Abstract
Human mitochondrial DNA (mtDNA) is believed to lack chromatin and histones. Instead, it is coated solely by the transcription factor TFAM. We asked whether mtDNA packaging is more regulated than once thought. To address this, we analyzed DNase-seq experiments in 324 human cell types and found, for the first time, a pattern of 29 mtDNA Genomic footprinting (mt-DGF) sites shared by ∼90% of the samples. Their syntenic conservation in mouse DNase-seq experiments reflect selective constraints. Colocalization with known mtDNA regulatory elements, with G-quadruplex structures, in TFAM-poor sites (in HeLa cells) and with transcription pausing sites, suggest a functional regulatory role for such mt-DGFs. Altered mt-DGF pattern in interleukin 3-treated CD34+ cells, certain tissue differences, and significant prevalence change in fetal versus nonfetal samples, offer first clues to their physiological importance. Taken together, human mtDNA has a conserved protein–DNA organization, which is likely involved in mtDNA regulation.
Global regulation of gene expression in the human genome is governed by a combination of chromatin structure, its availability to the transcription machinery, histone modifications, and DNA methylation status (Roadmap Epigenomics Consortium 2015; Zhu and Guohong 2016). However, this general scheme of gene expression regulation does not apply to the only component of the human genome that resides in the cytoplasm, within the mitochondrion—the mitochondrial genome (Bogenhagen 2012).
The mitochondrion has a pivotal role in cellular ATP production via the oxidative phosphorylation system (OXPHOS). In the vast majority of eukaryotes, OXPHOS protein-coding genes are divided between the mitochondrial and nuclear genomes (mtDNA and nDNA, respectively) with most (about 80) in the latter (nDNA), and 13 in the former (Calvo and Mootha 2010). Because of its centrality to life, OXPHOS dysfunction leads to devastating diseases, and mitochondrial DNA (mtDNA) variants and mutations are associated with a wide variety of common multifactorial disorders (Marom et al. 2017).
Unlike the nDNA, mtDNA gene expression is currently thought to be regulated by a relatively simple system with relic characteristics of the ancient bacterial ancestor of the mitochondria (Gustafsson et al. 2016). Accordingly, the core regulators of mtDNA transcription form an evolutionarily conserved set of factors, including two transcription factors (TFAM, TFB2M) (Falkenberg et al. 2002; Cotney et al. 2007; Shutt et al. 2010; Shi et al. 2012), one RNA polymerase (POLRMT) (Gaspari et al. 2004), one termination factor (MTERF1) (Yakubovskaya et al. 2010; Guja and Garcia-Diaz 2012), and a single known elongation factor TEFM (Minczuk et al. 2011). Second, some core mtDNA transcription regulators have bacterial characteristics, such as the phage ancestral structure of POLRMT (Ringel et al. 2011). Third, mtDNA genes are cotranscribed in strand-specific polycistrons (Aloni and Attardi 1971). Specifically, 12 mRNAs encoding protein subunits of the OXPHOS, 14 tRNAs, and two ribosomal RNA genes are encoded by the heavy strand; the mtDNA light strand encodes for a single mRNA (the ND6 subunit of OXPHOS complex I) and eight tRNA molecules. The polycistronic transcripts of both strands are cleaved, in turn, into individual transcripts following the tRNA punctuation model, or near alternative secondary structures, in cases in which genes are not separated by tRNAs, such as MT-CO3 and MT-ATP6 (Ojala et al. 1981; Montoya et al. 2006). Finally, it is currently thought that regulatory elements of human mtDNA gene expression are mainly located within the major mtDNA noncoding region, the D-loop. These include three strand-specific promoters: the light strand promoter (LSP), two heavy strand promoters (HSP1,2), the conserved sequence blocks (CSBI-III) including the transcription-replication transition site (CSBII), and the transcription termination associated sequence (TAS).
However, a growing body of evidence suggests that mtDNA transcriptional regulation is more complex than once thought. First, it has been found that known regulators of nuclear genes transcription, such as MEF2D (She et al. 2011), glucocorticoid receptor (Demonacos et al. 1993), the mitochondrial receptor for the thyroid hormone tri-iodothyronine (Wrutniak et al. 1995), bind the human mtDNA and regulate its gene expression (Leigh-Brown et al. 2010; Szczepanek et al. 2012; Barshad et al. 2018). Second, the usage of genomics tools, such as chromatin immunoprecipitation (ChIP)-seq, enabled us and others the identification of nuclear transcription regulators (JUN [cJUN], JUND, and CEBPB) that in vivo bind the human mtDNA (Blumberg et al. 2014; Marinov et al. 2014) and are imported into the mitochondria (Blumberg et al. 2014). As the binding sites of such factors occur within the mtDNA coding region, it is possible that human mtDNA coding sequences are written in two languages—the gene-coding one and the regulatory one. These pieces of evidence led us to hypothesize that the two billion years of endosymbiosis, and subsequent multiple adaptation events of the OXPHOS to changing energy requirements (Ellison and Burton 2010; Bar-Yaacov et al. 2015; Scott et al. 2015), were also accompanied by adaptation of mtDNA gene expression regulation to the eukaryote host environment.
In contrast to histone coating, and chromatin structure that modulates nuclear gene expression, the mtDNA higher-order organization, the nucleoid, is considered far less complex (Brown et al. 2011). Specifically, the mtDNA is known to be coated by a single HMG box protein, the transcription factor TFAM. There is a long-standing controversy regarding the mtDNA-binding specificity of TFAM. Whereas some findings imply lack of mtDNA sequence binding specificity (Kanki et al. 2004; Kaufman et al. 2007; Kukat et al. 2015), others proposed that TFAM has binding preferences to certain regions (Ghivizzani et al. 1994) and in vitro preferences to certain non-B mtDNA structures (Lyonnais et al. 2017). Recently, a study using a combination of high resolution microscopy and cell biology techniques revealed that TFAM coats the mtDNA in a dose-dependent manner, and that TFAM molecules bind the mtDNA approximately every 8 bp (Kukat et al. 2015). This observation is consistent with recent analysis of TFAM ChIP-seq experiments in HeLa cells (Wang et al. 2013). All of these findings support a very simple higher-order organization of the mammalian mtDNA. Nevertheless, recent reports showed direct involvement of the MOF Acetyl Transferase, a known chromatin structure modulator (Chatterjee et al. 2016) as well as members of the Sirtuin family (Nakamura et al. 2008), in mtDNA transcription regulation. Additionally, the mtDNA is thought to fold into transcription-related region-specific loops (Martin et al. 2005; Uchida et al. 2017). These pieces of evidence prompted us to hypothesize that the human mtDNA may have a chromatin-like packaging with a role in gene expression regulation.
Here, by analyzing DNase-seq experiments from multiple (more than 300) different human samples, we found mtDNA DNase Genomics Footprinting (DGF) sites that were common to ∼90% of the samples, which were conserved in mouse, colocalized with secondary DNA structures and with known mtDNA regulatory elements. The importance of this footprinting pattern for the organization and regulation of the mammalian mitochondrial genome is discussed.
Results
A common pattern of mtDNA DGF sites in a variety of human cell types
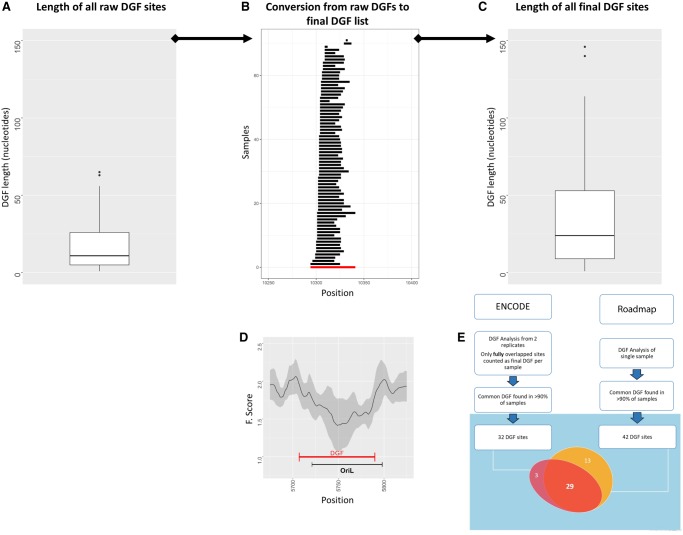
As a first step in characterizing mtDNA protein–DNA organization, we analyzed the comprehensive collection of DNase-seq experiments from the ENCODE Consortium (Fig. 1). We used only cells with experimental duplicates (70 cell samples) (Supplemental Table S1), separately analyzed each of the duplicates, and retained only those mtDNA DGF (mt-DGF) sites that were shared by the duplicates for further analyses. Briefly, to identify mt-DGF sites, we slightly modified a previously used approach (Mercer et al. 2011) and calculated an F-score for each mtDNA position in sliding sequencing read windows of variable size with a maximum of ∼124 bases and a minimum of 18 bases (see below); each sliding window was divided into one central and two flanking fragments (i.e., a left [L], right [R], and central [C] fragment) (Methods).
Figure 1.
Flow of mt-DGF analysis. (A) Distribution of raw mt-DGF lengths calculated for ENCODE samples. y-Axis represents length of each mt-DGF in nucleotides. (B) Conversion of a representative mt-DGF site from raw mt-DGF site data (black lines) collected from all analyzed samples to the final listed mt-DGF site (red line). Notice that for each mt-DGF, overlapping raw mt-DGFs were combined in all analyzed samples (for ENCODE and Roadmap, separately); the length of each combined mt-DGF was between the 5′ and 3′ nucleotide positions of the most proximal and distal overlapping raw DGFs, respectively. (C) Length distribution (in nucleotides; y-axis) of all final (combined) mt-DGF sites. (D) F-score graph of a representative mt-DGF site, which overlaps a well-known regulatory element—the light strand origin of replication (OriL): (x-axis) mtDNA position; (y-axis) calculated F-score; (black curve) mean F-score values, surrounded by their calculated SD (gray area), based on all ENCODE samples. (E) A flow chart of our mt-DGF analysis. The Venn diagram shows the actual number of common mt-DGF sites (i.e., identified in at least 90% of the samples) in the ENCODE and Roadmap data sets. The number of common sites whose mtDNA location overlapped between the two data sets is indicated.
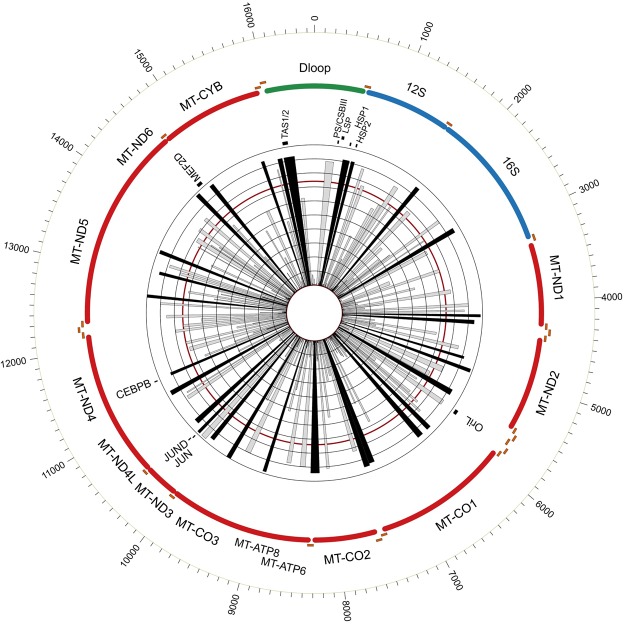
Our results revealed an average of 116.27 mt-DGF sites per cell line (SD = 22.65), encompassing a mean of 1868.36 bases (11.28%, ±1 SD = 403 bases) of the mtDNA sequence (Supplemental Data Set S1; Supplemental Fig. S1). Although a total of 246 mt-DGF sites were identified, covering more than half of the mtDNA sequence (8660 bases, representing 52.27% of the mtDNA), more than half of the sites (N = 135) were identified in ≤25% of the cell lines, suggesting that many mt-DGF sites are cell line-specific. While focusing on the most common mt-DGF sites, we found that 61 were shared by >75% of the tested samples, of which 32 were identified in >90% of the samples (Figs. 1, 2; Supplemental Table S2).
Figure 2.
Graphical overview of human mt-DGF sites. mtDNA location of all identified human mt-DGF sites (n = 246) according to their prevalence in the ENCODE sample collection. The inner ray histograms represent the prevalence of each site across ENCODE samples, with each concentric circle marking 10th percentile increments of the data set (from 0% to 100%). The red circle marks 74% of the samples, which refers to the average +1 SD of the samples tested. Black rays indicate the most prevalent mt-DGF sites. The mtDNA position of known regulatory elements and protein binding sites are indicated (JUN = cJUN).
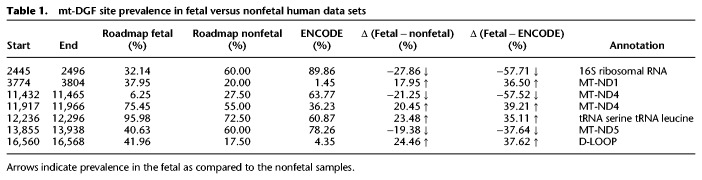
To further validate the most abundant mt-DGF sites, we extended our analysis to DNase-seq experiments generated by the NIH Roadmap Epigenomics Consortium, comprising 264 human samples from various tissues, mostly fetal (n = 224). Notably, the Roadmap data set enabled analysis of possible tissue specificity pattern of mt-DGF sites as well as comparison of fetal versus nonfetal samples (see details below). In total, we identified 221 mt-DGF sites in the Roadmap collection (Supplemental Tables S1, S2; Supplemental Data Set S2), of which 114 were in <25% of the samples and 64 were common to at least 75% of the tested samples. While focusing on the most abundant DGF sites, we found 42 sites that were common to >90% of the tested samples. Of the latter, 29 sites (Figs. 1, 2; Supplemental Table S2) were located throughout the mtDNA shared by the ENCODE and Roadmap data sets, suggesting the presence of a number of highly conserved mt-DGF sites across adult and fetal human cell types. Notably, one of the mt-DGF sites that was present in >90% of the fetal samples (Roadmap) was much less abundant in nonfetal samples (both in Roadmap and ENCODE data sets), suggesting some differences between fetal and nonfetal mt-DGF sites pattern (Table 1; and see below).
Table 1.
mt-DGF site prevalence in fetal versus nonfetal human data sets
As another validation of the identified mt-DGF sites, we analyzed Assay for Transposase-Accessible Chromatin (ATAC-seq) experiments, which, similar to DNase-seq, identify DNA sites that are occupied by proteins (Supplemental Table S1; Supplemental Data Set S3; Buenrostro et al. 2013). Our analysis of publicly available ATAC-seq experimental data from three different human cell types (GM12878, neural stem cell, CD34+) verified most (21) of the 29 common mt-DGF sites (Supplemental Table S3), thus supporting our approach and corroborating the true identification of the most common DGF sites in the human mtDNA.
The common mt-DGF sites are conserved from man to mouse, but do not differ in human SNP density
The high frequency of human mtDNA genetic variants, and the knowledge of their prevalence in human worldwide populations, enabled using SNP density as a tool to assess signatures of selection (Levin et al. 2013). Although mt-DGF sites were identified throughout the mtDNA, including protein-coding sequences, we limited the assessment of SNP density to third codon positions (n = 451) in the 23 mt-DGF sites within protein-coding genes; this focus was aimed to avoid misinterpretation due to selection acting on protein-coding sequences. Furthermore, in the frame of this analysis, we estimated the ratio between ancient mutations (nodal mutations, which were already exposed to long-term natural selection) and relatively recent mutations (present at the phylogenetic tree tips and that had less time to undergo selection) as previously performed (Blumberg et al. 2014). Although most mutational events in third codon mtDNA positions tend to be synonymous, some of these could be nonsynonymous (such as the replacement of GAG or GAA codons encoding Glu, by GAU, or GAC encoding Asp). Thus, we focused our analysis on third codon positions in mt-DGF sites that could generate only synonymous changes (number of positions = 219). This stringent approach revealed a slightly lower mt-DGF nodal/tip mutational events ratio (0.206) as compared to control (average ratio = 0.2182), yet this difference was not statistically significant (P = 0.32). Hence, analysis of mtDNA third codon silent SNPs that have accumulated during the course of human evolution did not reveal a significant difference between mt-DGFs and control simulations.
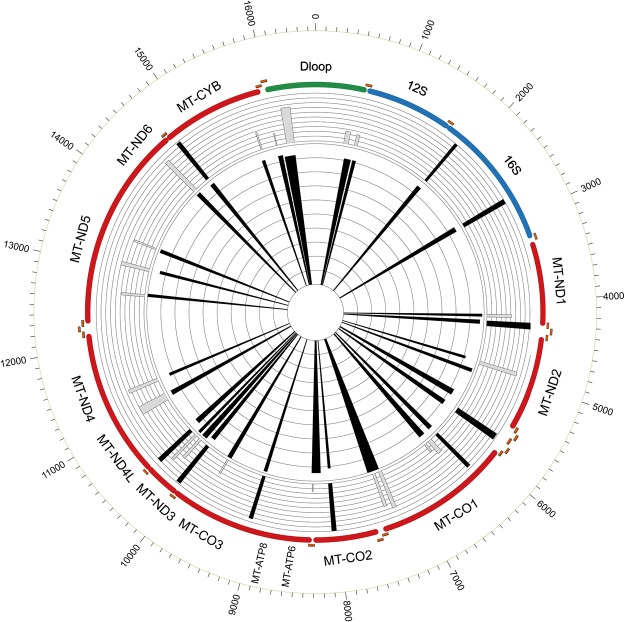
Since evolutionary conservation reflects functional importance, we assessed the conservation of mt-DGF sites from man to mouse by analyzing mt-DGF sites in mouse DNase-seq data generated by the ENCODE Consortium from 43 cell types in experimental duplicates (Supplemental Table S4; Supplemental Data Set S4). We found that 89.66% of the most common human mt-DGF sites were in the same mtDNA regions harboring mouse mt-DGF sites in at least 10% of the mouse cell lines tested (Fig. 3). Taken together, these data indicate conservation of the common mt-DGFs, supporting functional importance of mt-DGF sites during the course of evolution.
Figure 3.
Similarity of mouse mt-DGF sites to human common mt-DGF. Common human DGF sites across the human mtDNA map are presented, while superimposing mouse mt-DGF sites. The inner ray histogram corresponds to the 29 most common human mt-DGF sites, and the outer histogram corresponds to the mouse mt-DGF sites in grayscale, with black representing the most common sites.
Nuclear mitochondrial DNA fragments (NUMTs) are not enriched in mt-DGF sites
One could argue that many of the mtDNA DNase-seq reads are contaminated by mtDNA fragments that were gradually transferred into the nuclear genome during the course of evolution, known as Nuclear Mitochondrial pseudogenes (NUMTs) (Hazkani-Covo et al. 2003; Mishmar et al. 2004). Notably, DGF sites are defined by reduced number of reads at a given site, which might be affected by an excess of reads that were mapped to both the nucleus and to the active mtDNA. We therefore assessed the number of NUMT-associated reads in a comprehensive screen of mtDNA reads in the ENCODE DNase-seq data set, per sample per mtDNA position. To facilitate such a screen, we used the collection of NUMT variants in the entire human genome (n = 8031) (Methods), generated by Li et al. (2012). In general, the proportion of reads harboring NUMT-specific sequence variants comprised an average of only 0.165% of the reads (SD = 0.668%) (Supplemental Fig. S2). Furthermore, the proportion of NUMT reads was not statistically different between mt-DGF and non-DGF sites across the entire human mtDNA (DGF sites = 0.239%, SD = 1.676%; non-DGF = 0.163%, SD = 0.304%) (Supplemental Fig. S2). We conclude that NUMT reads had only negligible impact on our mt-DGF analysis.
The common mt-DGF sites colocalize with mtDNA regulatory elements
The existence of a common set of 29 mt-DGF sites shared between all available cell and tissue samples raise the possibility that these sites are functionally important. As an initial step toward assessing such possibility, we screened for association between the common mt-DGF sites and mtDNA elements with functional importance. This revealed that a subset of the mt-DGF sites colocalized with heavy strand promoters 1 and 2, origin of replication of the light strand (OriL), the termination associated sequence (TAS), and the recently identified protein-binding sites of JUN, JUND, and CEBPB (Fig. 2). Although this finding supported the importance of mt-DGF sites, it raised a question regarding the functionality of the remaining mt-DGF sites. Our recent identification of two transcription pausing sites that were common to 11 human cell lines tested (Blumberg et al. 2017), and their association with adjacent mt-DGF sites, urged us to extend our screen to additional such sites throughout the mtDNA. This screen revealed 20 pausing sites (PS) that were shared by at least eight of the 11 cell lines tested (Supplemental Fig. S3; Supplemental Table S5), of which 17 sites were in the light strand and three in the heavy strand. Notably, although only five of these pausing sites overlapped with the 29 common DGF sites, an additional 10 pausing sites overlapped with mt-DGF sites identified in >75% of the cell lines. This may imply that a subset of the mt-DGF sites, and the factors that bind them, are involved in mtDNA transcription. Notably, this result remains qualitative, and requires to be tested for statistical significance once additional pausing site experiments in more cell types become available.
G-quadruplex structures coincide with the common mt-DGF sites
It has been reported that stabilization of G-quadruplex (GQP) sequences in the human mtDNA affects mtDNA gene expression (Huang et al. 2015), associate with mtDNA deletions (Dong et al. 2014), and inhibit the mtDNA replication machinery (Bharti et al. 2014). With this in mind, and since our identified mt-DGFs associate with mtDNA regulatory elements, we asked whether the mt-DGF sites associate with G-quadruplex structures. Our results indicate that all 29 common mt-DGF sites overlap GQP sites (P = 2.16 × 10−9, two-tailed Fisher's exact test) (Supplemental Table S6).
The mt-DGF pattern correlates with TFAM-poor sites
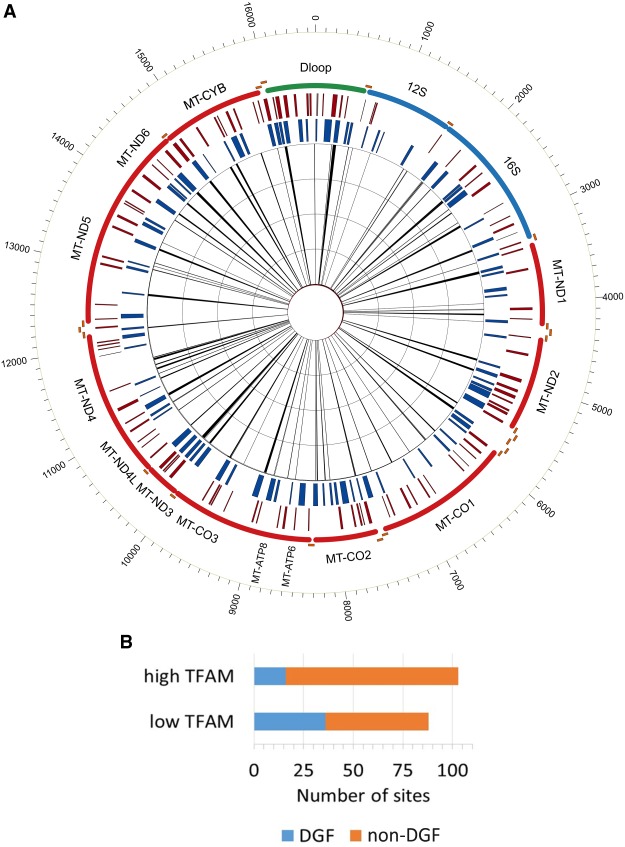
It is possible that most of the mt-DGF sites result from increased affinity of the coating protein TFAM to certain mtDNA sites. As a first step to test for this possibility, we took advantage of available TFAM ChIP-seq experiments performed in HeLa cells (Wang et al. 2013). Our analysis of HeLa DNase-seq revealed 92 mt-DGF sites, and TFAM ChIP-seq analyses revealed 103 human mtDNA sites enriched for TFAM binding, as well as 88 sites poor of TFAM ChIP-seq signals (marked as “high TFAM” and “low TFAM”, respectively) (Fig. 4; Supplemental Table S7; Methods). We found a significant overlap of the HeLa mt-DGFs (including the 29 common DGF sites) with the “low TFAM” sites (36 of 88 sites, P = 0.0059, two-tailed Fisher's exact test) as opposed to the “high TFAM” sites (18 of 103, P = 0. 89, two-tailed Fisher's exact test). These results indicate that most mt-DGF sites in HeLa cells are unlikely occupied by TFAM (Fig. 4). Twenty-five of the 29 most common mt-DGF sites colocalized with the “low TFAM” sites (P = 0.00065, two-tailed Fisher's exact test), whereas only 17 colocalized with the “high TFAM” sites (P = 0.69, two-tailed Fisher's exact test).
Figure 4.
TFAM binding and mt-DGF pattern in HeLa cells. (A) An mtDNA map of the DGF sites in HeLa cells (inner histograms), along with the low TFAM (blue bars) and high TFAM sites (red bars). (B) The proportion of high and low TFAM sites among mt-DGF sites in HeLa cells: (y-axis) the group of tested TFAM sites; (x-axis) number of sites. DGF sites are in blue, and non-DGF sites are in orange.
It has been suggested that TFAM preferentially binds GQP sites in vitro, although in 0.75M NaCl (Lyonnais et al. 2017), which is approximately five times higher than the physiological NaCl concentration (Li et al. 2016), but not at 0.25 M, which is approximately twofold higher than the physiological NaCl concentrations. We found that whereas “low TFAM” sites preferentially overlapped GQP sites (83/88 of the “low TFAM” sites, P = 3.1756 × 10−14, two-tailed Fisher's exact test as compared to control), the “high TFAM” sites did not (48/103 of the “high TFAM” sites, P = 0.19952, two-tailed Fisher's exact test as compared to control) (Supplemental Table S7). This supports correlation between in vivo non-B DNA structure and the mt-DGF sites.
Tissue-specific mt-DGF sites in the Roadmap collection
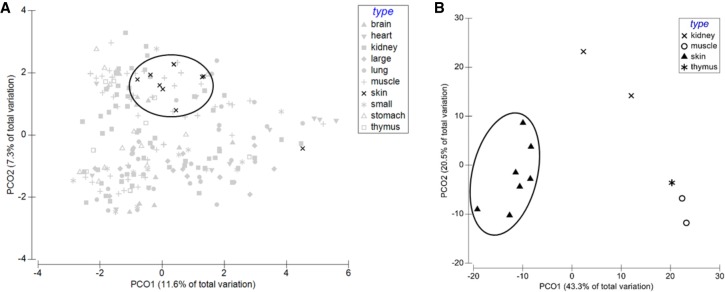
Because many of the identified mt-DGF sites were shared only by subsets of the tested cell lines, we hypothesized that at least part of those were tissue specific. To test for this hypothesis, we applied a PERMANOVA test and Principal Coordinates Analysis (PCO, using Primer-E; https://www.primer-e.com) to the Roadmap cell line data set. Inspection of the entire data set revealed strong clustering of samples from skin (Fig. 5); accordingly, most significant pairwise PERMANOVA analysis results (8/14) involved comparisons with skin (Supplemental Table S8). One may argue that our observed skin mt-DGF sites pattern could reflect a pattern in a certain embryo developmental stage rather than reflecting tissue specificity. To test for such a possibility, we analyzed the Roadmap DNase-seq data set while focusing on subgroups of samples which shared the same development day. Skin cluster was observed even while applying the PCO analysis to samples sharing the same developmental day (day = 97) (Supplemental Table S8; Fig. 5). Finally, further inspection of the PERMANOVA analysis reveal additional significant pairwise tissue comparisons, such as the comparisons involving muscle or kidney samples. Hence, certain cell types, such as the skin, may differ in mt-DGF patterns.
Figure 5.
Evidence for tissue specificity in the mt-DGF pattern. PCO analysis of the Roadmap collection. (A) All Roadmap samples (“large” and “small” refer to large intestine and small intestine, respectively). (B) All samples from fetal day 97 (total = 12; skin = 7). Encircled are the human skin samples.
mt-DGF sites pattern differs between fetal and nonfetal tissue samples
The most common mt-DGF sites have been shared between at least 90% of the samples, regardless of their tissue of origin and developmental stage. However, it is possible that mt-DGF site prevalence differs among developmental stages. As a step toward addressing this possibility, we divided the Roadmap data set into fetal (N = 224) and nonfetal (N = 40) samples and calculated mt-DGF site prevalence for each of the tested sample groups. Then, we calculated the difference in the prevalence of each identified mt-DGF site in fetal and nonfetal samples (here termed as “prevalence delta”—a mean value of −1 ± 8.53 SD), followed by identifying sites whose “prevalence delta” was significant (see details in Methods) (Supplemental Table S2). In brief, for each mt-DGF site, we calculated the prevalence delta by subtracting the occurrence of a given mt-DGF site in fetal samples (percentage) from its occurrence in nonfetal samples (percentage). First, 19 sites had a significant “prevalence delta” between fetal and nonfetal Roadmap samples (i.e., eight sites higher and 11 sites with lower values in the Roadmap fetal samples) (Supplemental Table S2). Second, 22 sites had a significant “prevalence delta” between fetal Roadmap and ENCODE samples (i.e., 11 sites higher and 11 sites with lower values in the Roadmap fetal samples, a mean value of −1.45 ± 16.86 SD) (Supplemental Table S2). While comparing the two previously described analyses, seven sites had significant “prevalence values” in both comparisons of fetal versus nonfetal data sets (Table 1). We conclude that although the most common mt-DGF sites are shared between samples regardless of their tissue or developmental stages, there are some mt-DGF sites whose prevalence differs between fetal and nonfetal samples, suggesting their physiological response.
mt-DGF site shift suggests function
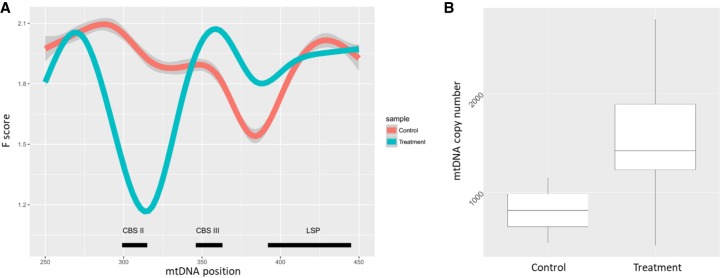
To gain more insight into the physiological importance of mt-DGF sites, we screened through ENCODE DNase-seq data from human cells exposed to diverse physiological conditions to identify conditions that were previously found to affect mitochondrial function. Treatment of mammalian cells by interleukin 3 (IL3) or erythropoietin was previously shown to lead to increase of mtDNA copy number, mitochondrial mass, as well as TFAM transcript levels (Carraway et al. 2010; Wellen et al. 2010). Our results indicate that treatment of human hematopoietic CD34+ lymphocytes by interleukin 3, hydrocortisone, succinate, and erythropoietin led to a 100-nt shift of the mt-DGF site at mtDNA positions 365–398 to nucleotide positions 279–347 (Fig. 6; Supplemental Table S1; Supplemental Data Set S5). As this treatment led to gain of a new mt-DGF site within Conserved Sequence Block II (CSBII), a known mtDNA transcription-to-replication transition point (Pham et al. 2006), we calculated mtDNA copy numbers in the treated and control cells. This analysis revealed a twofold increase in mtDNA copy numbers in the treated cells, suggesting a functional impact for the mt-DGF site shift (Fig. 6). We noticed that the experiments performed by ENCODE used control and treated cells having different mtDNA genetic backgrounds (haplogroups): Whereas the control cells belonged to either the U5b1 or K1a2a haplogroup, the treated cells belonged to the T2e haplogroup. To control for the possible inherent differences in mtDNA copy number between the mentioned mtDNA haplogroups, we reanalyzed the mtDNA copy number of B-lymphocytes from phylogenetically related genetic backgrounds, extracted from The 1000 Genomes Project (Cohen et al. 2016). Our results indicate no significant difference between the mtDNA copy number of cells belonging to mtDNA haplogroup U5 (N = 61 samples, mean mtDNA copy number = 421.77, SD = 88.36), haplogroup K1 (n = 15 samples, mean mtDNA copy number = 416.46, SD = 85.58), and haplogroup T2 (n = 18 samples, mean mtDNA copy number = 441.8, SD = 112.79 (T-test; U5 versus K1: P = 0.837 [NS]; U5 versus T2: P = 0.695 [NS]; T2 versus K1: P = 0.636 [NS]). Hence, mtDNA copy number does not significantly vary among the tested mtDNA haplogroups in B-lymphocytes; nevertheless, we cannot exclude such possible differences in other lymphocyte types. Taken together, our analyses offer first clues for physiological response of our discovered mt-DGF sites.
Figure 6.
Gain of mt-DGF site at the transcription-replication switch site, upon IL3-dependent activation of CD34+ cells. (A) Representation of the F-scores around the Light strand transcription start site: (y-axis) F-score; (x-axis) mtDNA position; (lowest F-score) most DNase I protected region; (thick line) average of F-scores across the analyzed samples; (dotted line) F-score calculated for single experiments; (red) control experiment; (gray) experiments of treated cells. (B) mtDNA copy number in control and treated cells.
Discussion
In the current study, we demonstrate for the first time, that the vast majority of human cell types display a conserved mtDNA footprinting pattern. Specifically, our analysis of DNase-seq experiments in more than 320 human cell types revealed 29 mt-DGF sites that were common to >90% of the tested samples. Since mt-DGF sites coincided with lower TFAM occupancy (in HeLa cells), higher-order organization of the human mitochondrial genome likely involves proteins other than TFAM. Therefore, although mtDNA condensation increases with elevated cellular concentration of TFAM (Kukat et al. 2015), mtDNA packaging is likely more organized and more complex than once thought. The finding of a common set of mt-DGF sites in multiple human cells, their colocalization with regulatory elements of mtDNA gene expression and some transcription pausing sites, along with the conservation of many mt-DGF sites from man to mouse, suggest that mt-DGF sites are functionally important.
One of our most intriguing findings is the colocalization of the mt-DGF sites with sequences that tend to adopt G-quadruplex structures. It was previously shown that in vivo stabilization of such structures in the mammalian mtDNA reduces the levels of mtDNA transcription and replication (Huang et al. 2015). This supports our interpretation that the novel mt-DGF sites are important for mtDNA regulation. Previously it was argued that such mtDNA structures are in vitro bound by TFAM (Lyonnais et al. 2017). Nevertheless, our analysis of ChIP-seq TFAM experiments in HeLa cells indicate that the mt-DGF sites are likely underoccupied by TFAM, and that the low TFAM sites, rather than the high TFAM sites, tend to harbor GQP sequences. As mt-DGF sites tend to harbor GQP sequences, mt-DGFs are likely bound by another protein(s), yet to be characterized. Notably, the apparent discrepancy with the in vitro GQP binding experiment of TFAM may be because, unlike the in vitro experiment, our observation was performed in living cells, throughout the mtDNA, and under physiological conditions. Previously, several proteins have been assigned to the mitochondrial nucleoid, yet only TFAM was clearly shown to participate in mtDNA packaging (Lee and Han 2017). Our highly ordered mt-DGF site pattern in human cells implies departure from current thought—it suggests a chromatin-like structure in the human mtDNA. Our observed correlation between the mt-DGF sites and mtDNA regulatory sites tempts us to suggest a conceptual similarity between the higher-order organization of the nuclear and mitochondrial genomes, hence reflecting a regulatory aspect of adaptation of the mitochondrion to its ancient host. The growing collection of transcription factors (and other DNA-binding proteins) that are used in ChIP-seq experiments increase the odds of identifying this unknown mtDNA-binding factor(s) in the future.
While considering all mt-DGF sites, profound differences in their prevalence and pattern among cell types was observed. In fact, most mt-DGF sites were identified in <25% of the tested samples (135/245 DGF sites). Notably, a skin-specific pattern of mt-DGF sites’ occurrence was the most prominent of all, yet other tissues (such as muscle and kidney) displayed significant pairwise differences. Such mt-DGF variation among tissue may impact the tissue variability of nucleoid structure and composition (Lee and Han 2017). One possibility to explain such variability stems from differences in physiological conditions. Indeed, by targeting bacterial DNA methyltrasferases to the mitochondria in human cells, it was found that the level of mtDNA protein occupancy differed among mtDNA regulatory sites in response to various physiological insults (Rebelo et al. 2009). It is thus plausible that sample variability in patterns of mt-DGF sites could, in part, reflect differential responses to physiological differences. This is supported by our observed shift in the mt-DGF site pattern in CD34+ cells treated with interleukin 3, hydrocortisone, succinate, and erythropoietin as compared to control (Fig. 6). This strongly suggests that our observed mt-DGF pattern cannot be attributed to recently suggested sequence-specific DNase I cutting bias (He et al. 2014), but rather has biological importance. Furthermore, our analysis revealed that the prevalence of several mt-DGF sites significantly differed between fetal and nonfetal samples, again supporting a physiological response of mt-DGF sites. Nevertheless, better understanding of this possibility awaits future controlled comparison of the mt-DGF sites pattern in cells exposed to a variety of physiological conditions as well as controlled assessment of their connection to mtDNA transcription and/or replication.
In summary, our comprehensive analysis of human mtDNA DNase Genomic Footprinting experiments revealed 29 mt-DGF sites that were common to >90% of approximately 320 different human cell types. This provided first clues for a systematic and regulated organization of this genome in the vast majority of human tissues. The striking colocalization of GQP motifs in mt-DGF sites along with overrepresentation of such DGF sites in mtDNA regions with lower TFAM occupancy (in HeLa cells), which also harbor GQP structures, further supports the potential functional importance of these sites and suggests that mtDNA-binding factors other than TFAM are involved in this mtDNA protein organization. The generality of this suggestion should be further tested once TFAM ChIP-seq experiments become available from additional human samples. Second, we found first evidence for the physiological response of mt-DGFs, suggesting their importance in living cells. Third, the colocalization of the mt-DGF sites with known mtDNA regulatory elements, with several transcription pausing sites throughout the mitochondrial genome and synteny conservation of the human mt-DGFs in the mouse mtDNA, again support their potential functionality. Hence, organization of the mitochondrial genome is likely far more regulated, and certainly more complex, than once thought.
Methods
DNase-seq and ATAC-seq data sets
ENCODE DNase-seq FASTQ files were downloaded from The ENCODE Project Consortium (The ENCODE Project Consortium 2012) website (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwDnase/) for human and for mouse (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/encodeDCC/wgEncodeUwDnase/). Roadmap DNase-seq SRA files were downloaded from the Roadmap Epigenomics project website (Roadmap Epigenomics Consortium 2015) (http://egg2.wustl.edu/roadmap/web_portal/index.html). Data of treated and control CD34+ cells was taken from The ENCODE Project Consortium website (https://www.encodeproject.org/search/?searchTerm=hematopoietic+multipotent+progenitor+cell&type=Experiment&assay_title=DNase-seq).
Guidelines of the DNase-seq experiments are available (https://www.encodeproject.org/documents/a549ac0a-c991-4f78-a0ba-36c81f232aed/@@download/attachment/DNase_experimental_guidelines_Jan2017.pdf).
ATAC-seq data was downloaded from Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra), from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), and from European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena). The ATAC-seq accession numbers are listed in Supplemental Data Set S3.
Sample-specific mtDNA sequence reconstruction and mapping, coverage calculation, and circular-like mapping of sample-specific mtDNA sequence
Analyses were applied as described previously (Blumberg et al. 2017). In brief, after mapping the DNase-seq reads (read length = 36 nt) the mtDNA reference genome (rCRS; NCBI Reference Sequence: NC_012920.1), sample-specific mtDNA reference genome was reconstructed and the DNase-seq reads were aligned while taking into account the circular organization of the mtDNA as recently performed (Blumberg et al. 2017). Read coverage for each position was calculated using the “genomecov” command in BEDTools (http://bedtools.readthedocs.org/en/latest/; version 2.25) (Quinlan and Hall 2010).
Analysis of mt-DGF sites
DGF sites were identified following the method outlined (https://github.com/StamLab/footprinting2012/). Briefly, for each mtDNA nucleotide position, an F-score was calculated in sliding read windows of variable size with a maximum of 124 bases and a minimum of 18 bases (see below), using the following equation: F = (C + 1)/L + (C + 1)/R, where “C” represents the average number of reads in the central fragment C; “L” represents the average read count in the proximal fragment, and “R” represents the average read count in the distal fragment. Following published specifications (https://github.com/StamLab/footprinting2012/) (Neph et al. 2012), the variable sliding window sizes was comprised of 6–100 bases for the “C” fragment and 6–12 bases for each of the “L” and “R” fragments. According to the preceding specifications, the calculated F-score values for a given position for all sliding windows enabled identifying the lowest calculated value as the optimal F-score. The sliding window was used to avoid bias for a certain window length, since the mt-DGF sites may reflect the binding of different proteins (sometimes in protein complexes) having different binding capacity, sequence preferences, and different length of sequence occupancy. Regions showing the lowest F-scores for all mtDNA positions in a given sample (mean – 1 SD), which reflected relative depletion of reads, were listed as mt-DGF sites. In the ENOCDE database, for which all analyzed samples had experimental duplicates, only sites that fully overlapped between the duplicates were considered a positive mt-DGF site (using the “intersect” command from BEDTools suite). mt-DGF from all ENCODE samples were merged in order to have a list of m-tDGF sites. This enabled subsequent comparison of the mt-DGF sites between samples (Fig. 1). Notably, since most Roadmap samples did not include experimental duplicates, for the sake of consistency, we listed all identified mt-DGFs while using only one experiment per sample.
Principal Coordinates Analysis
Principal Coordinates (PCO) Analysis is an approach to analyze and visualize similarities of data. Specifically, PCO analyses were performed to assess tissue-specific patterns of mt-DGF sites. In our case, we generated a matrix in which the x-axis constitutes the Roadmap samples (indicated according to their tissues of origin) and the y-axis constitutes the mtDNA coordinates of each identified mt-DGF site, while indicating presence/absence of each site per sample. This matrix enabled calculating the resemblance score between each of the analyzed samples using the D1 Euclidean Distance approach. To perform these analyses, we used Primer-E (version 6; http://www.primer-e.com/). Statistical details of the PCO analyses can be found in Supplemental Table S8.
PERMANOVA analysis
To assess the statistical significance of mt-DGF site sharing between tissues in the Roadmap data set, we performed a dissimilarity PERMANOVA analysis. This analysis assesses similarity in mt-DGF site sharing between all possible pairwise tissue available in the Roadmap data set. The P-values were corrected for multiple testing (Bonferroni correction).
Comparison of mt-DGF site prevalence between fetal and nonfetal samples
Differences in mt-DGF pattern between the fetal and nonfetal samples were calculated using the following consecutive steps: First, mt-DGF prevalence (i.e., the presence of each site in samples of a given data set) was calculated per site in the 224 fetal and 40 nonfetal Roadmap samples. Then, the difference in the prevalence of each identified mt-DGF site in fetal and nonfetal samples was calculated (here termed as “prevalence delta”). We considered an mt-DGF site prevalence to significantly differ between fetal and nonfetal sample data sets when the “prevalence delta” value exceeded 2 SD from the mean of “prevalence delta” values of all identified sites. Second, we applied the same logic while calculating and comparing the “prevalence delta” between Roadmap fetal samples and the ENCODE sample collection (all nonfetal samples). For the final set of significantly different fetal\nonfetal mt-DGF sites, we listed only the sites that showed a significant “prevalence delta” values in both comparisons (i.e., fetal versus nonfetal Roadmap samples, and fetal Roadmap versus Encoded samples).
NUMTs analysis
The proportion of NUMT reads was estimated by counting mtDNA-mapped DNase-seq reads (within BAM files) that contain NUMT variants using bam-readcount (https://github.com/genome/bam-readcount). For the sake of consistency and without compromising the comprehensive nature of our analysis, we used an updated collection of 8031 human NUMT variants published by Li et al. (2012). For each mtDNA position that harbors a NUMT-specific mutation, we recorded the proportion of reads harboring these mutations out of the reads covering such mtDNA position. For every analyzed sample, the sample-specific NUMT collection was generated by screening the reconstructed sample-specific mtDNA sequences.
Assessment of SNP frequency at the mt-DGF sites
Phylogenetic analysis of nearly 10,000 whole mtDNA sequences representing all major global populations allowed for the extraction of multiple ancient mutational events (Levin et al. 2013). The number of mutational events at the third codon position of coding region DGF sites was counted and compared to the number of mutational events in the entire set of third codon positions in all mtDNA protein-coding genes. The logic underlying this analysis was based on the following simple argument: A reduced number of mutational events at a given tested site reflects a signature of negative selection. To confirm that the lower number of variants in the third codon position in the DGF sites cannot be explained by chance, we applied 10,000 replicated simulations in which the sample size of the third codon position within mt-DGFs was retained but shuffled with other third codon positions throughout the mtDNA protein-coding regions.
Synteny mapping of human and mouse mt-DGF locations
Mouse and human mtDNAs share the same gene order and content but differ in two main characteristics: First, the length of human mtDNA is 16,569 bases, whereas mouse mtDNA constitute only 16,299 bases, mostly due to difference in the D-loop (245 bp shorter in the mouse). The rest of the 25 nt that are present in human but not in mouse mtDNAs are dispersed throughout this genome. Second, the nucleotide position “1” of the human mtDNA is located within the D-loop, whereas in the mouse nucleotide, “1” is the first position of tRNA phenylalanine. Thus, to correctly map in a single nucleotide resolution and correlate the DGF locations in human and mouse mtDNAs, we constructed a synteny map in a single base resolution, using each of the mtDNA genes as anchors, while taking into account the “indel” differences in the mouse versus human mtDNAs.
Prediction of G-quadruplex DNA structures
G-quadruplex DNA structures were predicted for each of light and heavy mtDNA strands using QGRS Mapper (http://bioinformatics.ramapo.edu/QGRS/index.php) (Kikin et al. 2008). We used the prediction parameters as in Dong et al. (2014) (GQP max. length = 33; minimum G-group size = 2; loop size = 0 to 36). The mtDNA site coordinates were listed after merging the predicted site coordinates for both mtDNA strands.
Pausing sites analysis
Pausing sites were identified as recently described (Blumberg et al. 2017). In brief, Pausing Index (PI) was calculated using the following equation: PI = (T + 1)/(GB + 1), where T represents density of reads in 20 bases of the tested position, and GB represents the density of reads in the gene body. Since the mtDNA molecule is cotranscribed in its entirety, with no gene specific promoters, for the sake of PI analysis we defined “gene body” as a maximum of a 1000 nt that flank the tested position. Accordingly, to minimize putative reciprocal influence of close internal pausing sites, “gene body” of each position was calculated in sliding windows of 10–1000 bases that flank each of the tested positions (both upstream and downstream). The highest PI value for each position was considered as the optimal value for the tested position. For each experiment, positions exhibiting higher PI values than the average + 1 SD were considered as pausing sites.
Identification of mtDNA sites with high and low TFAM occupancy
We reanalyzed the results of four biological replicates of TFAM ChIP-seq experiments in HeLa cells (Wang et al. 2013). Since TFAM binds throughout the mtDNA, we calculated F-scores for TFAM binding following the same logic that was used for the identification of the DGF sites. In brief, lowest F-scores (mean − 1 SD) were considered sites with low TFAM occupancy (similar to the identification of DGF sites). For TFAM enriched sites, similar approach but in the opposite direction was used, i.e., the top F-scores (mean + SD) were taken in account.
Statistical comparison between BED files
Statistical comparison between sets of sites represented in BED format was based on BEDTools (Quinlan and Hall 2010). Fisher's exact test on the number of overlaps between two sets of sites was measured using “fisher” command (http://bedtools.readthedocs.io/en/latest/content/tools/fisher.html).
mtDNA copy number estimation
mtDNA copy number of DNase-seq experiments was estimated as recently described (Cohen et al. 2016) with minor modifications. In brief, to control for over- or underrepresentation of sequencing reads in certain genomic regions, we compared the mtDNA read numbers to regions of 5 × 106 bases from each of the 22 autosomal chromosomes.
Haplogroups assignment
mtDNA haplogroup assignment of samples was performed by analyzing reconstructed whole mtDNA sequences using HaploGrep (Kloss-Brandstätter et al. 2011).
Visual representations of the mitochondrial genome
Circos was used for visualization of all circular mtDNA graphs (Krzywinski et al. 2009).
Data access
All of the listed Supplemental Data Sets include all BED files from which we extracted the mtDNA coordinates of the identified mt-DGFs in the relevant databases. For a detailed description of the data sets format, see ReadMe file (https://figshare.com/s/5efbf31aa6e34edeee2e).
Supplemental Data Set S1: Human mt-DGF sites from the ENCODE database:
Supplemental Data Set S2: Human mt-DGF site from RoadMap:
Supplemental Data Set S3: Human mt-DGF from ATAC-seq:
Supplemental Data Set S4: Mouse mt-DGF sites from ENCODE:
Supplemental Data Set S5: Human mt-DGF sites CD34+ treatment from ENCODE:
Supplementary Material
Acknowledgments
This work was funded by an Israel–US Binational Science Foundation research grant number 2013060 awarded to D.M. and A.K., by the US Army Life Sciences division grant number 67993-LS, and Israeli Science Foundation grant number 372/17 awarded to D.M. Finally, the authors thank the Harbor Foundation for a scholarship for excellent PhD students awarded to A.B.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.230409.117.
References
- Aloni Y, Attardi G. 1971. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci 68: 1757–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshad G, Marom S, Cohen T, Mishmar D. 2018. Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet 10.1016/j.tig.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Bar-Yaacov D, Hadjivasiliou Z, Levin L, Barshad G, Zarivach R, Bouskila A, Mishmar D. 2015. Mitochondrial involvement in vertebrate speciation? The case of mito-nuclear genetic divergence in chameleons. Genome Biol Evol 7: 3322–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti SK, Sommers JA, Zhou J, Kaplan DL, Spelbrink JN, Mergny JL, Brosh RM Jr. 2014. DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J Biol Chem 289: 29975–29993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg A, Sri Sailaja B, Kundaje A, Levin L, Dadon S, Shmorak S, Shaulian E, Meshorer E, Mishmar D. 2014. Transcription factors bind negatively-selected sites within human mtDNA genes. Genome Biol Evol 6: 2634–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg A, Rice EJ, Kundaje A, Danko CG, Mishmar D. 2017. Initiation of mtDNA transcription is followed by pausing, and diverges across human cell types and during evolution. Genome Res 27: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF. 2012. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta 1819: 914–920. [DOI] [PubMed] [Google Scholar]
- Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA. 2011. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol Cell Biol 31: 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK. 2010. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet 11: 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway MS, Suliman HB, Jones WS, Chen CW, Babiker A, Piantadosi CA. 2010. Erythropoietin activates mitochondrial biogenesis and couples red cell mass to mitochondrial mass in the heart. Circ Res 106: 1722–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Seyfferth J, Lucci J, Gilsbach R, Preissl S, Böttinger L, Mårtensson CU, Panhale A, Stehle T, Kretz O, et al. 2016. MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell 167: 722–738.e23. [DOI] [PubMed] [Google Scholar]
- Cohen T, Levin L, Mishmar D. 2016. Ancient out-of-Africa mitochondrial DNA variants associate with distinct mitochondrial gene expression patterns. PLoS Genet 12: e1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Wang Z, Shadel GS. 2007. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res 35: 4042–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonacos C, Tsawdaroglou NC, Djordjevic-Markovic R, Papalopoulou M, Galanopoulos V, Papadogeorgaki S, Sekeris CE. 1993. Import of the glucocorticoid receptor into rat liver mitochondria in vivo and in vitro. J Steroid Biochem Mol Biol 46: 401–413. [DOI] [PubMed] [Google Scholar]
- Dong DW, Pereira F, Barrett SP, Kolesar JE, Cao K, Damas J, Yatsunyk LA, Johnson FB, Kaufman BA. 2014. Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics 15: 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Burton RS. 2010. Cytonuclear conflict in interpopulation hybrids: the role of RNA polymerase in mtDNA transcription and replication. J Evol Biol 23: 528–538. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. 2002. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet 31: 289–294. [DOI] [PubMed] [Google Scholar]
- Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. 2004. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J 23: 4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghivizzani SC, Madsen CS, Nelen MR, Ammini CV, Hauswirth WW. 1994. In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol Cell Biol 14: 7717–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guja KE, Garcia-Diaz M. 2012. Hitting the brakes: termination of mitochondrial transcription. Biochim Biophys Acta 1819: 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson CM, Falkenberg M, Larsson NG. 2016. Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem 85: 133–160. [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Sorek R, Graur D. 2003. Evolutionary dynamics of large numts in the human genome: rarity of independent insertions and abundance of post-insertion duplications. J Mol Evol 56: 169–174. [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, et al. 2014. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods 11: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Tseng TY, Chen YT, Chang CC, Wang ZF, Wang CL, Hsu TN, Li PT, Chen CT, Lin JJ, et al. 2015. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res 43: 10102–10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. 2004. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol 24: 9823–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. 2007. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell 18: 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O, Zappala Z, D'Antonio L, Bagga PS. 2008. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res 36: D141–D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss-Brandstätter A, Pacher D, Schönherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. 2011. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat 32: 25–32. [DOI] [PubMed] [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, Joos F, Polosa PL, Park CB, Posse V, et al. 2015. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci 112: 11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Han J. 2017. Mitochondrial nucleoid: shield and switch of the mitochondrial genome. Oxid Med Cell Longev 2017: 8060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh-Brown S, Enriquez JA, Odom DT. 2010. Nuclear transcription factors in mammalian mitochondria. Genome Biol 11: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin L, Zhidkov I, Gurman Y, Hawlena H, Mishmar D. 2013. Functional recurrent mutations in the human mitochondrial phylogeny: dual roles in evolution and disease. Genome Biol Evol 5: 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schroeder R, Ko A, Stoneking M. 2012. Fidelity of capture-enrichment for mtDNA genome sequencing: influence of NUMTs. Nucleic Acids Res 40: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sun SR, Yap JQ, Chen JH, Qian Q. 2016. 0.9% saline is neither normal nor physiological. J Zhejiang Univ Sci B 17: 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Tarrés-Solé A, Rubio-Cosials A, Cuppari A, Brito R, Jaumot J, Gargallo R, Vilaseca M, Silva C, Granzhan A, et al. 2017. The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Sci Rep 7: 43992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov GK, Wang YE, Chan D, Wold BJ. 2014. Evidence for site-specific occupancy of the mitochondrial genome by nuclear transcription factors. PLoS One 9: e84713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom S, Friger M, Mishmar D. 2017. MtDNA meta-analysis reveals both phenotype specificity and allele heterogeneity: a model for differential association. Sci Rep 7: 43449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. 2005. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 123: 1227–1240. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. 2011. The human mitochondrial transcriptome. Cell 146: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. 2011. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res 39: 4284–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC. 2004. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat 23: 125–133. [DOI] [PubMed] [Google Scholar]
- Montoya J, López-Pérez MJ, Ruiz-Pesini E. 2006. Mitochondrial DNA transcription and diseases: past, present and future. Biochim Biophys Acta 1757: 1179–1189. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ogura M, Tanaka D, Inagaki N. 2008. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun 366: 174–179. [DOI] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- Pham XH, Farge G, Shi Y, Gaspari M, Gustafsson CM, Falkenberg M. 2006. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J Biol Chem 281: 24647–24652. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AP, Williams SL, Moraes CT. 2009. In vivo methylation of mtDNA reveals the dynamics of protein–mtDNA interactions. Nucleic Acids Res 37: 6701–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. 2011. Structure of human mitochondrial RNA polymerase. Nature 478: 269–273. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GR, Elogio TS, Lui MA, Storz JF, Cheviron ZA. 2015. Adaptive modifications of muscle phenotype in high-altitude deer mice are associated with evolved changes in gene regulation. Mol Biol Evol 32: 1962–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, Mao Z. 2011. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest 121: 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Dierckx A, Wanrooij PH, Wanrooij S, Larsson NG, Wilhelmsson LM, Falkenberg M, Gustafsson CM. 2012. Mammalian transcription factor A is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci 109: 16510–16515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. 2010. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci 107: 12133–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek K, Lesnefsky EJ, Larner AC. 2012. Multi-tasking: nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol 22: 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida A, Murugesapillai D, Kastner M, Wang Y, Lodeiro MF, Prabhakar S, Oliver GV, Arnold JJ, Maher LJ, Williams MC, et al. 2017. Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy-strand promoter. eLife 6: e27283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YE, Marinov GK, Wold BJ, Chan DC. 2013. Genome-wide analysis reveals coating of the mitochondrial genome by TFAM. PLoS One 8: e74513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev 24: 2784–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrutniak C, Cassar-Malek I, Marchal S, Rascle A, Heusser S, Keller JM, Fléchon J, Dauca M, Samarut J, Ghysdael J, et al. 1995. A 43-kDa protein related to c-Erb A α1 is located in the mitochondrial matrix of rat liver. J Biol Chem 270: 16347–16354. [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. 2010. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell 141: 982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Guohong L. 2016. Structural insights of nucleosome and the 30-nm chromatin fiber. Curr Opin Struct Biol 36: 106–115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.