). In consequence, we expect that the associations described in the present study are an underestimate of STR effects on phenotype. Moreover, our data are constrained by MIP technology, which limits the size and composition of STR alleles that we can ascertain (Fig. 1A; Supplemental Figs. S1, S2).
Considering next a mechanistic perspective, the association we observe between intronic STR expansions and splice disruptions may be an important mechanism by which STRs contribute to phenotypic variation. Intronic STR mutations can disrupt splicing, altering gene function (Li et al. 2004) and contributing to human disease (for review, see Ranum and Day 2002). In humans, unascertained diversity of splice forms contributes substantially to disease (Cummings et al. 2017), and this diversity is larger than commonly appreciated (Nellore et al. 2016). We demonstrate that this mechanism is common, at least for expansions, and argue that future work should evaluate the tolerance of introns to different magnitudes of STR variation. More generally, some patterns in highly variable STRs may be relevant for answering specific biological questions. For example, 3/24 hypervariable coding STRs encoded polyserines in F-box proteins, suggesting that STR variation may serve as a mechanism of diversification in this protein family, which shows dramatically increased family size and sequence divergence in some plant lineages (Clark et al. 2007; Xu et al. 2009).
The abundance of STRs in eukaryotic promoters (Sawaya et al. 2013), and their associations with gene expression (in cis) (Vinces et al. 2009; Bilgin Sonay et al. 2015; Gymrek et al. 2015), suggested that STRs affect transcription, possibly by altering nucleosome positioning (Vinces et al. 2009) or methylation (Quilez et al. 2016), among other mechanisms. Our preliminary analysis of STR effects on expression in A. thaliana found little evidence of strong effects, although this could be attributed to low sample size (about 30 lines in most cases). This smaller data set makes it difficult to compare expression phenotypes to organismal phenotypes. However, we found little evidence for STR selective constraint associated with cis-regulatory function.
The phenotypic contributions of loci with high mutation rates remain underappreciated, specifically in cases where such loci are difficult to ascertain with high-throughput sequencing. The results presented here argue that STRs are likely to play a substantial role in phenotypic variation and heritability. Accounting for the heterogeneity of different classes of genomic variation, and specifically variation in mutation rate, will advance our understanding of the genotype–phenotype map and the trajectory of molecular evolution.
Methods
STR identification, inclusion, and probe design
We used TRF (Benson 1999) (parameters: matching weight 2, mismatching penalty 5, indel penalty 5, match probability 0.8, indel probability 0.1, score ≥40, and maximum period 10) to identify STRs in the TAIR8 build of the Arabidopsis thaliana genome, identifying 7826 putative STR loci under 200 bp (Supplemental File S2). We restricted further analysis to the 2409 loci with repeat purity ≥89%. We chose 2307 STRs from among these, prioritizing STRs in coding regions, introns, or untranslated regions (UTRs), higher STR unit purity, and higher a priori expected variability (VARscore) (Legendre et al. 2007). We designed (Boyle et al. 2014) molecular inversion probes (MIPs) targeting these STR loci in 180-bp capture regions with 8-bp degenerate tags in the common MIP backbone. For this purpose, we converted STR coordinates to the TAIR10 build and used the TAIR10 build as a reference genome. We used single nucleotide variants (SNVs) in 10 diverse Arabidopsis thaliana strains (Gan et al. 2011) to avoid polymorphic sites in designing MIP targeting arms. We filtered out MIPs predicted to behave poorly according to previously established criteria (MIPGEN logistic regression score <0.7 for MIP capture success) (Boyle et al. 2014), discarded MIPs targeting duplicate regions, and substituted MIPs designed around SNVs as appropriate. We attempted to redesign filtered MIPs with 200-bp capture regions using otherwise identical criteria. This yielded a final set of 2046 STR-targeting MIP probes (Supplemental File S3). For all analyses, unless otherwise indicated, all genotyped STR loci are included (1968 STRs were genotyped in at least one strain).
MIP and library preparation
These 2046 probes were ordered from Integrated DNA Technologies as desalted DNAs at the 0.2 pmol scale and resuspended in Tris-EDTA pH 8.0 (TE) to a concentration of 2 µM and stored at 4°C. We pooled and diluted probes to a final stock concentration of 1 nM. We phosphorylated probes as described previously (Hiatt et al. 2013). We performed DNA preparation from whole aerial tissue of adult A. thaliana plants. We prepared MIP libraries essentially as described previously (Hiatt et al. 2013; Carlson et al. 2015) using 100 ng A. thaliana genomic DNA for each of 96 A. thaliana strains.
Sequencing
We sequenced pooled capture libraries essentially as previously described (Carlson et al. 2015) on NextSeq and MiSeq instruments collecting a 250-bp forward read sequencing the ligation arm and captured target sequence, an 8-bp index read for library demultiplexing, and a 50-bp reverse read sequencing the extension arm and degenerate tag for single-molecule deconvolution. In each run, 10% of the sequenced library pool consisted of high-complexity whole-genome library to increase sequence complexity. For statistics and further details of data acquired for each library see Supplemental Tables S8 and S9.
STR annotation
We annotated STRs according to Araport11 (Cheng et al. 2017), classifying all STRs as coding, intronic, intergenic, or UTR-localized, and indicating whether each STR overlapped with transposable element sequence. UTR-localized STR loci included all loci that overlapped with a gene annotation that were neither protein-coding nor introns, e.g., are transcribed but neither translated nor spliced. This group therefore included some noncoding RNA (ncRNA) genes, although in practice the UTR class was dominated by 5′ and 3′ untranslated regions of protein-coding genes (93%, or 718/772 total genome-wide STRs meeting selection criteria), and very few STRs were included on the basis of other annotations (e.g., 0.5% or 4/772 STRs overlap ncRNAs). STR loci overlapping with pseudogenes or transposable elements (TEs) >200 bp in length were classified as intergenic, to recognize their more neutral evolutionary trajectories (most TE STRs were intergenic). To identify regulatory DNA, we used the union of seven distinct DNase-seq experiments (Sullivan et al. 2014) covering pooled or isolated tissue types. For additional details, see the Supplemental Text.
Sequence analysis
Sequences were demultiplexed and output into FASTQ format using bcl2fastq2 v2.17 (Illumina). We performed genotype calling essentially as described previously (Carlson et al. 2015), with certain modifications (Supplemental Text; Supplemental Table S1). Missing data indicate failure to genotype confidently. Note that our A. thaliana strains are inbred, and more stringent filters and data processing would be necessary to account for heterozygosity. For information about comparison with the Bur-0 genome, see Supplemental Text. Updated scripts implementing the MIPSTR analysis pipeline used in this study are available at https://github.com/maximilianpress/MIPSTR.
Statistical analysis and data processing
We performed all statistical analysis and data exploration using R v3.2.1 (R Core Team 2016). For plant experiments, we used restricted maximum likelihood (REML) to fit linear mixed models using experiment and position as random effects and genotype as a fixed effect.
STR expansion inference
We inferred STR expansions where the maximum copy number of an STR is at least three times larger than the median copy number of that STR. Various alleles of STR expansions were inspected manually in BAM files. Selected cases were dideoxy-sequenced and analyzed as described.
Plant material and growth conditions
Plants were grown on Sunshine soil #4 under long days (16 h light:8 h dark) at 22°C under cool-white fluorescent light. T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center (ABRC) (Supplemental Table S11; Alonso et al. 2003). For flowering time experiments, plants were grown in 36-pot or 72-pot flats; days to flowering (DTF) and rosette leaf number at flowering (RLN) were recorded when inflorescences were 1 cm high. Results are combined across at least three experiments.
Gene expression and splicing analysis
We grew bulk seedlings of indicated strains on soil for 10 d, harvested at Zeitgeber time 12 (ZT12), froze samples immediately in liquid nitrogen, and stored samples at −80°C until further processing. We extracted RNA from plant tissue using the SV RNA Isolation kit (including DNase step; Promega), and subsequently treated it with a second DNase treatment using the Turbo DNA-free kit (Ambion). We performed cDNA synthesis on ∼500 ng RNA for each sample with oligo-dT adaptors using the RevertAid kit (Thermo Fisher). We performed PCR analysis of cDNA with indicated primers (Supplemental Table S10) and ∼25 ng cDNA with the following protocol: denaturation for 5 min at 95°; then 30 cycles of 30 sec at 95°, 30 sec at 55°, 90 sec at 72°, ending with a final extension step for 5 min at 72°. We gel-purified and sequenced electrophoretically distinguishable splice variants associated with STR expansions. Each RT-PCR experiment was performed at least twice with different biological replicates.
Population genetic analyses
For PCA, STRs with missing data across the 96 strains were omitted, leaving 987 STRs with allele calls for every strain. We estimated using the approximation (Haasl and Payseur 2010), where is the average frequency of all STR alleles at a locus. We computed multiallelic linkage disequilibrium estimates for SNP-SNP and SNP-STR locus pairs using MCLD (Zaykin et al. 2008). We downloaded array SNP data for the same lines (TAIR9 coordinates) from http://bergelson.uchicago.edu/wp-content/uploads/2015/04/call_method_75.tar.gz (Horton et al. 2012). For each locus, both SNP and STR, we computed linkage disequilibrium scores with 150 surrounding loci. To facilitate comparison, we computed lowess estimates of linkage only for those locus pairs in the plotted distance window in each case and only for locus pairs with r2 > 0.05.
Inference of conservation
STRs typed across 70 A. thaliana strains or fewer were dropped from this analysis, because the estimates of their variability were unlikely to be accurate, leaving 1821 STRs. We measured STR variation as the base-10 logarithm of the standard deviation of STR copy number (Supplemental Text). We used bootstrap aggregation (“bagging”) to describe a distribution of predictions as follows. An ensemble of 1000 support vector regression (SVR, fit using the ksvm() function in the kernlab package) (https://cran.r-project.org/web/packages/kernlab/index.html) models was used to predict expected neutral variation of each STR as quantified by each measure (Supplemental Text). We used this distribution of bootstrapped predictions for intergenic STRs to compute putative conservation scores (Z-scores) for each STR. Scores below the 2.5% (Z < −3.46) and above the 97.5% (Z > 3.65) quantiles of intergenic STRs were considered to be putatively constrained and hypervariable, respectively.
eQTL inference
We downloaded normalized transcriptome data for A. thaliana strains from NCBI GEO GSE80744 (Kawakatsu et al. 2016). We used the Matrix eQTL package (Shabalin 2012) to detect associations, applying quantile normalization to address expression outliers (http://www.bios.unc.edu/research/genomic_software/Matrix_eQTL/faq.html#out) and fitting also 10 principal components from SNP genotypes to correct for population structure. Following precedent (Gymrek et al. 2015), we fitted additive generalized linear mixed models assuming that STR effects on expression would be a function of STR copy number.
Genotype–phenotype associations
We downloaded phenotype data from https://github.com/Gregor-Mendel-Institute/atpolydb/blob/master/miscellaneous_data/phenotype_published_raw.tsv. We followed precedent (Atwell et al. 2010) in log-transforming certain phenotypes. In all analyses, we treated STRs as factorial variables (to avoid linearity assumptions) in a linear mixed-effect model analysis to fit STR allele effects on phenotype as fixed effects while modeling the identity-by-state kinship matrix between strains (computed from SNP data) as a correlation structure for strain random effects on phenotype. We performed this modeling using the lmekin() function from the coxme R package (https://cran.r-project.org/web/packages/coxme/index.html). We repeated every analysis using permuted STR genotypes as a negative control to evaluate P-value inflation and discarded traits showing such inflation. We used P < 10−6 as a genome-wide significance threshold commensurate to the size of the A. thaliana genome and the data at hand. For flowering time phenotypes, we used a more stringent P < 10−10 threshold, because these phenotypes showed somewhat shifted P-value distributions (which were nonetheless inconsistent with inflation, according to negative controls). We identified potentially confounding SNP associations using the https://gwas.gmi.oeaw.ac.at/#/study/1/phenotypes resource, using the criterion that a SNP association must have a P ≤ 10−4 and be within roughly 100 kb of the STR to be considered. We fit models including SNPs as fixed effects as before and performed model selection using AICC (Hurvich and Tsai 1989). Additional details about association analyses are in the Supplemental Text.
Data access
MIP sequencing data from this study have been submitted to the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA388228. Scripts and processed data for reproducing analyses are provided as Supplemental File S4, as well as at https://osf.io/5jm2c/?view_only=324129c85b3448a8bd6086263345c7b0.
Supplementary Material
Acknowledgments
We thank Keisha Carlson, Alberto Rivera, and members of the Queitsch laboratory for technical assistance and important conversations. We thank Evan Boyle, Choli Lee, Matthew Snyder, and Jay Shendure for assistance and advice concerning MIP design and use. We thank the Dunham laboratory and the Fields laboratory for access to and help with sequencing instruments. We thank UW Genome Sciences Information Technology for high-performance computing resources. This work was supported in part by National Institutes of Health (NIH) New Innovator Award DP2OD008371 to C.Q.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231753.117.
References
- The 1001 Genomes Consortium. 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna-Hidalgo R, Veltman JA, Hoischen A. 2016. New insights into the generation and role of de novo mutations in health and disease. Genome Biol 17: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre CM, Urton JR, Jean-Baptiste K, Dorrity MW, Cuperus JC, Sullivan AM, Bemm F, Jolic D, Arsovski AA, Thompson A, et al. 2017. Regulatory DNA in A. thaliana can tolerate high levels of sequence divergence. bioRxiv 10.1101/104323. [DOI]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Antignani V, Klocko AL, Bak G, Chandrasekaran SD, Dunivin T, Nielsen E. 2015. Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 27: 243–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin Sonay T, Carvalho T, Robinson M, Greminger M, Krutzen M, Comas D, Highnam G, Mittelman DA, Sharp AJ, Marques-Bonet T, et al. 2015. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. Genome Res 25: 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, O'Roak BJ, Martin BK, Kumar A, Shendure J. 2014. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics 30: 2670–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Stacey NJ, West CE, Zhao Y, Chory J, Tsukaya H, Azumi Y, Maxwell A, Roberts K, Sugimoto-Shirasu K. 2007. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell 19: 3655–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci 101: 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson KD, Sudmant PH, Press MO, Eichler EE, Shendure J, Queitsch C. 2015. MIPSTR: a method for multiplex genotyping of germline and somatic STR variation across many individuals. Genome Res 25: 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J Cell Mol Biol 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342. [DOI] [PubMed] [Google Scholar]
- Cummings BB, Marshall JL, Tukiainen T, Lek M, Donkervoort S, Foley AR, Bolduc V, Waddell LB, Sandaradura SA, O'Grady GL, et al. 2017. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med 9: eaal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert KA, Hile SE. 2009. Every microsatellite is different: intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog 48: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW III, Hammock EAD, Hannan AJ, King DG. 2008. Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci 31: 328–334. [DOI] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet 44: 445–477. [DOI] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, Daly MJ, Price AL, Pritchard JK, Sharp AJ, et al. 2015. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 48: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Willems T, Reich D, Erlich Y. 2017. Interpreting short tandem repeat variations in humans using mutational constraint. Nat Genet 49: 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. 2010. The number of alleles at a microsatellite defines the allele frequency spectrum and facilitates fast accurate estimation of θ. Mol Biol Evol 27: 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. 2013. Microsatellites as targets of natural selection. Mol Biol Evol 30: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan AJ. 2010. Tandem repeat polymorphisms: modulators of disease susceptibility and candidates for “missing heritability”. Trends Genet 26: 59–65. [DOI] [PubMed] [Google Scholar]
- Hannan AJ. 2018. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 19: 286–298. [DOI] [PubMed] [Google Scholar]
- Harpak A, Bhaskar A, Pritchard JK. 2016. Mutation rate variation is a primary determinant of the distribution of allele frequencies in humans. PLoS Genet 12: e1006489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt JB, Pritchard CC, Salipante SJ, O'Roak BJ, Shendure J. 2013. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res 23: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highnam G, Franck C, Martin A, Stephens C, Puthige A, Mittelman D. 2013. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res 41: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. 2012. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley MA, Clark AG. 2007. Evolutionary analysis of amino acid repeats across the genomes of 12 Drosophila species. Mol Biol Evol 24: 2598–2609. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai CL. 1989. Regression and time series model selection in small samples. Biometrika 76: 297–307. [Google Scholar]
- Jackson RJ. 1993. Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell 74: 9–14. [DOI] [PubMed] [Google Scholar]
- Karasov TL, Kniskern JM, Gao L, DeYoung BJ, Ding J, Dubiella U, Lastra RO, Nallu S, Roux F, Innes RW, et al. 2014. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L, Bergman A, Mrázek J, Gentles AJ. 2002. Amino acid runs in eukaryotic proteomes and disease associations. Proc Natl Acad Sci 99: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi Y, King DG. 2006. Simple sequence repeats as advantageous mutators in evolution. Trends Genet 22: 253–259. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DG. 2012. Indirect selection of implicit mutation protocols. Ann N Y Acad Sci 1267: 45–52. [DOI] [PubMed] [Google Scholar]
- King DG, Soller M, Kashi Y. 1997. Evolutionary tuning knobs. Endeavour 21: 36–40. [Google Scholar]
- Koornneef M, Meinke D. 2010. The development of Arabidopsis as a model plant. Plant J 61: 909–921. [DOI] [PubMed] [Google Scholar]
- Laidlaw J, Gelfand Y, Ng KW, Garner HR, Ranganathan R, Benson G, Fondon JW III. 2007. Elevated basal slippage mutation rates among the Canidae. J Hered 98: 452–460. [DOI] [PubMed] [Google Scholar]
- Lawson MJ, Zhang L. 2006. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol 7: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Pochet N, Pak T, Verstrepen KJ. 2007. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res 17: 1787–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. 2004. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21: 991–1007. [DOI] [PubMed] [Google Scholar]
- Lynch M. 1988. The rate of polygenic mutation. Genet Res 51: 137–148. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2010. Evolution of the mutation rate. Trends Genet 26: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Hanafey M, Powell W. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30: 194–200. [DOI] [PubMed] [Google Scholar]
- Moxon ER, Rainey PB, Nowak MA, Lenski RE. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol 4: 24–33. [DOI] [PubMed] [Google Scholar]
- Mularoni L, Ledda A, Toll-Riera M, Albà MM. 2010. Natural selection drives the accumulation of amino acid tandem repeats in human proteins. Genome Res 20: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellore A, Jaffe AE, Fortin JP, Alquicira-Hernández J, Collado-Torres L, Wang S, Phillips RA III, Karbhari N, Hansen KD, Langmead B, et al. 2016. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol 17: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Place M, Weber JL. 2008. Linkage disequilibrium between STRPs and SNPs across the human genome. Am J Hum Genet 82: 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramod S, Perkins AD, Welch ME. 2014. Patterns of microsatellite evolution inferred from the Helianthus annuus (Asteraceae) transcriptome. J Genet 93: 431–442. [DOI] [PubMed] [Google Scholar]
- Press MO, Queitsch C. 2017. Variability in a short tandem repeat mediates complex epistatic interactions in Arabidopsis thaliana. Genetics 205: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MO, Carlson KD, Queitsch C. 2014. The overdue promise of short tandem repeat variation for heritability. Trends Genet 30: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, Guilmatre A, Garg P, Highnam G, Gymrek M, Erlich Y, Joshi RS, Mittelman D, Sharp AJ. 2016. Polymorphic tandem repeats within gene promoters act as modifiers of gene expression and DNA methylation in humans. Nucleic Acids Res 44: 3750–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Ranum LP, Day JW. 2002. Dominantly inherited, non-coding microsatellite expansion disorders. Curr Opin Genet Dev 12: 266–271. [DOI] [PubMed] [Google Scholar]
- Rival P, Press MO, Bale J, Grancharova T, Undurraga SF, Queitsch C. 2014. The conserved PFT1 tandem repeat is crucial for proper flowering in Arabidopsis thaliana. Genetics 198: 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya SM, Lennon D, Buschiazzo E, Gemmell N, Minin VN. 2012. Measuring microsatellite conservation in mammalian evolution with a phylogenetic birth–death model. Genome Biol Evol 4: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S, Bagshaw A, Buschiazzo E, Kumar P, Chowdhury S, Black MA, Gemmell N. 2013. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One 8: e54710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S, Jones M, Keller M. 2015. Linkage disequilibrium between single nucleotide polymorphisms and hypermutable loci. bioRxiv 10.1101/020909. [DOI] [Google Scholar]
- Schlötterer C, Kauer M, Dieringer D. 2004. Allele excess at neutrally evolving microsatellites and the implications for tests of neutrality. Proc Biol Sci 271: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. 2012. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28: 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Jonge JD, Forsberg SKG, Pettersson ME, Sheng Z, Hennig L, Carlborg Ö. 2014. Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet 10: e1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. 2014. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep 8: 2015–2030. [DOI] [PubMed] [Google Scholar]
- Sun JX, Helgason A, Masson G, Ebenesersdóttir SS, Li H, Mallick S, Gnerre S, Patterson N, Kong A, Reich D, et al. 2012. A direct characterization of human mutation based on microsatellites. Nat Genet 44: 1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S, Todesco M, Schneeberger K, Harilal R, Balasubramanian S, Weigel D. 2009. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323: 1060–1063. [DOI] [PubMed] [Google Scholar]
- Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. 2002. Signature of balancing selection in Arabidopsis. Proc Natl Acad Sci 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, Davis SJ, Verstrepen KJ, Queitsch C. 2012. Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci 109: 19363–19367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K. 2008. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res 18: 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. 2009. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324: 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems TF, Gymrek M, Highnam G, Mittelman D, Erlich Y. 2014. The landscape of human STR variation. Genome Res 24: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems T, Gymrek M, Poznik GD, Tyler-Smith C, 1000 Genomes Project Chromosome Y Group, Erlich Y. 2016. Population-scale sequencing data enable precise estimates of Y-STR mutation rates. Am J Hum Genet 98: 919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems T, Zielinski D, Yuan J, Gordon A, Gymrek M, Erlich Y. 2017. Genome-wide profiling of heritable and de novo STR variations. Nat Methods 14: 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H. 2009. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci 106: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Sabeti PC, Hardenbol P, Fu Q, Fry B, Lu X, Ghose S, Vega R, Perez A, Pasternak S, et al. 2005. Positive selection of a pre-expansion CAG repeat of the human SCA2 gene. PLoS Genet 1: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Pudovkin A, Weir BS. 2008. Correlation-based inference for linkage disequilibrium with multiple alleles. Genetics 180: 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.