Abstract
Although many driver mutations are thought to promote carcinogenesis via abnormal splicing, the landscape of splicing-associated variants (SAVs) remains unknown due to the complexity of splicing abnormalities. Here, we developed a statistical framework to systematically identify SAVs disrupting or newly creating splice site motifs and applied it to matched whole-exome and transcriptome sequencing data from 8976 samples across 31 cancer types, generating a catalog of 14,438 SAVs. Such a large collection of SAVs enabled us to characterize their genomic features, underlying mutational processes, and influence on cancer driver genes. In fact, ∼50% of SAVs identified were those disrupting noncanonical splice sites (non-GT-AG dinucleotides), including the third and fifth intronic bases of donor sites, or newly creating splice sites. Mutation signature analysis revealed that tobacco smoking is more strongly associated with SAVs, whereas ultraviolet exposure has less impact. SAVs showed remarkable enrichment of cancer-related genes, and as many as 14.7% of samples harbored at least one SAVs affecting them, particularly in tumor suppressors. In addition to intron retention, whose association with tumor suppressor inactivation has been previously reported, exon skipping and alternative splice site usage caused by SAVs frequently affected tumor suppressors. Finally, we described high-resolution distributions of SAVs along the gene and their splicing outcomes in commonly disrupted genes, including TP53, PIK3R1, GATA3, and CDKN2A, which offers genetic clues for understanding their functional properties. Collectively, our findings delineate a comprehensive portrait of SAVs, novel insights into transcriptional de-regulation in cancer.
Comprehensive genomic characterization of multiple cancer types in large-scale genetic studies has increasingly broadened the catalog of somatic alterations that dictate cancer evolution, including single nucleotide variants (SNVs), small indels (insertions and deletions), and copy number alterations (Garraway and Lander 2013; Vogelstein et al. 2013; Martincorena and Campbell 2015). Moreover, it has also revealed disturbances in transcriptional regulation, such as expression changes and splicing defects that underlie cancer pathogenesis (Garraway and Lander 2013; Vogelstein et al. 2013). However, there has been only a little progress in the understanding of how somatic alterations in cancer genomes exert direct transcriptional consequences.
In cancer transcriptomes, splicing defects play important roles in many aspects of cancer development and progression (Venables 2004; Kalnina et al. 2005; Dvinge et al. 2016; Scotti and Swanson 2016; Singh and Eyras 2017). Discovery of somatic variants affecting RNA splicing factors, such as SF3B1 and U2AF1, which induce extensive alterations in RNA splicing (trans-acting regulation) in several kinds of cancers, highlights the relevance of RNA missplicing in cancer pathogenesis (Yoshida et al. 2011; Brooks et al. 2014; Dvinge et al. 2016). Another mechanism, which is the focus of this paper, is cis-acting regulation, in which somatic variants directly cause abnormal splicing of the affected gene. For example, somatic variants in canonical splice sites (highly conserved GT-AG dinucleotides at exon-intron boundaries) have long been reported to cause dysregulation of cancer-related genes (Venables 2004; Kalnina et al. 2005). These variants can induce different forms of abnormal splicing, such as exon skipping, intron retention, and activation of cryptic splice sites (SSs). Recent pan-cancer studies showed that SNVs causing aberrant intron retention in exon-intron boundaries are enriched in tumor suppressor genes (TSGs), especially TP53 (Supek et al. 2014; Jung et al. 2015). However, the complexity of splicing systems and the perplexing relationship between somatic variants and splicing alterations have limited the opportunities for systematic analyses of the extent and consequences of splicing-associated variants (SAVs): Due to the diversity of transcription across tissues as well as individuals, a huge number of transcripts have not been well defined, making it difficult to distinguish abnormal transcripts from normal variations. Together with this diversity, the rarity of SAVs (usually represented by just one sample in a cohort) hampers the sensitivity of conventional statistical methods to measure the association between variant status and splicing changes, unless there are some restrictions on the association to be considered (e.g., aberrant splicing caused by variants near exon-intron boundaries). In addition, there is not always a one-to-one relationship between them; a somatic variant occasionally generates different abnormal splicing events, whereas several different somatic variants sometimes cause the same splicing event.
To overcome these limitations, we have developed a novel algorithm, SAVNet (Splicing-Associated Variant detection by NETwork modeling), for detecting SAVs based on a list of somatic variants in a cohort and its matched RNA sequencing (RNA-seq) data using a rigorous statistical framework. One of the keys to success is that we carefully set the association rules between variants and abnormal splicing, including the restriction of relevant positional relationships between them. Furthermore, we have resolved the complex relationships between variants and splicing by utilizing network-based modeling and a Bayesian model averaging framework. Through this approach, we performed a comprehensive analysis of a large number of primary cancer samples across 31 cancer types from The Cancer Genome Atlas (TCGA), deciphering the landscape of splicing aberrations caused by cis-acting variants in human cancers.
Results
Overview of the SAVNet framework
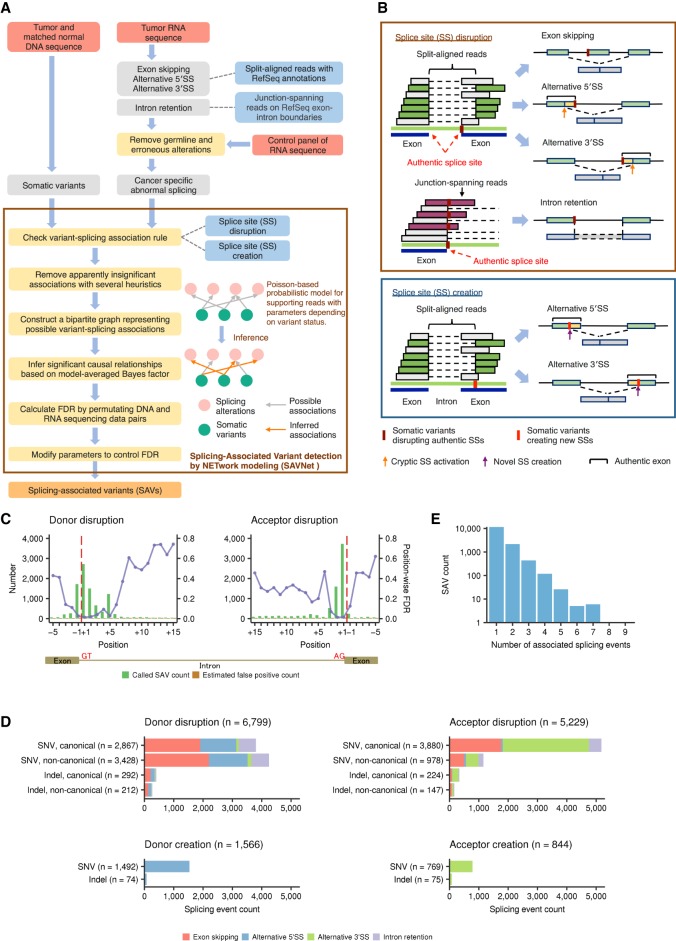
The overview of the proposed framework (SAVNet) is summarized in Figure 1A. First, we collected evidence of abnormal splicing from tumor-derived RNA-seq data. Exon skipping and alternative 5′ SS and 3′ SS usage (defined by RefSeq transcript annotation) were extracted by capturing splicing junctions demarcated by split-aligned sequencing reads, whereas intron retention was identified by detecting sequencing reads spanning exon-intron boundaries (Fig. 1B). To obtain reliable and interpretable results, we focused exclusively on either (1) a somatic variant located at or close to an authentic exon-intron boundary (registered in the RefSeq database), in which normal splicing is disrupted (SS disruption), or (2) a somatic variant located within a newly created SS inferred by an alternative SS usage event (SS creation). To represent the complex relationships, we constructed a bipartite graph showing all potential associations between somatic variants and abnormal splicing events for each gene. Next, based on a probabilistic model for the number of abnormal splicing-supporting reads and the presence of a somatic variant, we deduced significant causal relationships through the evaluation of a Bayes factor incorporating a Bayesian model averaging framework (Supplemental Fig. S1A; Flutre et al. 2013; Stephens 2013). A simulation study investigating the effect of the number of variant-splicing associations validated that the proposed framework can utilize the information from multiple associations for the sensitive identification of SAVs (Supplemental Fig. S1B,C).
Figure 1.
Workflow and evaluation of SAVNet and overview of SAVs. (A) Workflow for detecting SAVs by SAVNet from matched WES and RNA-seq data. (B) Schematics depicting quantification methods of exon skipping and alternative 5′ SS or 3′ SS usage (by split-aligned reads) and intron retention (by junction-spanning reads) and examples of somatic variants associated with abnormal splicing. SAVs within authentic SSs that disrupt normal splicing (SS disruption) and those outside authentic SSs that create alternative SSs (SS creation) were evaluated separately. (C) Evaluation of position-wise numbers of SAVs (green) and estimated false positives (brown) between the fifth exonic base (−5) and the 15th intronic base (+15) for splicing donor and acceptor sites. Purple points with lines show estimated position-wise FDRs. Red dashed lines represent exon-intron boundaries. (D) Number of each type of abnormal splicing events for each SAV type, stratified by (1) donor or acceptor, (2) disruption or creation, (3) SNVs or indels, and (4) canonical or noncanonical sites. Numbers in parentheses indicate the number of each type of SAV. (E) Histogram of the number of SAVs according to the number of associated abnormal splicing events. See also Supplemental Figures S1 and S2, A and B.
In the TCGA cohort, we compiled a total of 4,825,046 SNVs and 523,236 indels from 8976 samples across 31 cancer types that underwent both whole-exome sequencing (WES) and RNA-seq using our in-house pipeline (see Methods; Supplemental Tables S1, S2). Initially, to determine the relevant positions within authentic SSs, we applied SAVNet to these sequencing data and assessed the accuracy of SAVNet for each position by calculating position-wise false discovery rates (FDRs) using a permutation of combinations of WES and RNA-seq data. Within authentic SSs, SS-disrupting variants at positions −3 through +6 of donor sites and −1 through +6 (except for position +4) of acceptor sites had low FDR values (below 20%), whereas much higher FDRs were observed at other positions (Fig. 1C). This observation prompted us to focus on somatic variants at these positions in the subsequent analysis. In addition, to control the overall FDR at these positions below 5%, we employed a threshold of e3.0 or greater for the Bayes factor, depending on cancer type (Supplemental Fig. S1D). To evaluate the sensitivity of SAVNet under these settings, we compared our framework with two studies using the TCGA data (Jung et al. 2015; Jayasinghe et al. 2018). In the overlapping patient population (n = 929 [Jung et al. 2015] and 8247 [Jayasinghe et al. 2018], respectively), SAVNet detected a markedly higher number of SAVs, including more than a half of those found in the previous studies (Supplemental Fig. S1E–H). These results demonstrate the excellent detectability and satisfactory accuracy of SAVNet.
Landscape of SAVs in human cancers
With this optimized setting, we identified 14,438 somatic variants (13,414 SNVs and 1024 indels) responsible for 18,036 splicing alterations in the TCGA samples (Fig. 1E; Supplemental Table S3). A total of 11,153 SNVs and 875 indels disrupted splicing donor (n = 6799) or acceptor (n = 5229) motifs, of which 4406 SNVs and 359 indels were not located within GT-AG canonical sites. In addition, 2261 SNVs and 149 indels were detected to create novel splicing donor (n = 1566) and acceptor (n = 844) sites. Thus, 7175 (49.7%) somatic variants would not be expected to be identified by conventional methods that concentrate on SAVs involving canonical sites. Although the number of SAVs per sample was generally low (median of 1), there were quite a few samples with more instances of SAVs, particularly in cancer types with high somatic variant rates, such as lung and skin cancers (Supplemental Fig. S2A).
Overall, these splicing alterations included exon skipping (n = 6873), intron retention (n = 1917), and alternative 5′ SS and 3′ SS usage (n = 4522 and 4724, respectively) (Fig. 1D). Although the vast majority of SAVs caused a single splicing alteration, 2778 (19.2%) variants induced multiple splicing alteration events (Fig. 1E; Supplemental Fig. S2B). The transcriptional consequences substantially differed according to the somatic variant pattern (donor vs. acceptor and disruption vs. creation). Exon skipping and intron retention were caused by variants disrupting both donor and acceptor sites (Fig. 1D). As expected, donor disruptions tended to generate an alternative 5′ SS (n = 2783), whereas acceptor disruptions more frequently gave rise to an alternative 3′ SS (n = 3625). Exon skipping was the most frequent consequence of donor disruptions (n = 4442), whereas alternative 3′ SSs accounted for more than one-half of acceptor disruptions. Many new splice donor and acceptor sites were created by variants outside authentic SSs. Aberrant splicing events associated with variants in trans-acting splicing factors (Dvinge et al. 2016) showed no overlap with those detected by SAVNet (Supplemental Tables S4, S5).
Positional effects of SAVs disrupting authentic SSs
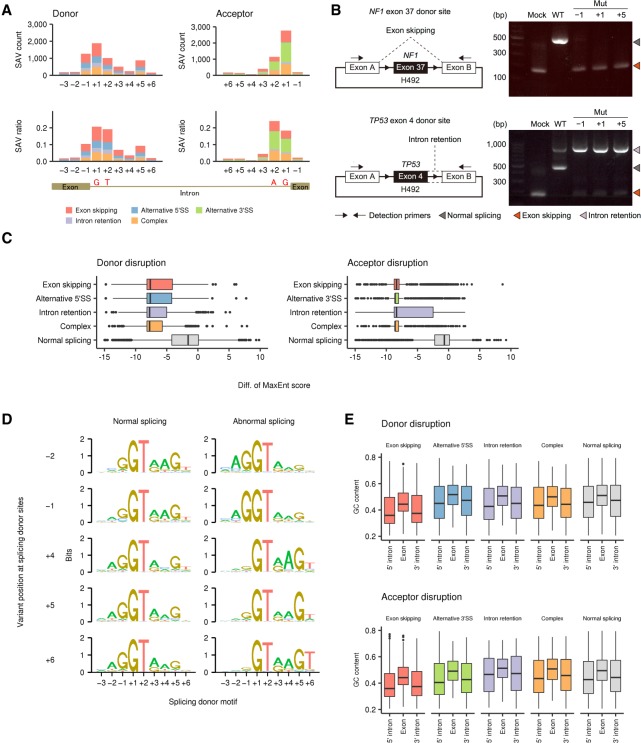
To investigate the positional effects of somatic variants on splicing, we evaluated the number of SAVs disrupting authentic SSs and their ratio to overall variants according to the distance from the exon-intron boundary. This analysis revealed a substantial difference among SS positions, although the proportion of splicing outcomes was nearly consistent within donor and acceptor SSs, respectively (Fig. 2A; Supplemental Fig. S2C). As previously reported (Jung et al. 2015), canonical GT-AG sites (at positions +1 and +2) had the highest ratios of splicing aberrations (18.4%–24.2%). In donor SSs, noncanonical sites showed a comparable total number of SAVs (n = 3428) with canonical sites (n = 2867), whereas the majority of SAVs in acceptor SSs were present at canonical sites (n = 3880, 79.9%). Together with the last exonic bases (−1) of donor sites, whose relevance was pointed out in the earlier study (Jung et al. 2015), the fifth intronic bases (+5) also had a relatively high ratio of abnormal splicing, followed by the third intronic bases (+3). Besides GT dinucleotides, these bases are well conserved and relevant to the interaction with U1 and U5 small nuclear RNAs (Lee and Rio 2015; Sibley et al. 2016). In fact, using minigene splicing assays (Nishida et al. 2011), we experimentally demonstrated that not only canonical but also noncanonical site variants cause abnormal splicing (Fig. 2B). The transcripts harboring variants at positions +5 as well as −1 showed abnormal splicing, such as exon skipping or intron retention, with comparable efficiency to canonical site variants (+1), while the wild-type transcripts were largely normally spliced.
Figure 2.
Genomic features of SS-disrupting variants generating distinct splicing alterations. (A) Number of SNVs disrupting splice donor and acceptor sites (SAV count, upper) and their fraction relative to total SNVs (SAV ratio, lower) at each position in the entire cohort. See also Supplemental Figure S2C for indels. (B) In vitro splicing analyses using H492 minigene constructs (left) showing exon skipping or intron retention (right) caused by SAVs at positions −1, +1, and +5 of NF1 exon 37 or TP53 exon 4 donor sites, respectively. (WT) Wild type, (Mut) mutated. (C) Change in splicing strength (based on MaxEnt scores) triggered by somatic variants at authentic splicing donor (left) and acceptor (right) sites according to splicing outcomes. “Complex” represents samples showing more than one splicing alteration, and “Normal splicing” represents samples lacking the relevant splicing alterations despite the presence of somatic variants in genes with detectable expression (fragments per kilobase of exon per million fragments mapped [FPKM] ≥ 10). See also Supplemental Figure S3B–E. (D) Sequence motifs of splicing donor sites at which somatic SNVs lead to normal (left) or abnormal splicing (right; identified by SAVNet) according to the variant position. See also Supplemental Figure 3, F and G. (E) GC contents of exons affected by SAVs, and their flanking 5′ and 3′ introns were compared among the five splicing groups. See also Supplemental Figure S4.
Features of genomic sequences associated with SAVs
Splicing outcomes mediated by SAVs appear to be context-dependent: Somatic variants within authentic SSs can cause different forms of splicing aberration, while the same substitutions at the same relative position frequently do not alter splicing. To elucidate the factors determining the potential of somatic variants within authentic SSs to alter splicing, we compared the genomic features of SSs between normal (those not identified as SAVs) and abnormal splicing groups (SAVs) (Supplemental Fig. S3A). Generally, SS-disrupting SAVs attenuated the splicing strength more than variants that induced no abnormal splicing, regardless of the substituted position and consequent splicing alteration type (Fig. 2C; Supplemental Fig. S3B–E). A sequence motif analysis revealed a distinctive feature of SSs disrupted by SAVs, especially those at positions other than canonical GT-AG sites. As for variants occurring at the penultimate (−2) and last (−1) exon bases in the donor SSs, splicing motifs with abnormal splicing showed more conserved exonic bases but less conserved intronic bases when compared with normal splicing motifs, except for the universally conserved canonical GT dinucleotides (Fig. 2D; Supplemental Fig. S3G). This difference was opposite for SAVs at intronic bases, in which consensus sequences were more conserved in introns, especially at positions +4 through +6, but not in exons. These findings are compatible with the proposed mutually repressive relationship between the exonic and intronic regions of donor sites (Burge and Karlin 1997; Carmel 2004). Analysis of the disrupted acceptor sites revealed that thymine (T) at position +4 was overrepresented in samples with SAVs at position +3, which may be due to the frequent C > G substitutions at TpC dinucleotides attributed to APOBEC activity (Supplemental Fig. S3F; Alexandrov et al. 2013; Shiraishi et al. 2015).
Consistent with the previous report (Jung et al. 2015), inspection of the exon-intron architecture revealed that exon skipping was characterized by a lower GC content in both exons and flanking introns, shorter exon and longer intron length, and stronger splicing strength (Fig. 2E; Supplemental Fig. S4A–F). These features are characteristic of SSs governed by the exon definition mechanism, in which exons are initially recognized by splicing factors (Keren et al. 2010; Naftelberg et al. 2015). In contrast, intron retention and alternative SS usage were associated with longer exon length, suggesting that these SSs are regulated in common by the intron definition mechanism.
Mutational signatures associated with SAV generation
Despite the expansion of our understanding on the signatures of mutational processes (Alexandrov et al. 2013; Shiraishi et al. 2015), the effect of these signatures on a specific type of somatic variants have not been fully elucidated. Here, we noticed occasional discrepancies between the efficiency of somatic variants to cause abnormal splicing and the actual number of SAVs. For instance, position +2 of acceptor sites showed only a moderate number of SAVs, albeit the highest SAV ratio (Fig. 2A). These discrepancies may be attributed to the overall number of somatic variants (including those not associated with splicing alterations) and their substitution patterns at each position, which reflect both the unique base composition at SSs and mutational signatures. In fact, positions at −1, +1, and +5 of donor sites as well as +1 of acceptor sites, which were dominated by G bases, showed frequent G > A and G > T substitutions, suggestive of age- and smoking-related mutational processes, respectively (Fig. 3A, upper). In contrast, position +2 of donor and acceptor sites, which predominantly consist of A/T bases, showed a relatively low frequency of somatic variants. Among them, variants at canonical GT-AG sites caused splicing alterations, regardless of their base substitution pattern, whereas almost all SAVs at positions −1 and +5 of donor sites occurred at G bases, indicating almost no effect of substitutions from other bases on splicing (Fig. 3A, lower). In addition, positions having a smaller fraction of abnormal splicing were more strongly affected by the base substitution pattern. For example, G > A substitutions were the most common at position +3 of donor sites but did not result in splicing aberrations. Moreover, despite their low frequency of overall variants, C > G substitutions (compatible with the APOBEC cytidine deaminase mutational pattern as shown below) accounted for a considerable proportion of SAVs at position +3 of acceptor sites. These findings are consistent with relatively limited conservation of splicing motifs at these positions.
Figure 3.
Mutational signatures underlying the generation of SS-disrupting SAVs. (A) Base substitution patterns of total somatic variants (upper) and SAVs (lower) at each exonic and intronic position of splice donor and acceptor sites. Different colors are used to display different types of alternative bases. The x-axes represent different reference bases, and the y-axes represent the numbers of variants. Fractions of SAVs relative to total somatic variants (purple points) with Bayesian confidence intervals (5% to 95% posterior quartiles) are also shown. (B) Fraction of estimated SAVs relative to estimated total variants attributed to each mutational signature. Red dashed line represents the overall fraction of SAVs relative to total variants. See also Supplemental Figure S5, A and B. (C) Scatter plot showing the relationship between SAV and total variant counts in 31 cancer types. A linear regression line (red) is fitted to the data points for each cancer, excluding those for COAD, SKCM, and UCEC. The truncated mean is used to exclude the samples with extremely large numbers of somatic variants. See also Supplemental Figure S5C–H.
To evaluate the underlying mutational process operative in SAV occurrence, we estimated the extent of contribution (posterior probability) by each mutational signature for all the variants found in the current sample set using a pmsignature algorithm (Shiraishi et al. 2015) and calculated the fraction of SAVs to total variants for each mutational signature (see Method). Among the five major mutational signatures (processes generating a large number of somatic variants) (Supplemental Fig. S5A), the smoking signature (C > A substitutions) showed the largest contribution to SAV generation, followed by APOBEC (C > T and C > G substitutions at TpC sites) and aging signatures (C > T substitutions at CpG sites). Signatures related to ultraviolet exposure (C > T substitutions at YpC sites) and altered activity of the error-prone polymerase POLE (C > A substitutions at TpCpT sites and C > T substitutions at TpCpG sites) had less impact (Fig. 3B; Supplemental Fig. S5B). These differences can partly be explained by the predominance of G bases at highly affected positions (−1, +1, and +5 of donor sites and +1 of acceptor sites), and the transcriptional strand bias of several mutational signatures, i.e., the smoking signature, preferentially affecting C bases on the noncoding strand (G bases on the coding strand), was strongly enriched, whereas the ultraviolet signature, which frequently alters C bases on the coding strand, was underrepresented. Reflecting these differences among mutational processes, lung squamous cell carcinomas (LUSC) and lung adenocarcinomas (LUAD) had more SAVs than expected from the overall somatic variant rate, whereas cancers frequently affected by POLE alterations, such as uterine corpus endometrioid carcinomas (UCEC) and colon adenocarcinomas (COAD), as well as ultraviolet-associated skin cutaneous melanomas (SKCM), showed a relatively lower number of SAVs (Fig. 3C). The effects of smoking status and POLE alterations were similarly observed within LUAD as well as UCEC and COAD (Supplemental Fig. S5C–H).
Characteristics of SAVs creating alternative SSs
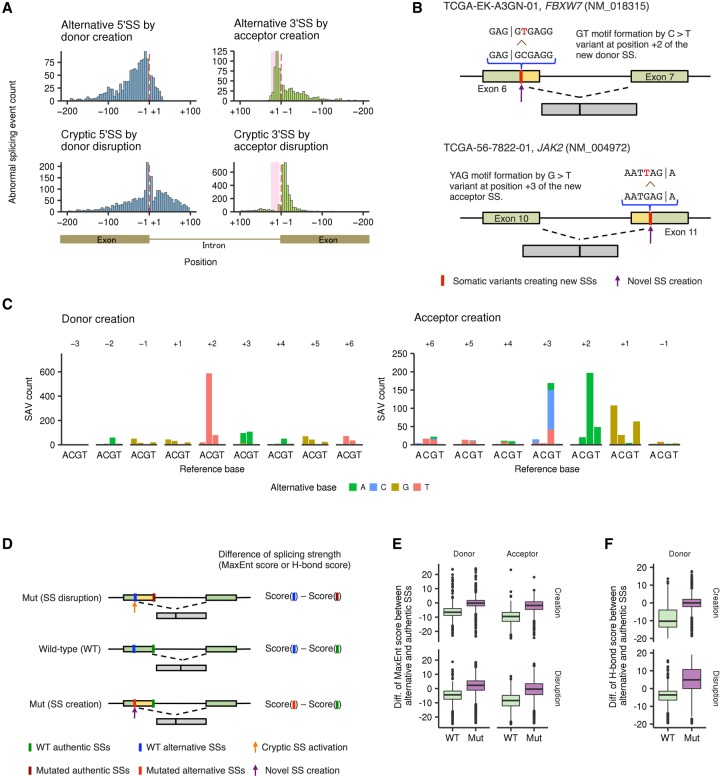
Our analysis also revealed the positional distribution of SAVs creating alternative donor and acceptor sites. Newly created donor sites were widely distributed in both exons and introns, whereas abnormal acceptor sites were created predominantly within the polypyrimidine tract (Fig. 4A, upper), likely reflecting the involvement of additional conserved elements in introns, such as branchpoint sequences and polypyrimidine tracts (Lee and Rio 2015; Sibley et al. 2016). Apparently, similar distributions were also seen for cryptic SSs activated by variants disrupting the authentic SSs (Fig. 4A, lower). However, unlike newly created acceptor sites, a biased localization of cryptic acceptor sites toward exons was observed, which can be plausibly explained by a depletion of AG dinucleotides in the polypyrimidine tract.
Figure 4.
Genomic features and positional differences of SS-creating SAVs. (A) Histogram showing the distribution of newly created alternative SSs (upper) and cryptic SSs caused by SS disruption (lower). Red dashed lines and pink shading represent exon-intron boundaries and polypyrimidine tract regions (positions +5 through +25), respectively. (B) Two typical examples of SS-creating SAVs (through formations of GT and YAG motifs, respectively) are displayed. (C) Base substitution patterns of SAVs creating alternative SSs according to the distance from the newly created exon-intron boundaries. Colors and axes are the same as in Figure 3A. (D) The effects of SAVs on splicing strength (based on MaxEnt or H-bond scores) for authentic or alternative SSs were assessed. For SS-disrupting SAVs, the difference in splicing strength between alternative and unsubstituted authentic SSs (WT) was compared with that between alternative and substituted authentic SSs (Mut). For SS-creating SAVs, the difference between unsubstituted alternative and authentic SSs (WT) was compared with that between substituted alternative and authentic SSs (Mut). (E,F) Box plots showing the differences of MaxEnt scores for alternative 5′ SSs and 3′ SSs (E) and H-bond scores for alternative 5′ SSs (F).
We also evaluated the substitution pattern of somatic variants creating new splicing sites based on their relative position within the newly created SSs. Most newly created donor sites resulted from GT canonical site generation through C > T substitutions at position +2, whereas variants associated with acceptor creation tended to form a new YAG (Y, pyrimidine) motif at positions +3 through +1 (Fig. 4B,C). These results suggest that, showing a strong bias toward particular base substitutions, these SAVs generate additional consensus donor or acceptor motifs that are more efficient for splicing than those in authentic SSs, as implicated by stronger splicing strength (assessed by MaxEnt [Yeo and Burge 2004] or H-bond [Freund et al. 2003] scores) (Fig. 4D–F).
Enrichment of SAVs in TSGs
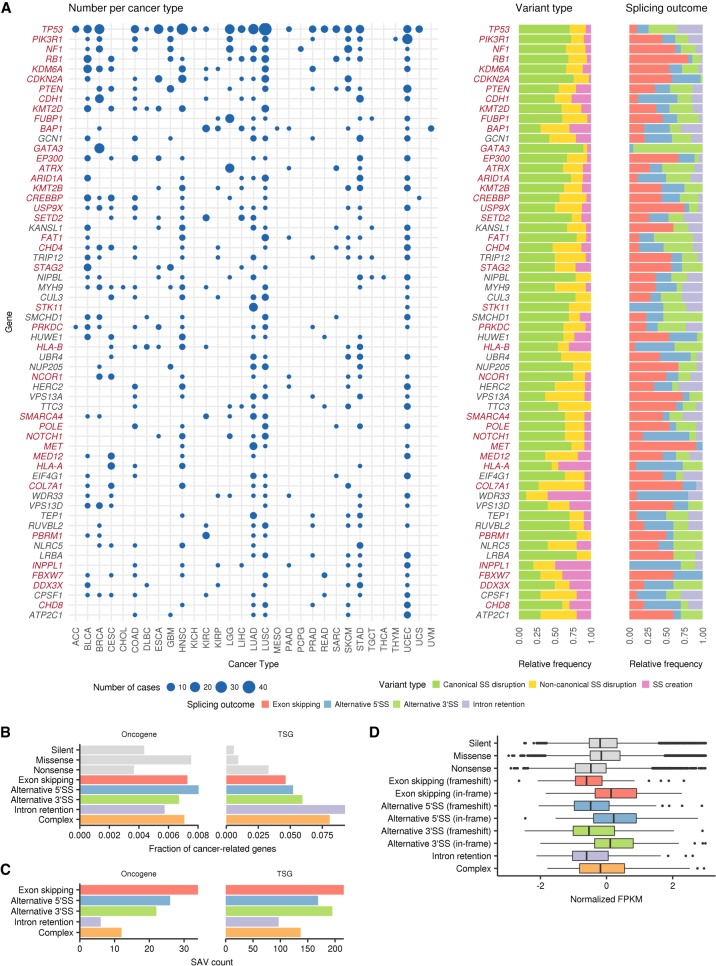
To evaluate the role of SAVs during cancer development, we investigated which genes are frequently altered by SAVs. Thirty-eight (63.3%) out of 60 frequently affected genes (present in ≥10 samples across the entire cohort) were well-established TSGs (Fig. 5A,B; Supplemental Fig. S6A,B). In agreement with this study (Jung et al. 2015), in which intron retention was argued to be a major mechanism of SAV-induced TSG inactivation, SAVs that caused intron retention showed the strongest enrichment of TSGs, regardless of the cancer gene sets (Vogelstein et al. 2013; Lawrence et al. 2014; Ye et al. 2016). However, SAVs associated with exon skipping and alternative SS usage also had a greater proportion of TSGs, even when compared with nonsense variants, accounting for 88% of SAVs affecting TSGs (Fig. 5C). These findings suggest that, together with intron retention, exon skipping and alternative SS usage play crucial roles in TSG inactivation. In contrast, oncogenes were less frequently affected by SAVs, comparable to missense variants. In total, 1684 SAVs in candidate cancer-related genes (Ye et al. 2016) were identified in 14.7% of the TCGA samples (1315 of 8976). Particularly, as many as 914 SAVs found in 9.3% of samples targeted well-known TSGs (Vogelstein et al. 2013), of which 341 were not located at canonical sites. Moreover, SAVs accounted for 9.5% of loss-of-function variants in these genes. Therefore, SAVs represent an important but previously underestimated mechanism for TSG inactivation, irrespective of splicing outcome.
Figure 5.
Entire spectrum of SAVs across cancer types. (A) Left: Landscape of SAVs in frequently altered genes (total number ≥10) across cancer types. The point size indicates the number of affected samples. Genes are sorted by the total number of SAVs in all cancer types, and known cancer-related genes (Ye et al. 2016) are shown in red. Right: Relative frequencies of variant types and splicing outcomes of SAVs. For SAVs causing multiple splicing alterations, splicing outcomes with the largest number of supporting reads are selected. (B) The fractions of SAVs affecting oncogenes or TSGs (based on Vogelstein et al. 2013) relative to total SAVs according to splicing outcomes were compared with other types of somatic variants (silent, missense, and nonsense). See also Supplemental Figure S6. (C) The number of SAVs affecting oncogenes or TSGs (based on Vogelstein et al. 2013). (D) Box plots showing changes in normalized (z-scored) mRNA expression (FPKM) for each splicing outcome, as compared to other types of somatic variants.
Like nonsense variants, splicing alterations are thought to trigger nonsense-mediated decay (NMD), a surveillance mechanism that selectively degrades abnormal transcripts containing a premature termination codon (Jung et al. 2015; Scotti and Swanson 2016). To clarify the effects of SAVs on gene expression through NMD in TSGs, we investigated the whole transcript level across different types of abnormal splicing. In line with the previous report (Jung et al. 2015), transcripts with intron retention showed a substantially lower expression level than normal transcripts, which was comparable to those with nonsense variants (Fig. 5D). The expression of transcripts with exon skipping or alternative SS usage was also reduced when their splicing alterations caused frameshift changes.
Genes frequently altered by SAVs
Among genes frequently targeted by SAVs, TP53 was the most frequently altered gene, affecting 233 samples in 22 cancer types (Fig. 5A). Although the last bases of exons 4, 6, and 9 were reported to be frequently mutated (Supek et al. 2014; Jung et al. 2015), we identified a number of recurrent variants at splice donor and acceptor sites of introns 3 through 9, with prominent base-level and/or SS-level hotspots at donor and acceptor sites of intron 4 (Supplemental Tables S6, S7). Approximately one-half of recurrent SAVs simultaneously produced different types of abnormal splicing, while identical abnormal splicing events were generated by different SAVs, such as retention of introns 7, 8, and 9 caused by donor and acceptor SAVs of each intron (Fig. 6, upper left). Most of these SAVs induced frameshift splicing alterations, likely leading to mRNA degradation through NMD, whereas other SAVs generated in-frame exon skipping or alternative SS usage, such as exon 5 acceptor variants activating cryptic 3′ SS, followed by a 15-amino acid (aa) deletion. PIK3R1, encoding the p85 regulatory subunit of phosphatidylinositol 3-kinase, ranked second (39 samples), approximately one-half of which were found in UCEC (Fig. 5A). The majority of these SAVs caused splicing alterations resulting in in-frame deletions of the iSH2 domain, which is also affected by the small deletions typically observed in this gene (Fig. 6A, upper right; Cheung et al. 2011).
Figure 6.
Genes frequently affected by SAVs in human cancers. (A) Distribution of SAVs and their resultant splicing outcomes for TP53 (upper left), PIK3R1 (upper right), GATA3 (lower left), and CDKN2A (lower right). SS-disrupting and SS-creating SAVs are aggregated according to the authentic and alternative SSs, respectively. The numbers in circles or triangles represent the number of SS-disrupting and SS-creating SAVs for each SS, respectively. See also Supplemental Figure S7A. (B) Fraction of the most frequent relative to total associated splicing outcomes for each SS-level SAV hotspot (found in ≥8 samples). The most frequent splicing outcome is noted in parentheses for each SS. The same color indicates the identical SAVs in terms of position and substitution or indel patterns. See also Supplemental Figure S7B.
In many genes, particularly NF1 and RB1, most SAVs and consequent splicing alteration events were diverse and widely distributed throughout the entire gene (Supplemental Fig. S7A, upper and middle), whereas several genes displayed prominent hotspots of SAVs (Supplemental Tables S6, S7). Among the latter genes, SAVs affecting the same SSs tended to generate identical splicing consequences (Fig. 6B; Supplemental Fig. S7B). A typical example was CDKN2A, a well-known TSG that encodes the p16INK4A and p14ARF proteins, which was recurrently affected by SAVs targeting exon 2 common to both proteins (Fig. 6A, lower right). Other instances included GATA3 SAVs found in breast invasive carcinomas (BRCA), in which most of them were the identical CA dinucleotide deletion at the acceptor site of exon 5, thus activating a cryptic 3′ SS (7 nucleotides [nt] downstream) (Fig. 6A, lower left). Utilization of this cryptic splice acceptor caused a reading frameshift, resulting in loss of the second zinc finger (ZnF2) domain (Usary et al. 2004). As was the case with GATA3, several genes showed tissue specificity, such as FUBP1 and ATRX in lower grade gliomas (LGG), probably reflecting the organ-specific growth advantage conferred by these alterations. In contrast, most of the frequently altered genes were relevant across multiple cancer types (Fig. 5A).
Together with these well-established TSGs, SAVNet identified several recurrently altered genes (found in ≥10 samples) which had not been included in the cancer-related gene list (Ye et al. 2016) but reported or predicted to function in a tumor-suppressive manner, including KANSL1 (Yoshida et al. 2013), NIPBL (Barber et al. 2008), CUL3 (Ooi et al. 2013), MYH9 (Schramek et al. 2014), SMCHD1 (Leong et al. 2013), and HUWE1 (Fig. 5A; Inoue et al. 2013). Thus, SAVNet may have potential to identify putative TSGs that are more prone to be affected by splicing aberrations. Conversely, MET, which encodes a hepatocyte growth factor receptor, was the only frequently affected oncogene, whose variants in the exon 14 donor site caused in-frame exon skipping known to activate c-Met (Fig. 5A; Supplemental Fig. S7A, lower left; Ma et al. 2005). Additionally, SAV hotspot analysis also identified recurrent SAVs occurring at the donor site of exon 3 of MIEN1, a putative oncogene located on the ERBB2 (also known as HER2) amplicon (Fig. 6B; Supplemental Fig. S7A, lower right; Dasgupta et al. 2009). Although the underlying mechanisms need to be clarified, SAVs may contribute to the activation of several oncogenes.
Discussion
The development and application of SAVNet have led to the systematic detection of a substantial number of SAVs that had been overlooked by earlier studies (Supek et al. 2014; Jung et al. 2015), although we focused only on those disrupting or creating splicing donor or acceptor motifs. Following previous studies (Xu and Lee 2003; Supek et al. 2014; Jung et al. 2015), our comprehensive and thorough analysis revealed the landscape of cis-acting somatic variants affecting splicing and characterized their positional differences, genomic features, and underlying mutational processes in detail, showing their enrichment in cancer driver genes, especially in TSGs. In particular, we demonstrated that exon skipping and alternative SS usage were more frequently involved in SAV-mediated TSG inactivation than intron retention. In addition, we clarified the relevance of SAVs at noncanonical sites, including the previously unrecognized position +3 and +5 of donor sites. The proposed framework with FDR control, which can dissect complex variant-splicing associations based on the Bayesian approach, is applicable to identify additional classes of somatic variants that disrupt splicing regulatory elements, including exonic and/or intronic splicing enhancers and silencers, although further elaboration of association rules will be required. Based on our findings, not only exonic but also intronic SNVs near exon-intron boundaries should be carefully evaluated as pathogenic variants, irrespective of the presence of amino acid changes. In the era of precision medicine, our framework and the acquired list of SAVs will constitute helpful resources to capture more driver variants, including previously overlooked SAVs, in cancer patients.
Methods
Download of TCGA WES and RNA-seq data
WES and RNA-seq data were downloaded from Cancer Genomic Hub (currently hosted at NCI Genomic Data Commons [https://portal.gdc.cancer.gov/legacy-archive]). We used samples whose tumor and matched control WES and RNA-seq data are all available. We excluded LAML (acute myeloid leukemia) and OV (ovarian serous cystadenocarcinoma), because most of their DNA samples underwent whole-genome amplification, leading to a large amount of artifactual variants.
Alignment of TCGA WES data
As a reference genome, we used the sequences of assembled chromosomes, unlocalized and unplaced scaffolds from GRCh37 (human reference assembly), as well as NC_007605 (Epstein-Barr virus) and hs37d5 (decoy from The 1000 Genomes Project Phase II) sequences. Our preliminary comparison showed the choice of GRCh37 and GRCh38 had almost no impact on the detection of SAVs. In WES analysis, for downloaded sequence data in BAM format, we first convert it to FASTQ format using bamtofastq command (with collate=1 exclude=QCFAIL,SECONDARY,SUPPLEMENTARY options) of biobambam (https://github.com/gt1/biobambam). FASTQ-formatted sequences were aligned with BWA-MEM version 0.7.8 (Li and Durbin 2009) with −T0 option and sorted by biobambam bamsort command (with index=1 level=1 inputthreads=2 outputthreads=2 calmdnm=1 calmdnmrecompindentonly=1 options). Then, PCR duplicates were removed by biobambam bammarkduplicates command (with markthreads=2 rewritebam=1 rewritebamlevel=1 index=1 options).
Detection of somatic SNVs and short indels
Our approach for detecting somatic SNVs and short indels consists of the following five steps:
Identification of candidate somatic SNVs and short indels using the approach based on Fisher's exact test (as previously described [Yoshida et al. 2011]), which is currently implemented in GenomonFisher (https://github.com/Genomon-Project/GenomonFisher);
Excluding candidates present in pooled control samples by using EBFilter (https://github.com/Genomon-Project/EBFilter), a variant filtering algorithm based on a rigorous empirical Bayesian framework (Shiraishi et al. 2013);
Local realignment of short reads around candidate variants, which is implemented in GenomonMutationFilter (https://github.com/Genomon-Project/GenomonMutationFilter);
Removal of putative OxoG artifacts; and
Annotation of the variants using Annovar (Wang et al. 2010).
For step (1), we tallied up the numbers of mismatched bases at each position using short reads with mapping quality of ≥20 (for SNVs and indels) and with base quality of ≥15 (for SNVs). First, we roughly extracted the candidates satisfying the following criteria: (1) sequence depth ≥ 10; (2) mismatch ratio in tumor samples ≥ 0.05; (3) number of variant-supporting reads ≥ 4; and (4) mismatch ratio in matched control samples < 0.03. Next, we performed Fisher's exact test to assess the differences in the ratios of the numbers of reference-supporting to variant-supporting reads between tumor and matched control samples, and candidate variants with P-value ≤ 0.1 were adopted.
For step (2), we performed filtering of all the remaining candidates, based on a beta-binomial error model, as described previously (Shiraishi et al. 2013). Briefly, we estimated the parameters of the beta-binomial error model using nonmatched control samples (20 samples in this paper), obtained the predictive distributions of the mismatch ratios, and compared them with the observed mismatch ratio of tumor samples to quantify the statistical significance. We adopted candidate variants with P-value < 10−4.
For step (3), we performed local realignment of all short reads surrounding the candidate variants and their paired reads to the reference and variant-containing sequences and counted the numbers of reference- and variant-supporting read pairs for tumor and matched control samples. We used “read pair-based” count to avoid double counting of a variant located in both reads of a single read pair with a small insert size. Then, we adopted candidates satisfying the following criteria: (1) number of variant-supporting read pairs in tumor samples ≥4; (2) number of variant-supporting read pairs in matched control samples ≤1; and (3) P-value of Fisher's exact test comparing the ratios of the numbers of reference- and variant-supporting read pairs between tumor and matched control samples ≤0.1.
For step (4), to remove putative OxoG artifacts (Costello et al. 2013), we calculated ALT_F1R2 (the number of variant-supporting read pairs whose first and second parts are aligned in the forward and reverse directions, respectively) and ALT_F2R1 (the number of variant-supporting read pairs whose first and second parts are aligned in the reverse and forward directions, respectively) for C > A and G > T substitutions. Then, C > A substitutions were removed if ALT_F1R2 < 2 or ALT_F2R1/(ALT_F1R2 + ALT_F2R1) > 0.9, and G > T substitutions were removed if ALT_F2R1 < 2 or ALT_F1R2/(ALT_F1R2 + ALT_F2R1) > 0.9.
Alignment of TCGA RNA-seq data
Genome indexes were generated using STAR version 2.5.2a (Dobin et al. 2013) with the GRCh37 release 19 GTF file (ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_human/release_19/gencode.v19.annotation.gtf.gz) and –sjdbOverhang 100 option. For each sample, alignment to the reference genomes was performed by STAR version 2.5.2a with the following options: --runThreadN 6 --outSAMtype BAM Unsorted --outSAMstrandField intronMotif --outSAMunmapped Within --outSJfilterCountUniqueMin 1 1 1 1 --outSJfilterCountTotalMin 1 1 1 1 --outSJfilterOverhangMin 12 12 12 12 --outSJfilterDistToOtherSJmin 0 0 0 0 --alignIntronMax 500000 --alignMatesGapMax 500000 --alignSJstitchMismatchNmax -1 -1 -1 -1 --chimSegmentMin 12 --chimJunctionOverhangMin 12. Then, sorting and indexing of BAM files were performed using SAMtools version 1.2 (Li et al. 2009).
Quantification of expression values for each gene from RNA-seq data
To quantify gene expression, we used our in-house software GenomonExpression (Supplemental Code S1; https://github.com/Genomon-Project/GenomonExpression), which calculates a slightly modified version of FPKM (fragments per kilobase of transcript per million mapped reads) measures (Shiraishi et al. 2014). Briefly, after excluding improperly aligned or low-quality read pairs (mapping quality <20), sequence depth in the exonic regions was calculated, and normalized as per kilobase of exon as well as per million of aligned bases for each RefSeq gene. For genes with multiple transcript variants, their expression values were determined by selecting a transcript variant with the maximum FPKM value.
Identification of splicing-associated variants (SAVNet)
To identify splicing-associated variants, we developed and applied the novel approach, SAVNet (Supplemental Code S2; https://github.com/friend1ws/SAVNet), which consists of the following steps.
1. Collection of evidences of different types of abnormal splicing
We consider four types of abnormal splicing: exon skipping, alternative 5′ splice site, alternative 3′ SS usage, and intron retention. The first three types (exon skipping, alternative 5′ SS, alternative 3′ SS) are extracted using splicing junctions (defined as pairs of start and end positions demarcated by spliced-aligned reads). We first extract abnormal splicing junctions (not registered in RefSeq genes) (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/refGene.txt.gz) with ≥2 supporting reads (number of uniquely mapped reads crossing the junction) in at least one sample in the cohort by processing SJ.out.tab files generated as by-products of the STAR alignment step. Then, using our in-house program (junc_utils) (Supplemental Code S3; https://github.com/friend1ws/junc_utils), we classify each splicing junction into exon skipping, alternative 5′ SS, or alternative 3′ SS by the following criteria:
Exon skipping: Two ends of the splicing junction correspond to annotated intron start (splicing donor) and end (splicing acceptor) sites of a gene, respectively.
Alternative 5′ SS: One end of the splicing junction corresponds to an annotated intron end (splicing acceptor) site of a gene, whereas the other end is located within the gene but not at an annotated intron start (splicing donor) site of the gene.
Alternative 3′ SS: One end of the splicing junction corresponds to an annotated intron start (splicing donor) site of a gene, whereas the other end is located within the gene but not at an annotated intron end (splicing acceptor) site of the gene.
Splicing junctions that do not meet any of the above criteria are removed.
Intron retentions are identified by our in-house program (intron_retention_utils simple_count command) (Supplemental Code S4; https://github.com/friend1ws/intron_retention_utils). For each exon-intron boundary, the number of putative intron retention reads (those covering ≥10 bp of both sides of the exon-intron boundary) as well as that of normally spliced reads covering the last exonic base of the exon-intron boundary is counted. In this paper, to remove events observed in noncancer samples, we used a panel of 742 control samples (collected from the TCGA cohort) and filtered out splicing junctions with ≥2 supporting reads in ≥8 control samples, and intron retentions whose intron retention fraction (the number of intron retention reads divided by total reads (intron retention reads + normally spliced reads) covering the exon-intron boundary) is ≥0.05 in ≥8 control samples.
2. Association of splicing alterations with somatic variants to construct possible variant-splicing bipartite graphs
In this step, we list candidate combinations of somatic variants and possibly associated splicing alterations for each gene, which are subject to further investigation in the later step. For each gene, let g = (g1, g2, …, gN) ∈ {0, 1, …, M}N denote the status of somatic variants of N samples in the cohort, where M denotes the number of distinct somatic variants, gn = 0 represents that the nth sample does not have any somatic variants in the target gene, and gn = m represents that the nth sample has the mth somatic variant. Also, let denote the number of supporting reads (the number of putative intron retention reads for intron retention) for the jth splicing alteration, and let denote the weight for each sample used for normalization to negate variations in the amount of total sequence reads. We set wn = Un/107, where Un is the number of uniquely aligned read pairs of the nth sample.
The mth somatic variant is considered to be associated with the jth splicing alteration if the following three conditions are satisfied:
-
Their positional relationship implicates that the abnormal splicing can be a consequence of disruption of authentic SSs (those registered in the RefSeq database) or creation of novel SSs caused by the somatic variant. More specifically, we check the following relationship:
Abnormal splicing junction events caused by authentic SS disruption: (1) A somatic variant occurs at authentic splicing donor (between positions −3 [the third exonic base] through +6 [the sixth intronic base]) or acceptor sites (between −1 through +6), and (2) an abnormal splicing junction event (exon skipping and alternative 5′ SS and 3′ SS) encompasses or is located within 100 bp of the variant.
Abnormal intron retention caused by authentic SS disruption: (1) A somatic variant occurs at authentic splicing donor or acceptor sites, and (2) an intron retention occurs at the disrupted SS or its opposite site of the same intron.
Alternative SS usage caused by new SS creation: A somatic variant occurs within the newly created SS of an unannotated junction end of an abnormal splicing event (alternative 5′ SS or 3′ SS).
- The average number of supporting reads for the jth splicing alteration in samples with the mth somatic variant is at least three times larger than those in samples without any somatic variants of the gene in consideration
The median number of supporting reads for the jth splicing alteration in samples without any somatic variants of the gene in consideration is zero.
The second and third criteria are incorporated to reduce the computational cost (which increases exponentially as the number of associations). The third criterion also can help improve both sensitivity and accuracy. In our preliminary observation, the numbers of supporting reads showing abnormal splicing are typically zero for almost all samples without any splicing-associated variants, whereas they are nonzero (sometimes as small as 2 or 3) for those harboring SAVs. Adopting this criterion in fact resulted in a slight improvement of the false discovery ratio.
We create a bipartite graph (VM, VS, E) for the entire structure of variant-splicing associations, where vertices (VM,VS) represent somatic variants and splicing alterations and edges (E) represent combinations of associated somatic variants and splicing alterations. We then split the association graphs into several subgraphs through checking the connectivity, and performed the pruning procedure described in the next section.
3. Pruning of edges to select the best model explaining the data
Here, we choose a subgraph of the bipartite graph constructed in the previous step, which most effectively explain the status of somatic variants and their impacts on splicing alterations (quantified by the numbers of supporting reads). We use the idea of “configuration” from previous eQTL and GWAS studies performed in complicated situations (Flutre et al. 2013; Stephens 2013). The configuration here is a |E|-dimensional binary vector = (γm,j)(m,j)∈E, where γm,j ∈ {0,1} indicates whether the mth variant and the jth splicing alterations have a causal relationship (1) or not (0). When there is no causal relationship between any somatic variants and splicing alterations (which we call the null model henceforth), = , where is a vector whose elements are all zero . Under a configuration γ, we classify somatic variants into “active” and “inactive” for the jth splicing junction.
For each configuration γ, we assume that the supporting reads yj are generated by Poisson distributions whose parameters are dependent on the activity status of somatic variants and multiplied by sample weights. The parameter of the Poisson distribution for the nth sample is set to wnλ0 when it has only inactive variants on the jth splicing alteration , whereas it is set to wnλ1 for active variants . Additionally, we assume that λ0 and λ1 are generated by a gamma distribution with shape and rate parameters (α0,β0), (α1,β1), respectively. In this study, we set (α0,β0) = (1,1) and (α1,β1) = (1,0.01). Therefore, the likelihood of yj given γ is
and the likelihood of the whole data (Y = {yj}j=1, 2, …, J) is . Also, the likelihood of yj under the null model = 0, which can be calculated as a special case of the above, is
For each variant m, we perform Bayesian model comparison to determine whether the somatic variant has any causal relationships with any splicing alterations or not . Typically, to perform model comparison, we evaluate the Bayes factor between the two distinct models. Here, as there are often many distinct null and nonnull models, we aggregate these models through Bayesian model averaging and evaluate the Bayes factor between the aggregated null and nonnull models
where Pr(γ) is set to be the uniform distribution. The variant m is identified as a SAV if its logarithm of the Bayes factor is above the threshold (the default value is set to 3). Also, the splicing alterations caused by the variant m are identified by selecting the best model by
4. Evaluation of FDR by permutation
To evaluate FDR, we permute the pairs of genomic and transcriptome data so that somatic variants and splicing alterations from different patients are coupled and perform the same procedures (step 1 to 3). Assuming that Dtarget and are the numbers of SAVs identified in the original step (correct combinations) and in the ith permutation procedure, respectively, then FDR is estimated as
In this paper, we performed 100 permutation trials (I = 100).
5. Postprocessing and rescuing SAVs
SAVs causing alternative intronic 5′ or 3′ SSs are generally accompanied with intron retention at the original authentic SSs. Therefore, in these cases, we removed intron retention and retained only alternative 5′ SSs or 3′ SSs in this paper. To sensitively detect recurrent SAVs, we performed additional screening and adopted variants satisfying the following criteria: (1) The combination of the same somatic variants (the same substitution at the same position) and the same splicing alterations was identified in other samples by the SAVNet procedure described above; (2) variant mismatch ratio in tumor samples ≥0.05; (3) number of variant-supporting reads ≥3; (4) mismatch ratio in tumor samples ≥10-fold of that in matched control samples; and (5) number of reads supporting associated splicing alterations ≥2.
Evaluation of influences of spliceosome variants on abnormal splicing
First, in the TCGA cohort, we searched for previously known somatic variants of splicing factors, including missense variants at K700, K666, H662, R625, E622, G740, G742, N626, and E902 of SF3B1, S34 and Q157 of U2AF1, and P95 of SRSF2, as well as missense, nonsense, and frameshift variants of ZRSR2 (Dvinge et al. 2016). First, for each cancer type, we extracted splicing alterations with ≥2 supporting reads in at least one sample. Then, we identified splicing factors affected in ≥1% samples within each cancer type and compared the number of RNA-seq reads supporting each splicing alteration between samples with and without the splice factor variants to derive the P-value using the t-test. Finally, we calculated the Q-value for each splicing alteration using the qvalue R package (http://github.com/jdstorey/qvalue), and splicing alterations with Q-value < 0.05 were considered to be associated with splice factor variants.
Estimation of mutational signatures and membership of SAVs
We used pmsignature for estimating the signatures of mutational processes operative in the entire cohort and each cancer cohort as described in the previous paper (Shiraishi et al. 2015). Then, the extracted mutation signatures were classified into any of the COSMIC signatures (http://cancer.sanger.ac.uk/cosmic/signatures) using minimum centered cosine similarity. Mutation signatures with centered cosine similarities to all the COSMIC signatures <0.75 were classified to “other.” The estimates of membership (conditional probabilities attributed to each mutation signature) for each variant are provided by the following equation:
where the notation for each variable is described in the previous paper (Shiraishi et al. 2015). Finally, variant-level membership estimates were aggregated according to the COSMIC signatures and the presence of association with splicing alterations, so that the total numbers of variants and SAVs caused by each mutation signature (e.g., tobacco, ultraviolet) were estimated.
Quantification of splicing-related features
MaxEnt (Yeo and Burge 2004) and H-bond (Freund et al. 2003) scores were calculated using the spliceSites R package (https://bioconductor.org/packages/release/bioc/html/spliceSites.html). To derive lengths and GC contents of exons affected by SAVs and their flanking introns, we extracted exonic nucleotides and adjacent upstream and downstream 150 intronic nucleotides. Then, we discarded 10 exonic and 20 intronic nucleotides from the exon-intron boundaries since they constitute splicing signals with specific nucleotides. Here, we excluded SS-disrupting SAVs affecting short exons (≤30 bp), exons with multiple annotated start and end positions to avoid ambiguity.
Cell line
HEK293T cells were obtained from the RIKEN Cell Bank. Cell lines were authenticated by the provider and routinely tested for mycoplasma infection.
Minigene splicing assay
For each region of interest, exonic and ∼300-bp flanking fragments containing either wild-type or variant sequences were synthesized (GeneArt, Thermo Fisher Scientific) and cloned into the NheI and BamHI sites of the plasmid H492 (a kind gift from Prof. Masafumi Matsuo, Kobe University) using the In-Fusion HD cloning kit (TaKaRa). Each construct was transiently transfected into HEK293T cells using X-tremeGENE 9 DNA Transfection Reagent (Roche) in six-well tissue culture plates. Forty-eight hours after transfection, total RNA was isolated with an RNeasy Mini kit (QIAGEN) and used to synthesize cDNA with ReverTra Ace qPCR RT Kit (TOYOBO). Each cDNA was amplified with KOD FX Neo DNA polymerase (TOYOBO) using the primers (forward, 5′-ATTACTCGCTCAGAAGCTGTGTTGC-3′, and reverse, 5′-AAGTCTCTCACTTAGCAACTGGCAG-3′), which correspond to sequences of exons of H492. PCR products were separated by electrophoresis on 2% agarose gels and visualized with a UV transilluminator (UVP). To confirm the sequence of each band, the PCR products were gel-purified and analyzed by Sanger sequencing (Supplemental Data).
Data analysis
All analyses were performed in Python 2.7.8 and R 3.3.2 and most figures were generated using the ggplot2 R package (Wickham 2016). In all box plots, the center line and lower and upper hinges correspond to the median and the first and third quartiles (25 and 75 percentiles), respectively. The upper and lower whiskers extend from the upper and lower hinges to the largest or smallest values no further than 1.5 × IQR from the hinges, respectively, where IQR represents inter-quartile range, or distance between the first and third quartiles. Sequence logos were drawn via our in-house program (Supplemental Code S5; https://github.com/friend1ws/ggseqlogo).
Data access
All Sanger sequencing reads of minigenes generated in this study are available as Supplemental Data. The processed data and scripts for generating figures are available via GitHub (https://github.com/friend1ws/savnet_paper) and as Supplemental Codes S1–S5.
Supplementary Material
Acknowledgments
This work was supported by Grant-in-Aid from the Japan Agency for Medical Research and Development (Advanced Genome Research and Bioinformatics Study to Facilitate Medical Innovation [17km0405207h0002]), Grant-in-Aid for Scientific Research (KAKENHI 15H05912, 15H05909, and 15K00398), National Cancer Center Research and Development Funds (29-E-3), and Post-K Research and Development (R&D) projects (hp170227). We thank André Kahles, Cameron Soulette, Gunnar Rätsch, Angela Brooks, and PCAWG-3 splicing team members for helpful discussions and feedback on the performance of SAVNet. We thank Naoyuki Kataoka for expert opinion, Miki Sagou for technical assistance, and Yuki Saito for manuscript proofreading. The supercomputing resources were provided by the Human Genome Center, the Institute of Medical Science, The University of Tokyo. The results shown here are partly based upon data generated by TCGA Research Network (http://cancergenome.nih.gov/).
Author contributions: Y.S. and K.K. designed the study. Y.S., K.C., A.O., H.T., and S.M. developed bioinformatics pipelines. Y.S. designed and implemented SAVNet. Y.S., K.K., and Y.K. analyzed and interpreted the data. K.K. performed functional assays. Y.S. and K.K. generated figures and tables and wrote the manuscript. S.O. and S.M. supervised the entire project. All authors participated in discussion and interpretation of the data and results.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231951.117.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. 2008. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci 105: 3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. 2014. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One 9: e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Carmel I. 2004. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA 10: 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, et al. 2011. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 1: 170–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, Fostel JL, Friedrich DC, Perrin D, Dionne D, et al. 2013. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 41: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Wasson LM, Rauniyar N, Prokai L, Borejdo J, Vishwanatha JK. 2009. Novel gene C17orf37 in 17q12 amplicon promotes migration and invasion of prostate cancer cells. Oncogene 28: 2860–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. 2016. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 16: 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutre T, Wen X, Pritchard J, Stephens M. 2013. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet 9: e1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund M, Asang C, Kammler S, Konermann C, Krummheuer J, Hipp M, Meyer I, Gierling W, Theiss S, Preuss T, et al. 2003. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res 31: 6963–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. 2013. Lessons from the cancer genome. Cell 153: 17–37. [DOI] [PubMed] [Google Scholar]
- Inoue S, Hao Z, Elia AJ, Cescon D, Zhou L, Silvester J, Snow B, Harris IS, Sasaki M, Li WY, et al. 2013. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev 27: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe RG, Cao S, Gao Q, Wendl MC, Vo NS, Reynolds SM, Zhao Y, Climente-González H, Chai S, Wang F, et al. 2018. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep 23: 270–281.e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. 2015. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 47: 1242–1248. [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Line A. 2005. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer 42: 342–357. [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong HS, Chen K, Hu Y, Lee S, Corbin J, Pakusch M, Murphy JM, Majewski IJ, Smyth GK, Alexander WS, et al. 2013. Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer Res 73: 1591–1599. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, Hansen M, Schaefer E, Naoki K, Lader A, et al. 2005. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non–small cell lung cancer. Cancer Res 65: 1479–1488. [DOI] [PubMed] [Google Scholar]
- Martincorena I, Campbell PJ. 2015. Somatic mutation in cancer and normal cells. Science 349: 1483–1489. [DOI] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, Kornblihtt AR. 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 84: 165–198. [DOI] [PubMed] [Google Scholar]
- Nishida A, Kataoka N, Takeshima Y, Yagi M, Awano H, Ota M, Itoh K, Hagiwara M, Matsuo M. 2011. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat Commun 2: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi A, Dykema K, Ansari A, Petillo D, Snider J, Kahnoski R, Anema J, Craig D, Carpten J, Teh BT, et al. 2013. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res 73: 2044–2051. [DOI] [PubMed] [Google Scholar]
- Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. 2014. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343: 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nat Rev Genet 17: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, et al. 2013. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res 41: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Fujimoto A, Furuta M, Tanaka H, Chiba K, Boroevich KA, Abe T, Kawakami Y, Ueno M, Gotoh K, et al. 2014. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One 9: e114263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Tremmel G, Miyano S, Stephens M. 2015. A simple model-based approach to inferring and visualizing cancer mutation signatures. PLoS Genet 11: e1005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CR, Blazquez L, Ule J. 2016. Lessons from non-canonical splicing. Nat Rev Genet 17: 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Eyras E. 2017. The role of alternative splicing in cancer. Transcription 8: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. 2013. A unified framework for association analysis with multiple related phenotypes. PLoS One 8: e65245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. 2014. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156: 1324–1335. [DOI] [PubMed] [Google Scholar]
- Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, et al. 2004. Mutation of GATA3 in human breast tumors. Oncogene 23: 7669–7678. [DOI] [PubMed] [Google Scholar]
- Venables JP. 2004. Aberrant and alternative splicing in cancer. Cancer Res 64: 7647–7654. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer, New York. [Google Scholar]
- Xu Q, Lee C. 2003. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res 31: 5635–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Wang J, Jayasinghe R, Lameijer EW, McMichael JF, Ning J, McLellan MD, Xie M, Cao S, Yellapantula V, et al. 2016. Systematic discovery of complex insertions and deletions in human cancers. Nat Med 22: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park MJ, Terui K, Suzuki H, et al. 2013. The landscape of somatic mutations in Down syndrome–related myeloid disorders. Nat Genet 45: 1293–1299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.