Abstract
Characterization of intratumoral heterogeneity is critical to cancer therapy, as the presence of phenotypically diverse cell populations commonly fuels relapse and resistance to treatment. Although genetic variation is a well-studied source of intratumoral heterogeneity, the functional impact of most genetic alterations remains unclear. Even less understood is the relative importance of other factors influencing heterogeneity, such as epigenetic state or tumor microenvironment. To investigate the relationship between genetic and transcriptional heterogeneity in a context of cancer progression, we devised a computational approach called HoneyBADGER to identify copy number variation and loss of heterozygosity in individual cells from single-cell RNA-sequencing data. By integrating allele and normalized expression information, HoneyBADGER is able to identify and infer the presence of subclone-specific alterations in individual cells and reconstruct the underlying subclonal architecture. By examining several tumor types, we show that HoneyBADGER is effective at identifying deletions, amplifications, and copy-neutral loss-of-heterozygosity events and is capable of robustly identifying subclonal focal alterations as small as 10 megabases. We further apply HoneyBADGER to analyze single cells from a progressive multiple myeloma patient to identify major genetic subclones that exhibit distinct transcriptional signatures relevant to cancer progression. Other prominent transcriptional subpopulations within these tumors did not line up with the genetic subclonal structure and were likely driven by alternative, nonclonal mechanisms. These results highlight the need for integrative analysis to understand the molecular and phenotypic heterogeneity in cancer.
Intratumor heterogeneity is a common feature across diverse cancer types. Dynamic changes among intratumoral subpopulations over time and following therapy present a key challenge to current standards of cancer treatment (Ding et al. 2012; Gerlinger et al. 2012; Shah et al. 2012; Wu 2012; Mroz et al. 2015). Genetic variation, such as copy number alterations, is a well-studied source of intratumoral heterogeneity (Vogelstein et al. 2013; Melchor et al. 2014). The extent to which such alterations are able to drive tumor development typically relies on specific expression dysregulation of tumor cells. While some of the alterations have been tied to perturbations of known oncogenes and tumor suppressors, such as MYC and TP53 (Sekiguchi et al. 2014; Glitza et al. 2015), the process by which many genetic alterations impact transcriptional processes to drive disease progression and drug resistance, particularly in combination, is not well understood (Lohr et al. 2014). In that regard, the ability to examine the transcriptional states of genetically distinct intratumoral subclones would be helpful for evaluating the likely functional impact of associated subclonal mutations. Such knowledge could help design rational strategies for development of new treatments, identify cellular pathways responsible for variable patient response or resistance to treatment, and improve prognostic stratification and evaluation of therapeutic approaches (Walker et al. 2014).
While some insights into the relationship between genetic and transcriptional heterogeneity have been gained from bulk analysis, further characterization on the single-cell level is needed to more accurately dissect the pathway and regulatory features associated with distinct genetic subclones. Single-cell RNA-sequencing (scRNA-seq) methods can provide detailed information on the transcriptional state of the cancer cells. However, integration with genotypic information at the single-cell level is necessary to establish correspondence between transcriptionally distinct subpopulations and genetic subclones. At the same time, simultaneous unbiased assessment of DNA and RNA from an individual cell remains challenging (Dey et al. 2015; Macaulay et al. 2015; Wang et al. 2017).
Computational approaches for bulk sequencing data detect copy number variations (CNVs) based on consistent deviations in read coverage as well as allelic imbalance within the region (Wang et al. 2007; Boeva et al. 2012; Chen et al. 2013). In the context of scRNA-seq, recent publications have used deviations in average expression magnitude within affected regions from a normal tissue reference to illustrate the presence of chromosome-scale CNVs (Patel et al. 2014; Tirosh et al. 2016). Likewise, analysis of allelic imbalance may also be informative about CNVs in the context of scRNA-seq. Here, we propose a computational approach called HoneyBADGER to quantitatively infer the presence of subclone-specific focal CNV and loss-of-heterozygosity (LOH) events in individual cells using allele and expression information from scRNA-seq data.
Results
Prevalence of monoallelic detection in scRNA-seq data presents challenges
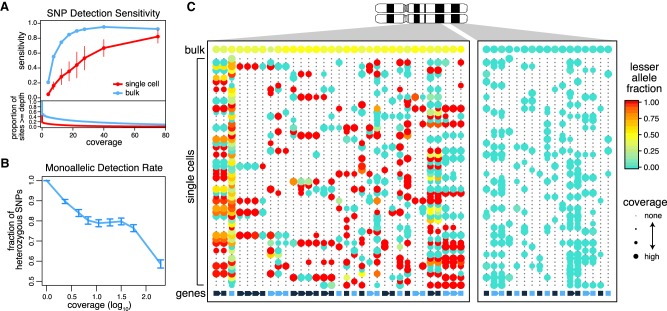
To evaluate whether sequence variant information available in the RNA reads can be used to distinguish subclones, we first examined the ability to detect single-nucleotide variants from scRNA-seq data. By using whole-exome sequencing (WES) to identify heterozygous single-nucleotide polymorphisms (SNPs) in the K562 cell line, we evaluated the sensitivity of detecting such SNPs from both bulk and single-cell RNA-seq K562 data (Fig. 1A). The average sensitivity for detecting covered SNPs (three or more reads) in single-cell data was only 0.34 compared with 0.76 for the bulk RNA-seq data. Much of this difference can be attributed to the lower read coverage in single-cell data. However, sensitivity remains significantly lower even for well-covered SNPs (Fig. 1B). In such cases, all of the detected transcript reads originate from only one of the alleles. Like others, we find such monoallelic detection to be prevalent in scRNA-seq data (Deng et al. 2014; Borel et al. 2015; Wang et al. 2017). Although the likelihood of observing both alleles generally increases with increasing level of gene expression, it remains low even for highly expressed genes (Fig. 1B). The prevalence of monoallelic detection—a consequence of transcriptional stochasticity, sparse sampling of mRNA molecules, and subsequent uneven amplification by the scRNA-seq protocols (Deng et al. 2014)—limits the confidence with which we can deduce the absence of a variant in a cell. This, together with the sparse coverage characteristic of scRNA-seq data, suggests that joint statistical analysis of many of variant sites is necessary to achieve genotype classification of cells.
Figure 1.
Prevalence of monoallelic detection and sparse signals in scRNA-seq data. (A) Sensitivity of heterozygous SNP detection as a function of coverage in single-cell and bulk RNA-seq data for the K562 cell line. Sensitivity was calculated as the proportion of sites that are called heterozygous in the RNA-seq sample among the sites that were called heterozygous in the WES data. Error bars, SD. The coverage distribution (bottom) is shown for bulk and an average of the individual cells. (B) Prevalence of monoallelic detection in scRNA-seq data. Lowly expressed genes are nearly exclusively detected in a monoallelic manner. The monoallelic detection rate generally goes down with expression magnitude; however, it remains high even for well-covered polymorphisms. Error bars, 95% confidence interval of the binomial proportions. (C) Lesser allele fraction profile visualizes patterns of allelic imbalance for germline heterozygous SNPs identified from scRNA-seq. The dot plot illustrates coverage (size) and allele bias (color) for germline heterozygous SNPs (rows) detected in different cells (columns). The bottom row designates genes with alternating color labels. Single cells commonly exhibit stretches of monoallelic detection within genes, as noted by the same color dots. However, across genes in a single cell, both alleles can be observed, suggesting that both alleles are present. In contrast, within a deletion region (right), single cells can only express from the nondeleted allele.
HoneyBADGER identifies CNVs in single cells
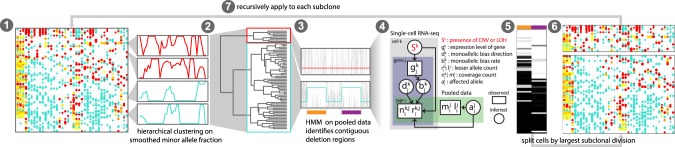
Contiguous regions of variant sites are affected by focal heterozygous chromosomal deletions, amplifications, and LOH events in a coordinated manner (Fig. 1C). For example, if many SNPs across multiple genes within a putative deletion region are consistently expressed from the same allele in a given cell, then the cell likely harbors that deletion. Thus, joint analysis of heterozygous SNPs encompassed by such regions can overcome the uncertainty of individual SNPs detection. However, the number of SNPs and their associated read coverage must be sufficient to rule out the possibility of such allelic imbalance being observed by chance because of monoallelic detection. We therefore developed a hidden Markov model (HMM) integrated Bayesian approach for detecting CNVs and LOHs from single-cell RNA-seq data (HoneyBADGER) to identify candidate CNV regions and perform joint statistical analysis of multiple SNPs within these regions to achieve genotype classification of cells (see Methods). HoneyBADGER employs a Bayesian approach to quantify the posterior probability of a CNV in each cell based on the observed allele ratios within the affected region, taking into consideration the expected prevalence of monoallelic detection (Methods; Fig. 2).
Figure 2.
Overview of HoneyBADGER. CNVs and LOHs are identified from scRNA-seq data in the following seven steps: (1) Cells are first clustered on smoothed lesser-allele frequencies; (2) cells are split into two main groups and pooled; (3) a hidden Markov model on the pooled lesser-allele fraction identified regions with potential CNVs or LOHs; (4) a Bayesian hierarchical model assessed the posterior probability of a CNV or LOH for each region in each cell; (5) cells are clustered by their posterior probabilities of CNV or LOH for each region; (6) cells are split into putative subclones; and (7) the approach is recursively applied to each subclone until no new subclones can be detected.
As an initial step, HoneyBADGER identifies candidate CNV regions by a recursive HMM approach (Fig. 2). Briefly, the allele frequency data are pooled across cells, and CNV-affected regions are identified by the HMM based on a consistent deviation of the allele fraction of heterozygous variants away from the expected 0.5 allele fraction. The presence of genetically distinct subpopulations—such as mixture of tumor and microenvironment, or different tumor subclones—decreases the sensitivity of the CNV detection step and the accuracy of the identified CNV boundaries. To handle such heterogeneous subpopulations, HoneyBADGER recursively clusters cells by similarity of their smoothed lesser allele fraction profile, where the lesser allele is defined as the allele that is less frequently observed across our population of cells. In the presence of a deletion, we expect to see persistent depletion of this lesser allele across our population of cells harboring the deletion. Where candidate CNVs of interest are known based on genomic sequencing or biological knowledge, such as common deletions spanning TP53, this candidate CNV discovery step with HMM may be skipped altogether. We note that the identity of the lesser allele implies phasing of haplotypes across multiple genes, beyond the phasing signal apparent from the monoallelic detection within individual genes.
Then, for each candidate CNV region identified, Honey BADGER evaluates the posterior probability that an individual cell harbors the alteration based on observed patterns of allelic imbalance using a Bayesian hierarchical framework (Methods; Fig. 2). This second layer of evaluation protects against false positives introduced in the HMM phase and takes into account potential uncertainty in phasing by reassessing the identity of the lesser allele using allele counts from individual cells rather than pooled frequencies. Based on the posterior probabilities for these deletions, we then separate cells into genetic branches and recursively search for additional subclonal alterations within each branch.
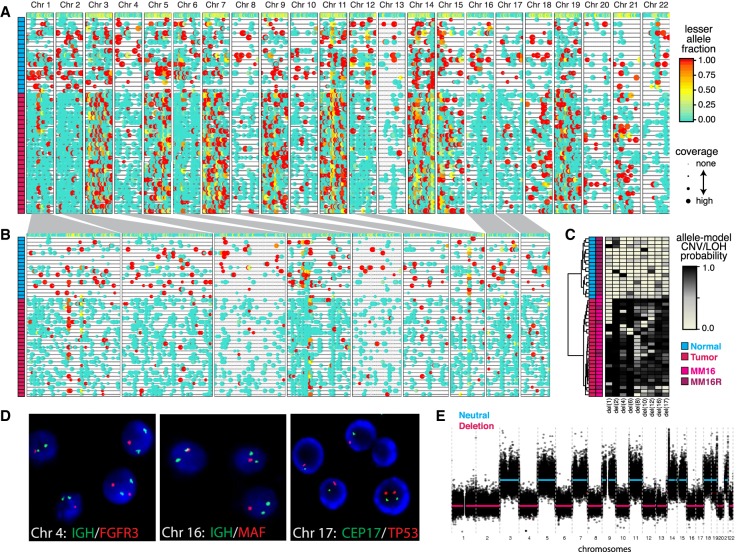
At a basic level, the analysis of CNV/LOH occurrence by HoneyBADGER enables separation of tumor cells from karyotypically normal cells. To demonstrate HoneyBADGER, we first examined 44 cells from serial bone marrow (BM) biopsies of a multiple myeloma (MM) patient. Twenty-three cells were analyzed from a biopsy obtained at diagnosis (MM16) with an estimated 90% purity, and 21 cells were analyzed from a biopsy obtained 6 mo later in a minimal disease state after chemotherapy (MM16R) with an estimated 10% purity based on SDC1+ (also known as CD138+) expression. By using known heterozygous SNPs from previous bulk sequencing efforts, we identified multiple clonal whole-chromosome deletions (Fig. 3A,B). As expected due to the low purity in sample MM16R, only five of 21 cells (24%) originating from MM16R are inferred to be tumor cells, harboring all of the identified CNVs with high posterior probability (Fig. 3C). Similarly, 22 out of 23 cells (96%) originating from MM16 are inferred to be tumor. Thus, the percentage of putative normal and MM cells in MM16 and MM16R are consistent with bulk purity estimates. We validate identified deletions using FISH and cytogenetics (Fig. 3D) and bulk WES (Fig. 3E).
Figure 3.
HoneyBADGER analysis of 44 multiple myeloma (MM) cells. (A) Lesser allele profiles where each column is a heterozygous SNP and each row is a single cell. Points are colored by the lesser allele fraction, with yellow suggesting equal detection of both alleles and red and blue indicating monoallelic detection in either direction. Points are sized by coverage at the SNP site in the given cell. Cells are ordered based on row dendrogram in C. (B) Allele profiles for regions identified by HoneyBADGER as potential CNV or LOH regions. Width corresponds to size of region. Cells are ordered based on row dendrogram in C. (C) Heatmap of posterior probability of CNVs or LOHs in each identified region where each column is a region and each row is a cell. Row side-colors annotate cells as originating from MM16 or MM16R and as classified as normal or tumor. (D) Interphase FISH and cytogenetics of cells from MM16. Of the 200 cells analyzed, 82.5% had a single D17Z1 and TP53 signal; 79.5%, a single MAFB (20q12) signal. The sample analyzed is estimated to have 81%–95% tumor purity by SDC1+. Representative cells shown. (E) Copy number inference by bulk WES for MM16.
HoneyBADGER can further resolve focal CNVs previously that are not detectable by expression-based karyotyping approaches (Patel et al. 2014). By using scRNA-seq data from Patel et al. (2014), we applied HoneyBADGER to examine 65 glioblastoma (GBM) cells mixed with 10 normal cells from patient MGH31. We took advantage of the contamination of normal cells to identify heterozygous SNPs without reliance on additional sequencing data, such as WES. Briefly, we pooled all single cells from MGH31 and identified sites exhibiting multiple alleles. To avoid somatic alterations, we restricted SNP sites to known common population SNPs (MAF > 10%) from the ExAC database (Lek et al. 2016). HoneyBADGER recovers known deletions on Chromosomes (Chr) 10, 13, and 14 (Supplemental Fig. 1A,B). Furthermore, it identifies an additional focal deletion (15 Mb) on Chr 19 with equal clonality to the deletion on Chr 10 (Supplemental Fig. 1A–C; Supplemental Table 1). Given the relatively small size of this deletion, it could not be detected using expression-based karyotyping (Supplemental Fig. 1D), highlighting increased sensitivity of the allele-based approach. We note that even without the presence of karyotypically normal cells to assist with the identification of heterozygous SNPs, our approach is still able to identify clonal deletions based on a significant depletion of common heterozygous SNPs from the ExAC database on Chr 10 compared with other regions of comparable size and gene density (Supplemental Fig. 2A,B).
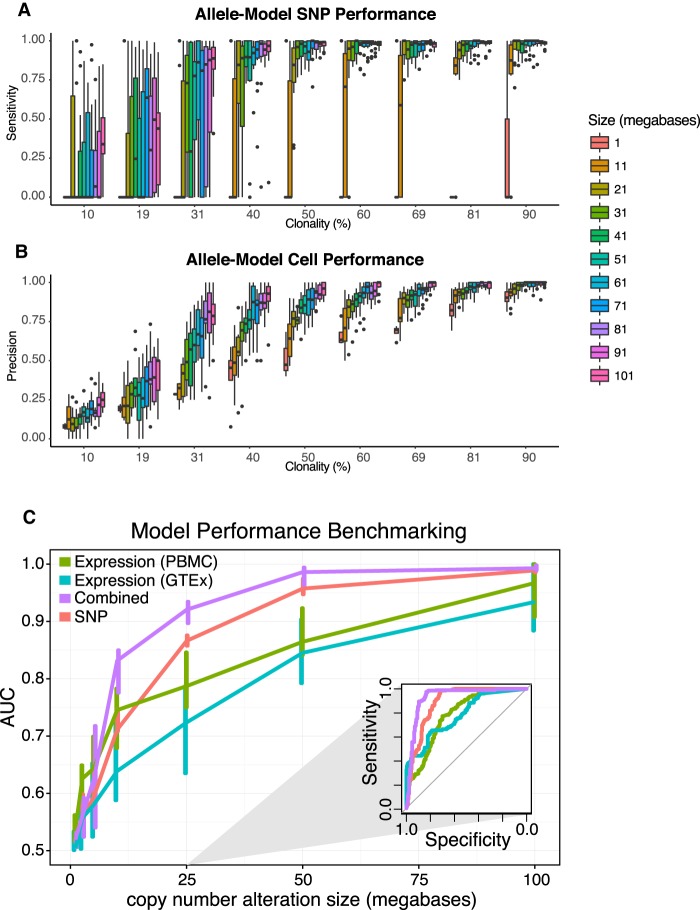
To assess the performance of HoneyBADGER on CNVs of varying size and clonality, we simulated deletions of varying size and clonality by inserting fragments of known deletions into CNV-neutral regions in MGH31 (see Methods). These simulations suggest that HoneyBADGER can accurately identify and resolve clonal deletions as small as 10 Mb in size (Fig. 4A), as well as chromosome-arm-level subclonal deletions present in as few as 30% of cells (Fig. 4B; Supplemental Fig. 2C,D).
Figure 4.
HoneyBADGER performance as a function of clonality and CNV size. (A) Allele-model sensitivity for identifying SNPs affected by deletion. (B) Allele-model precision for distinguishing tumor (cells with deletion) from normal (cells without). (C) Prediction performance of HoneyBADGER's posterior probability estimates as a function of deletion size. Four different HoneyBADGER models are shown using different colors: expression-only model with PBMC and CD19+ expression normalization reference (green); expression-only model with normal blood GTEx expression normalization reference (blue); expression and SNP combined model (purple); and SNP-only model (red). Inner quartile range is indicated by the vertical lines. Performance was quantified by ROC AUC. (Inset) Representative ROC curves for a simulated 25-Mb deletion.
While the benchmarks thus far have focused on full-transcript-coverage scRNA-seq data produced using the Smart-seq2 protocol (Picelli et al. 2014), newer droplet microfluidic protocols that sequence only the 3′-end of transcripts are becoming increasingly common (Klein et al. 2015; Macosko et al. 2015). To assess the utility of our allele-based approach with such protocols, we analyzed acute myeloid leukemia (AML) BM mononuclear cells measured using 10× chromium, taken from a patient (AML035) before and after hematopoietic stem cell transplant (HSCT) (Zheng et al. 2017). Without a WES reference, we again leveraged common heterozygous variants from ExAC to identify potential heterozygous variants from pre- and post-HSCT samples. The increased number of cells enhances our ability to identify heterozygous SNPs. However, compared with the Smart-seq2, we were able to identify less than half as many SNPs in the 10× chromium data (Supplemental Fig. 3A). Performance simulations show that with such 3′-tag data, the allele-based approach will be able to detect full chromosome and chromosome-arm-level alternations but will be substantially limited in identifying more focal alterations (Supplemental Fig. 3B). While we were not able to identify any such large-scale copy number alterations in the AML sample (Supplemental Fig. 3C,D), when both pre- and post-HSCT samples were examined together, the allele-based approach clearly identified allelic patterns indicative of the presence of two distinct genotypes (Supplemental Fig. 3E). Consistent with observations from the original publication (Zheng et al. 2017), we find that cells from the post-HSCT sample were genotypically distinct from the pre-HSCT sample, reflecting successful engraftment of donor stem cells from the HSCT treatment. Thus, an allele-based approach can distinguish cellular genotypes before and after HSCT using patterns of common natural genetic variation from 3′-tag scRNA-seq data, even without external genotype information (Kang et al. 2018).
Integration of expression data enhances power and enables identification of copy-neutral LOH
In addition to allelic imbalance, the presence of deletions, on average, also leads to diminished expression of genes within affected loci compared with copy-neutral expression references of the same cell type (Mayshar et al. 2010; Macaulay et al. 2015). Similarly, the presence of amplifications, on average, leads to increased expression of genes within affected loci compared with copy-neutral expression references of the same cell type. Assessment of these expression-based karyotyping approaches has so far been qualitative in nature (Patel et al. 2014; Tirosh et al. 2016), and the extent to which they are able to capture smaller, focal deletions remains to be quantified. To provide such a quantitative evaluation, we implemented an expression-based HMM to identify regions potentially affected by CNVs as well as a Bayesian hierarchical model to assess the posterior probabilities of alterations using normalized expression data (Supplemental Fig. 4A). As before, we simulated deletions of varying size and clonality by inserting fragments of known deletions into CNV-neutral regions in MGH31 (see Methods). We find that the quantitative expression-based approach is able to identify chromosome-arm-level clonal and nearly clonal alterations with high sensitivity and precision (Supplemental Fig. 4C–E) but has difficulties resolving smaller, subclonal alterations as accurately as the allele-based approach. We find that the expression-based approach is particularly sensitive to the normalization reference used (Supplemental Fig. 4B). With modern scRNA-seq data sets often capturing many diverse cell types, independent normalization of different cell types by corresponding references may be necessary.
Joint consideration of both allele- and expression-based evidence should increase predictive power. It should also allow distinguishing deletions from copy-neutral LOH events. We therefore extended HoneyBADGER to incorporate both types of evidence in inferring the posterior probability of affected regions identified by either the allele- or expression-based HMMs. Indeed, we find that an integrated model offers improved performance in distinguishing regions of deletion from neutral regions (Fig. 4C; Supplemental Fig. 4F). While high copy number amplifications are common in cancer, the measurements of gene expression as well as allelic imbalance are too variable to confidently infer the exact copy number. Our approach, therefore, does not infer the precise copy number but is aimed at distinguishing deletion, amplification, and LOH regions from the unaffected regions.
To demonstrate the utility of our integrated approach, we applied Honey BADGER to 55 breast cancer cells from patient BC09 from Chung et al. (2017). Chung et al. (2017) previously identified several cells to harbor known breast cancer–related point mutations, including mutations in LRPAP1, MARCH6, ANKFY1, DNMT1, GTPBP3, BLZF1, POLA2, TMEM189, AGO3, NNT, PLK4, and CPSF1 (Supplemental Table 2). However, these cells were inferred to be normal based on expression-based karyotyping, suggesting a likely misclassification by the expression-based approach. Reanalysis with the allele-based model of Honey BADGER shows that these cells harbor multiple chromosome-arm and chromosome-level abnormalities (Supplemental Fig. 4). We confirm using bulk WES (Supplemental Table 2) that such misclassification arose due to copy-neutral LOH where copy number is maintained, thus resulting in limited changes in normalized expression but detectable allelic imbalance. Our allele and expression–combined approach is thus able to identify copy-neutral LOH events and segregate tumor in a way consistent with the point mutation evidence (Supplemental Fig. 5).
To further evaluate the utility of our integrated approach with 3′-tag droplet-based scRNA-seq measurements, we applied HoneyBADGER to 1340 single cells from an unsorted BM biopsy from a MM patient (MM135) prepared using the 10× chromium protocol. To determine the appropriate normalization for the expression data, we first applied our allele-based approach to identify a deletion on Chr 13, which separated a set of putative normal cells lacking the deletion (Supplemental Fig. 6A). We confirmed using expression-based clustering analysis that these putative normal cells did not express known MM marker genes (Supplemental Fig. 6B). Expression profiles of these putative normal cells were then averaged to serve as a normal expression reference. We then applied our integrated approach to identify a number of chromosome-arm and chromosome-level abnormalities on Chr 1, 8, 11, 13, and 22 (Supplemental Fig. 6C–E; Supplemental Table 3). Unbiased hierarchical clustering on the posterior probabilities of these alterations effectively separated MM from non-MM cells (Supplemental Fig. 6E). We confirmed the identified chromosomal aberrations using FISH and cytogenetics (Supplemental Table 3). In addition to the chromosomal abnormalities identified by HoneyBADGER, FISH, and cytogenetics identified an additional Chr 18 deletion that was missed by our computational approach due to the low number of expressed gene and detected SNPs within the region, resulting in high uncertainty. Thus, while we were able to accurately identify most chromosome-arm and chromosome-level abnormalities in this 3′-tag scRNA-seq data set, lower SNP density in such data results in low sensitivity, as expected from previous benchmarks (Supplemental Fig. 4F).
Analysis of progressive MM identifies genetic subclones with distinct transcriptional signatures
To examine the interaction of genetic and transcriptional heterogeneity in a context of MM progression, we applied Honey BADGER to analyze tumor samples, collected at two distinct time points, from a treatment-refractory MM patient (MM34). The initial MM sample (MM34) was collected from the BM at the time of diagnosis, and a second extramedullary MM (MM34A) sample was collected from an ascites dissemination following two months of unsuccessful thalidomide/dexamethazone and bortezomib treatment.
We first applied HoneyBADGER to identify regions of CNV in 63 extramedullary MM cells from MM34A. Our allele-based HMM identified clonal deletions on multiple chromosomes, including Chr 1, 2, 3, 8, 13, 16, and 17, and our expression-based HMM identified a clonal amplification on Chr 3 (Fig. 5A; Supplemental Fig. 7A; Supplemental Table 4). We confirm these CNVs by bulk WES (Supplemental Fig. 7A).
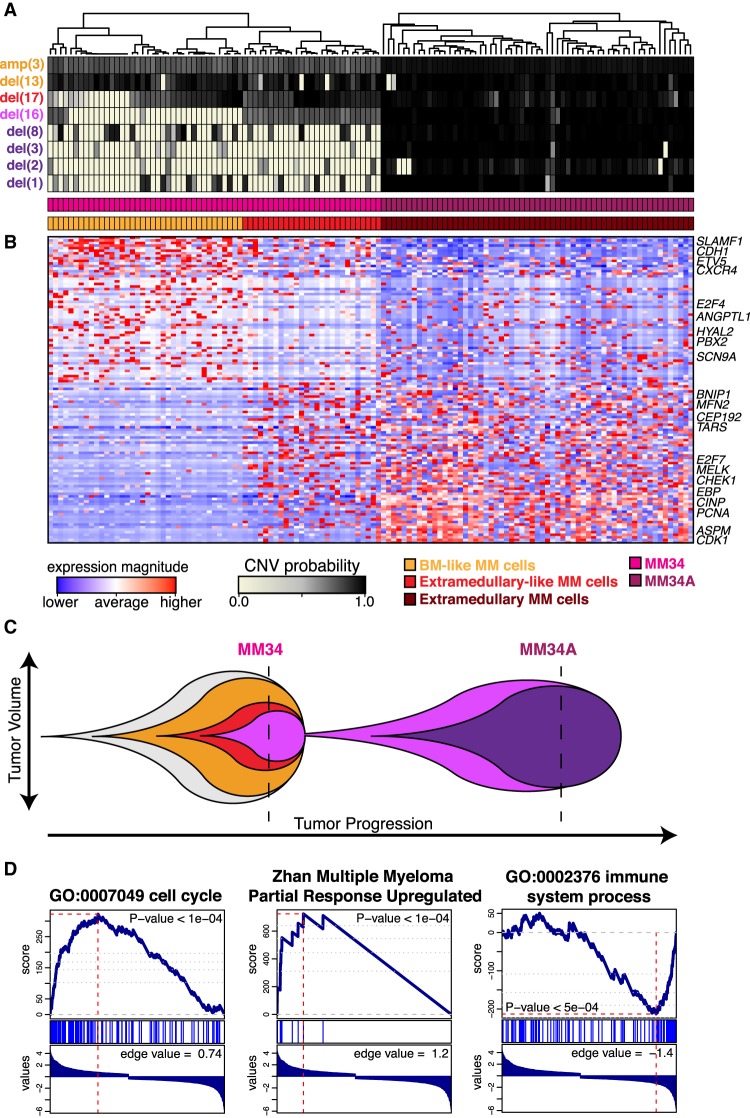
Figure 5.
Transcriptional characterization of MM34 and MM34A. (A) Posterior probability of alterations in MM34 and MM34A. (B) Heatmap of 120 consistently significantly differentially expressed genes in comparing the BM-like MM cells versus extramedullary-like MM cells in MM34, and BM-like MM cells versus extramedullary-like MM cells in MM34 and MM34A (Supplemental Table 4). Select genes of relevance to MM or cancer based on the literature search are annotated. (C) Proposed linear pattern of subclonal evolution. (D) Gene set enrichment analysis shows enrichment in cell cycle processes (left) and a known MM partial response signature (middle) for genes up-regulated in the extramedullary-like MM34 subclone, whereas enrichment in immune response processes (right) is seen for genes up-regulated in the BM-like MM34 subclone.
Next, we sought to identify these deletions in 65 BM MM cells from MM34 using our integrated approach. We find that while nearly all cells from MM34 harbor the Chr 13 deletion, only a fraction harbor the Chr 16 and 17 deletions, indicative of a linear subclonal expansion (Fig. 5A; Supplemental Fig. 7B). Consistent with HoneyBADGER's findings, in the initial BM MM sample, CNV analysis from bulk WES identified a deletion on Chr 13 (Supplemental Fig. 7B), while FISH and cytogenetics analysis of 200 interphase cells also identified a deletion on Chr 13 in 61% cells in addition to deletion of MAF (16q23) in 38% and TP53 (17p13.1) in 11.5% cells (Supplemental Fig. 7C). The percentage of MM cells harboring each deletion inferred from Honey BADGER was found to be consistent with the estimates from FISH and cytogenetics and bulk WES in both samples (Supplemental Fig. 7D). Based on these findings, we speculate that a genetic subclone harboring deletions on Chr 13, Chr 16, and Chr 17 most likely expanded to seed the extramedullary MM dissemination, acquiring additional alterations during this process (Fig. 5C).
Having identified this extramedullary-like subclone in the initial BM biopsy, we next examined its transcriptional signature. We identified 132 consistently differentially expressed genes (P-value <0.05) when comparing the extramedullary-like subclone with other BM-specific MM cells in MM34 as well as jointly with the extramedullary MM cells in MM34A (Fig. 5B; Supplemental Fig. 8; Supplemental Table 5). Among the down-regulated genes, E2F4, DPEP2, and CDH1 are located in the deleted region of Chr 16, indicating direct effects on gene expression from the genotype. These genes function in the suppression of cell cycle progression, activation of proinflammatory cytokines through leukotrienes, or cell adhesion events commonly suppressed during the tumor progression and metastasis (Ren et al. 2002; Thiery 2002). Among the rest of transcriptional changes, up-regulation of cell cycle–associated genes are likely conferred by the release of E2F4 repressor complexes from their promoters. It is noteworthy that Chr 17 deletion preceded that of Chr 16, suggesting that down-regulation of TP53 on Chr 17 and E2F4 has cooperated for the cell cycle progression during tumor evolution. As CDH1 and DPEP2 function in protein networks, the downstream effects are less visible in the transcriptional changes. Gene set enrichment analysis (GSEA) (Subramanian et al. 2005) of genes up-regulated (P < 0.1) in the extramedullary-like subclone showed significant enrichment (q-value <0.05) in the genes associated with cell cycle and a known partial response signature in MM (Zhan et al. 2006), while genes down-regulated (P < 0.1) showed significant enrichment (q-value <0.05) in immune response processes (Fig. 5D; Supplemental Table 6; Milacic et al. 2012; Fabregat et al. 2016). Thus, by identifying genetic subclones from scRNA-seq data, we can assess the functional impact of subclonal alterations at the transcriptional level.
Unbiased analysis of transcriptional heterogeneity identifies aspects independent of the subclonal structure
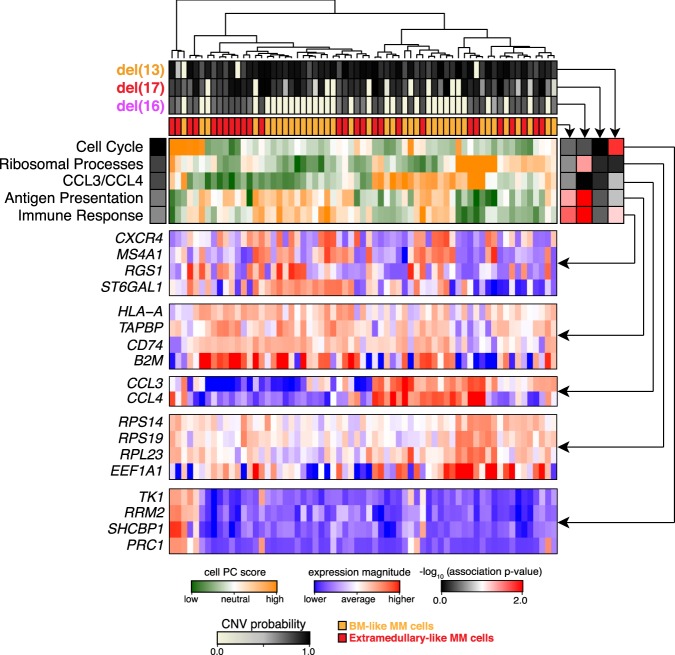
Despite significant transcriptional differences between genetic subclones, alternative sources of heterogeneity, such as differences in epigenetic state or cellular microenvironment, may impact transcriptional state and ultimately phenotypic heterogeneity. Our inference of genetic information from scRNA-seq data provides a unique opportunity to assess the relative impact of these mechanisms on transcriptional state. To do so, we first characterized transcriptional heterogeneity in MM34 using pathway and gene set overdispersion analysis (PAGODA) (Fan et al. 2016), which identifies nonredundant aspects of significant coordinated variability within annotated pathways and correlated gene sets (Fig. 6). PAGODA identified prominent aspects of transcriptional heterogeneity driven by ribosomal processes marking key transcriptional subpopulations. Other aspects of transcriptional heterogeneity were driven by expression of T-cell chemokines CCL3 and CCL4, as well as B2M and genes involved in antigen presentation. CCL3 and CCL4 have been previously implicated in MM tumor growth through regulation of the MM microenvironment (Roodman 2002; Vallet et al. 2011). Likewise B2M has been used to predict MM progression (Rossi et al. 2010). Previously, anti-B2M monoclonal antibodies have been also shown to overcome bortezomib resistance in MM (Zhang et al. 2015), thus providing potential therapeutic implications for early discovery of these subpopulations. When we compare these key transcriptional subpopulations with the inferred subclonal cell populations, we find that many of the identified aspects of transcriptional heterogeneity are independent of the subclonal structure. The extramedullary-like subclone was best matched by a less prominent aspect of transcriptional heterogeneity involved in immune response. Thus, while the aspect of transcriptional heterogeneity corresponding to the genetic subclonal structure is apparent from the unbiased transcriptional analysis alone, alternative nonclonal mechanisms can drive more prominent aspects of transcriptional variation.
Figure 6.
Pathway and gene-set overdispersion analysis of MM34. Unbiased transcriptional analysis of the initial bone marrow biopsy sample (MM34). Hierarchical clustering of cells (columns) is shown based on their overall transcriptional similarity. Top five most significant (P-value <0.05) aspects of transcriptional heterogeneity (rows) are shown by the green–orange heatmap in the center. Expression patterns of subsets of genes underlying each identified aspect of transcriptional heterogeneity are shown in the blue–red heatmaps below. Top panels show posterior probabilities of different deletions, and the consensus similarity to the extramedullary dissemination for each cell. Correspondence of different transcriptional subpopulations to the underlying subclonal structure is shown by the association heatmap (right; black–red). In particular, strong correspondence is observed between genetic subclones and the immune response aspect of transcriptional heterogeneity. However other, more prominent transcriptional subpopulations (CCL3/CCL4, antigen presentation) appear independently of the subclonal structure.
Discussion
Altogether, our results demonstrate the ability to integrate genetic and transcriptional information using scRNA-seq data to identify and characterize transcriptional programs driving distinct genetic subclones. We show that compared with an expression-based approach, an allele-based analysis offers substantially greater sensitivity and precision in identifying deletions that are smaller on the chromosome scale or present at lower subclonal fraction within the measured cell population. Combining allele- and expression-based approaches further improves performance and enables to identification of copy-neutral LOH events. Our approach accurately recapitulates expected cancer cell fractions in single cells compared with bulk estimates can robustly distinguish tumor from normal cells based on identified CNVs and is suitable for both full-transcript-length and 3′-tagging scRNA-seq protocols. By examining MM patient data, we find that while key genetic subclones do exhibit distinct transcriptional signatures that likely contribute to cancer progression, other more prominent aspects of transcriptional heterogeneity can be independent of the genetic subclonal structure and are most likely driven by alternative mechanisms, including potentially variation in epigenetic state or microenvironment. By inferring genotype information from scRNA-seq data, our approach can help unravel the impact of genetic and transcriptional heterogeneity and their interplay in cancer progression.
Methods
Patient samples and library generation
This study was approved by the institutional review board (IRB) of Samsung Medical Center (IRB approval no. SMC2013-09-009-012) and carried out in accordance with the principles of the Declaration of Helsinki. The study subjects were Korean patients diagnosed with MM at Samsung Medical Center, Seoul, Korea. BM aspirates or ascites were subjected to Ficoll Paque Plus (GE Healthcare) gradient and magnetic separation with anti-CD138 antibody microbeads (Miltenyi Biotech). From the CD138− enriched cells, genomic DNA and RNA was purified using the AllPrep kit (Qiagen). Matching blood DNA was isolated by the QIAamp DNA blood kit (Qiagen). Normal control RNA was collected from CD19+ microbead-purified blood B cells from four healthy volunteers. For bulk WES, genomic DNA (1 µo) from the BM and matching blood samples was sheared by Covaris S220 (Covaris) and used for library construction with SureSelect XT human all exon v5 and SureSelect XT reagent kit, HSQ (Agilent Technologies) according to the manufacturer's protocols. After multiplexing, the libraries were sequenced on the HiSeq 2500 sequencing platform (Illumina), using the 100-bp paired-end mode of the TruSeq Rapid PE cluster kit and TruSeq rapid SBS kit (Illumina). For scRNA-seq, CD138-enriched cells were subjected to single cell capture and cDNA amplification using the C1 single-cell auto prep system (Fluidigm) with the SMARTer kit (Clontech). Sequencing libraries were generated and multiplexed using Nextera XT DNA sample prep kit (Illumina) and sequenced on the HiSeq 2500 in the 100-bp paired-end mode of the TruSeq rapid PE cluster kit and TruSeq rapid SBS kit following the Smart-seq2 protocol (Picelli et al. 2014).
Bulk WES analysis
Reads from the FASTQ files were mapped against the human reference genome (GRCh37) using BWA MEM v0.7.8 (Li and Durbin 2010). Duplicates were removed using Picard tools v1.87 (https://broadinstitute.github.io/picard/). Indel realignment and quality score recalibration were performed using GATK v3.3.0 based on the GATK best practices guidelines (DePristo et al. 2011). Germline heterozygous variants were then identified using GATK's UnifiedGenotyper followed by variant quality score recalibration. To identify copy number alterations, mapped BAMs were analyzed by FREEC v7.2 with parameters recommended for WES data analysis by the authors (coefficientOfVariation = 0.062, window = 500, step = 250, breakPointThreshold = 1.5, readCountThreshold = 50, noisyData = TRUE) (Boeva et al. 2011). Genomic coordinates for copy number alternation were identified from the outputted text summaries and used for downstream analysis. To estimate subclonal deletion frequencies from bulk WES, the following equation was used based on an assumption of 100% purity: b = f*0 + (1 − f)*0.5, where b is the average LAF (lesser allele fraction), and f is subclonal fraction for deletion. This leads to f = 1 − 2b. The LAF value of a deletion region was obtained by averaging LAF values over the segments within the region. The segments and their LAF values were determined by FREEC.
Evaluating SNP detection rates from bulk and single-cell RNA-seq data
Single-nucleotide variants were called on bulk and single-cell K562 paired-end RNA-seq data using TopHat2 (Kim et al. 2013) and genome analysis toolkit (GATK) (McKenna et al. 2010; DePristo et al. 2011). Bulk K562 data (Deng et al. 2011) was downloaded from the Sequence Read Archive (accession number SRR315337) using SRAToolkit v2.5.7. Single-cell K562 RNA-seq measurements were carried out using the C1 single-cell auto prep system, in the same way as MM cells. Variants were called separately on the individual cell (instead of joint calling) for a fair comparison to bulk data, which was also separately fed to the same variant calling pipeline. Later, to match precision, single-cell variants that occurred in only one cell were discarded. Alignment was performed using TopHat v2.0.10 (along with Bowtie 2 v2.1.0) (Langmead and Salzberg 2012) against human genome v37 with decoy, allowing two mismatches and two gaps and the --max-multihits = 2 option to report up to two alignments per read. Then only uniquely aligned reads were kept. Human GRCh37.73 transcriptome annotation was used to guide spliced mapping. Aligned reads were sorted by coordinates using SAMtools v0.1.19 (Li et al. 2009) and duplicates were removed using Picard v1.107 (https://broadinstitute.github.io/picard/) MarkDuplicates. GATK 3.0.0 was used for the subsequent processing, including indel realignment, base quality score recalibration (BQSR), unified genotyper and variant quality score recalibration (VQSR). The -U ALLOW_N_CIGAR_READS option was used to handle spliced reads. To provide known polymorphic sites to GATK, dbSNP 138 was used for single-nucleotide substitutions and Mills_and_1000G_gold_standard.indels for known indel sites. After VQSR, only variants marked as “PASS” were kept. Likewise, for previously published scRNA-seq data from Patel et al. (2014), SRA files were downloaded from GEO (accession GSE57872) and converted to FASTQs using SRAToolKit v2.3.5. Alignment was performed using TopHat2 v2.0.10 (along with Bowtie 2 v2.1.048) against human genome v37 with decoy, allowing two mismatches and two gaps and the --max-multihits = 2 option to report up to two alignments per read. Then only uniquely aligned reads were kept. Human GRCh37.73 transcriptome annotation was used to guide spliced mapping. Aligned reads were sorted by coordinates using SAMtools v0.1.1949, and duplicates were removed using Picard v1.107 (https://broadinstitute.github.io/picard/) MarkDuplicates.
Heterozygous SNP identification
Where bulk WES data are available, heterozygous SNPs were called directly from bulk WES using GATK 3.0.0. Where bulk WES was not available, common heterozygous variants were identified from the Exome Aggregation Consortium (ExAC) variant sites database. ExAC variants were filtered to include single-nucleotide variants only with minor allele frequency >10%. Variants were further filtered based on presence within the data set of interest. Variants were considered heterozygous if reads from both the annotated reference and alternate alleles were present and distributed according to Bin(P = 0.5, n) > 1 × 10−8, where n is the total read coverage at that SNP. The resulting putative heterozygous SNPs were used to generate allele count matrices to assess the reference and alternative allele counts at each position using Rsamtools v1.28.0 (http://bioconductor.org/packages/release/bioc/html/Rsamtools.html).
Single-cell analysis
For gene expression quantification, reads from the FASTQ files were mapped against the USCS hg19 human reference genome using TopHat2 v2.1.0 (Kim et al. 2013) and quantified using featureCounts v1.4.4 (Liao et al. 2014). We do not anticipate realigning reads to GRCh38 will affect conclusions as coding SNPs relevant to our analysis remain largely consistent between the two builds. For the previously published scRNA-seq data from Patel et al. (2014), expression matrices were downloaded from GEO (accession GSE57872). Differential expression analysis on the two identified subclones was performed using SCDE (v1.99.1) (Kharchenko et al. 2014; Fan et al. 2016) with default parameters following recommended protocols (http://hms-dbmi.github.io/scde/diffexp.html). Significantly differentially expressed genes were identified using an absolute noncorrected Z-score cut-off of 1.96, corresponding to P-value <0.05, for heatmap visualization, and 1.28, corresponding to P-value <0.2, for GSEA. GSEA was performed using the LIGER (https://github.com/JEFworks/liger) package with input values as sorted MLE estimates of fold-change limited to significantly differentially expressed genes. In total, 10,593 curated (C2), GO (C5), oncogenic (C6), and immune (C7) gene sets from MSigDB (Liberzon et al. 2015) were tested. Gene sets with less than five genes or more than 500 genes were omitted. Pathway and gene-set overdispersion analysis to identify transcriptional subpopulations was performed using PAGODA (SCDE v1.99.1) (Kharchenko et al. 2014; Fan et al. 2016) with the same gene sets.
HMM
HoneyBADGER implements an expression-based HMM as well as an allele-based HMM to identify regions potentially affected by CNVs. For the expression-based HMM, a transition matrix is defined on three hidden states representing deletion, neutral, and amplification:
where t = 1 × 10−5 by default. Emission probabilities are defined by a normal distribution with means and variance estimated from the normalized expression data (see Supplemental Methods). For the allele-based HMM, a transition matrix is defined on two hidden states representing deletion or LOH, and neutral:
where t = 1 × 10−5 by default. Emission probabilities are defined by a binomial distribution with the size parameter given by the pooled coverage at the SNP position and an expected P = 0.1 for the lesser allele in the case of deletion or LOH and P = 0.45 for neutral. Default transition probabilities transition have been set based on the size of the regions expected to be able to detect. We find that both the expression-based HMM and allele-based HMM is robust to choices of transition probability t (Supplemental Fig. 9). However, for genomic regions and protocols with high rates of erroneous SNP detection or high normalized expression variance due to technical noise, we anticipate that these transition probabilities may need to be tuned.
Hierarchical Bayesian model
HoneyBADGER contains implementations of an expression-based approach, an allele-based approach, and an integrative approach for assessing the posterior probability of CNVs in given regions. All Bayesian hierarchical models were written in BUGs for Gibbs sampling. Simulation from the models using MCMC was accomplished through rJAGS. Four chains were initialized specifying starting values for Sk and ddk as 0 or 1 in all possible permutations where appropriate. The MCMC chains were allowed to run for 1000 iterations, with an adaptation of 100 and a burn-in of 100. Trace plots were used to ensure appropriate mixing on the hyper parameters, and Gelman plots were used to diagnose convergence of chains (Supplemental Fig. 10A,B).
For a particular region of interest, our goal is to make inference on the copy number status of a cell for that region given its observed allelic imbalance for germline heterozygous SNPs within the region and gene expression in the region relative to a putative diploid expression reference of comparable cell type. For a candidate region, let Sk = 1 if cell k has a CNV and Sk = 0 if cell k is copy number neutral.
In both allele- and expression-based models, we seek to estimate the posterior distribution of Sk given the observations. We can accomplish this through a hierarchical Bayesian framework, modeling the observed gene expression as a function of the variables of interest:
where ddk = 1 for a copy number gain and ddk = 0 for a copy number loss, such that Sk and ddk together capture the copy number status for cell k, and is the observed average normalized gene expression for genes within the tested region of interest in cell k. Likewise, for the allele-based model, we model observations at both the individual cell and bulk or pooled SNP-level information integrated into an additional hierarchical level involving observed gene-level monoallelic expression rates. An additional combined model approach makes inference on Sk using both gene expression and allele information.
Data access
The scRNA-seq and WES data for the MM cells have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE110499. HoneyBADGER is freely available under the GPL and is available as an R package (R Core Team 2017) with the source code available in the Supplemental Material and on GitHub (https://github.com/JEFworks/HoneyBADGER). Additional tutorials and documentation are available at http://jef.works/HoneyBADGER/.
Supplementary Material
Acknowledgments
We thank Patrik Ernfors for helpful feedback on the manuscript. J.F. was supported by an NIH grant F99 CA222750-01. P.V.K. was supported by NIH R01HL131768 from NHLBI and CAREER (NSF-14-532) award from the NSF. H.-O.L. was supported by Korea Basic Science Research Program grant NRF-2017R1D1A1B03032194. W.-Y.P. was supported by the Korea Health Technology R&D project HI13C2096 through KHIDI, Ministry of Health & Welfare, Korea.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.228080.117.
References
- Boeva V, Zinovyev A, Bleakley K, Vert J-P, Janoueix-Lerosey I, Delattre O, Barillot E. 2011. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 27: 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E. 2012. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28: 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel C, Ferreira PG, Santoni F, Delaneau O, Fort A, Popadin KY, Garieri M, Falconnet E, Ribaux P, Guipponi M, et al. 2015. Biased allelic expression in human primary fibroblast single cells. Am J Hum Genet 96: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Gunel M, Zhao H. 2013. SomatiCA: identifying, characterizing and quantifying somatic copy number aberrations from cancer genome sequencing data. PLoS One 8: e78143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, Ryu HS, Kim S, Lee JE, Park YH, et al. 2017. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 8: 15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet 43: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramsköld D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. 2015. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 33: 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, et al. 2016. The reactome pathway knowledgebase. Nucleic Acids Res 44: D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, Kaper F, Fan J-B, Zhang K, Chun J, et al. 2016. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods 13: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glitza IC, Lu G, Shah R, Bashir Q, Shah N, Champlin RE, Shah J, Orlowski RZ, Qazilbash MH. 2015. Chromosome 8q24.1/c-MYC abnormality: a marker for high-risk myeloma. Leuk Lymphoma 56: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, et al. 2018. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. 2014. Bayesian approach to single-cell differential expression analysis. Nat Methods 11: 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161: 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr J, Stojanov P, Carter S, Cruz-Gordillo P, Lawrence M, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, et al. 2014. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. 2015. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods 12: 519–522. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti J-C, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. 2010. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7: 521–531. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchor L, Brioli A, Wardell CP, Murison A, Potter NE, Kaiser MF, Fryer RA, Johnson DC, Begum DB, Hulkki Wilson S, et al. 2014. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia 28: 1705–1715. [DOI] [PubMed] [Google Scholar]
- Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H, D'Eustachio P, Stein L. 2012. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 4: 1180–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Tward AM, Hammon RJ, Ren Y, Rocco JW. 2015. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 12: e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9: 171–181. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev 16: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodman GD. 2002. Role of the bone marrow microenvironment in multiple myeloma. J Bone Miner Res 17: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Rossi D, Fangazio M, De Paoli L, Puma A, Riccomagno P, Pinto V, Zigrossi P, Ramponi A, Monga G, Gaidano G. 2010. β-2-microglobulin is an independent predictor of progression in asymptomatic multiple myeloma. Cancer 116: 2188–2200. [DOI] [PubMed] [Google Scholar]
- Sekiguchi N, Ootsubo K, Wagatsuma M, Midorikawa K, Nagata A, Noto S, Yamada K, Takezako N. 2014. Impact of C-Myc gene-related aberrations in newly diagnosed myeloma with bortezomib/dexamethasone therapy. Int J Hematol 99: 288–295. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. 2002. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet S, Pozzi S, Patel K, Vaghela N, Fulciniti MT, Veiby P, Hideshima T, Santo L, Cirstea D, Scadden DT, et al. 2011. A novel role for CCL3 (MIP-1α) in myeloma-induced bone disease via osteocalcin downregulation and inhibition of osteoblast function. Leukemia 25: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, Mirabella F, Lopez-Corral L, Humphray S, Murray L, et al. 2014. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia 28: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, Hakonarson H, Bucan M. 2007. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fan J, Francis JM, Georghiou G, Hergert S, Li S, Gambe R, Zhou CW, Yang C, Xiao S, et al. 2017. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res 27: 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ. 2012. CLL clonal heterogeneity: an ecology of competing subpopulations. Blood 120: 4117–4118. [DOI] [PubMed] [Google Scholar]
- Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, et al. 2006. The molecular classification of multiple myeloma. Blood 108: 2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, He J, Liu Z, Lu Y, Zheng Y, Li H, Xu J, Liu H, Qian J, Orlowski RZ, et al. 2015. Anti-β2-microglobulin monoclonal antibodies overcome bortezomib resistance in multiple myeloma by inhibiting autophagy. Oncotarget 6: 8567–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.