Abstract
Detection of DNA methylation in the genome has been possible for decades; however, the ability to deliberately and specifically manipulate local DNA methylation states in the genome has been extremely limited. Consequently, this has impeded our understanding of the direct effect of DNA methylation on transcriptional regulation and transcription factor binding in the native chromatin context. Thus, highly specific targeted epigenome editing tools are needed to address this. Recent adaptations of genome editing technologies, including fusion of the DNMT3A DNA methyltransferase catalytic domain to catalytically inactive Cas9 (dC9-D3A), have aimed to alter DNA methylation at desired loci. Here, we show that these tools exhibit consistent off-target DNA methylation deposition in the genome, limiting their capabilities to unambiguously assess the functional consequences of DNA methylation. To address this, we developed a modular dCas9-SunTag (dC9Sun-D3A) system that can recruit multiple DNMT3A catalytic domains to a target site for editing DNA methylation. dC9Sun-D3A is tunable, specific, and exhibits much higher induction of DNA methylation at target sites than the dC9-D3A direct fusion protein. Importantly, genome-wide characterization of dC9Sun-D3A binding sites and DNA methylation revealed minimal off-target protein binding and induction of DNA methylation with dC9Sun-D3A, compared to pervasive off-target methylation by dC9-D3A. Furthermore, we used dC9Sun-D3A to demonstrate the binding sensitivity to DNA methylation for CTCF and NRF1 in situ. Overall, this modular dC9Sun-D3A system enables precise DNA methylation deposition with the lowest off-target DNA methylation levels reported to date, allowing accurate functional determination of the role of DNA methylation at single loci.
DNA methylation has been shown to play a critical role in development and pathogenesis of various disease states and is frequently associated with transcriptional repression (Li et al. 1992; Okano et al. 1999; Jackson-Grusby et al. 2001; Egger et al. 2004; Bernstein et al. 2007; Smith and Meissner 2013; Perino and Veenstra 2016; Gao and Teschendorff 2017). In recent years, single base resolution methylome maps have been generated for different cell types and organisms, providing insights into the many possible functions of DNA methylation in the genome (Cokus et al. 2008; Irizarry et al. 2009; Lister et al. 2009, 2013; Maunakea et al. 2010; Stadler et al. 2011). Although comparative analyses of DNA methylation patterns with other genomic data such as transcription factor (TF) binding, gene expression, and chromatin state have been used to infer the functions of this DNA modification, these techniques only provide correlative information. Therefore, a growing body of research is aimed at disentangling the cause or consequence of gene repression by DNA methylation, including its potential role in the shaping of the TF binding landscape.
Traditional approaches to directly assess the relationship between CG methylation (mCG) within TF binding motifs and the occupancy of potentially mCG-sensitive TFs have relied on either the artificial insertion of DNA sequences that contain a TF binding motif with differing mCG states, biochemical experiments leveraging gel shift properties, pharmacological inhibition, or genetic perturbation of DNA methyltransferases (DNMTs) (Renda et al. 2007; Stadler et al. 2011; Maurano et al. 2015). DNMT inhibitors, such as 5-azacytidine, were previously used to alter DNA methylation states and infer the influence upon TF binding (Wang et al. 2012; Maurano et al. 2015). However, these pharmacological inhibitors of DNMTs alter the DNA methylation landscape globally, induce broad transcriptional changes, and result in highly pleiotropic off-target effects. Therefore, these are not suitable to understand the nuanced impact of DNA methylation on TFs at specific binding sites. More recently, a systematic evolution of ligands (methylated and unmethylated short double stranded DNA) by exponential enrichment (SELEX)–based investigation of the effect upon TF binding capacity in vitro of mCG in the core DNA-binding site for hundreds of TFs and TF binding domains revealed that the majority of TFs are sensitive to mCG, resulting in either increased or reduced DNA-binding affinity (Yin et al. 2017). However, in vitro binding assays lacking the native chromatin context and the artificial insertion of DNA sequences cannot accurately recapitulate an actual biological process, while pharmacological and genetic perturbations of DNMTs cause undesired global depletion of mCG. Thus, these techniques suffer from confounding secondary effects due to a lack of target selectivity and are therefore unable to accurately address the role of mCG in regulating mCG-sensitive TF binding. Therefore, there is a need to develop epigenome editing tools that can achieve targeted and highly specific modulation of mCG states at TF binding motifs in order to clarify the roles of mCG in shaping the mCG-sensitive TF occupancy landscape.
A major challenge has been the development of precise, adaptable tools that are capable of directing targeted DNA methylation to individual loci in different genomic contexts. Advances have been made by directly fusing DNMT enzymes (e.g., the de novo mammalian DNMT 3a [DNMT3A] or the prokaryotic CG methyltransferase, M.SssI/MQ1) to programmable DNA-binding domains, including zinc fingers, transcription activator-like effectors (TALEs), and deactivated Cas9 domains (dCas9) to induce targeted deposition of DNA methylation (Rivenbark et al. 2012; Siddique et al. 2013; Nunna et al. 2014; Bernstein et al. 2015; Stolzenburg et al. 2015; Amabile et al. 2016; Liu et al. 2016; McDonald et al. 2016; Ford et al. 2017; Lei et al. 2017; O'Geen et al. 2017; Stepper et al. 2017; Xiong et al. 2017). Thus, in theory, it is now possible to examine context-dependent transcriptional changes in response to localized epigenomic changes, but this varies greatly between target sites and DNA-binding domains (Jurkowski et al. 2015; Köferle et al. 2015). Furthermore, we currently understand very little about the off-target effects of these systems upon methylation throughout the genome. Given the importance of target specificity to these systems, gaining a comprehensive understanding of specificity, and developing approaches to improve it, is critical for the implementation and progression of accurate epigenome engineering.
Recently, attempts at investigating TF occupancy and mCG binding sensitivity have focused on the CCCTC-binding factor (CTCF), a TF involved in DNA looping and chromatin architecture (Phillips and Corces 2009; Hashimoto et al. 2017). These studies have utilized direct fusion constructs between dCas9 and DNMT3A (Liu et al. 2016) or M.SssI (Lei et al. 2017). The latter achieved highly specific, but only low level, induction of methylation at a limited number of CG sites across their target region (Lei et al. 2017), while the former attained higher levels of methylation induction but also reported off-target methylation (Liu et al. 2016). This suggests limited versatility and specificity of these single fusion constructs, despite both studies reporting reduced CTCF occupancy upon changes in mCG state. Furthermore, only recently has attention begun to be paid to potential off-target DNA methylation or demethylation delivered by these epigenome editing tools, with widespread off-target effects being observed (Galonska et al. 2018; Liu et al. 2018), but no satisfactory solution to this problem has been reported so far. Hence, it is paramount to study the sensitivity of TF binding to methylated DNA within their native chromatin context at endogenous loci, with the highly precise targeted DNA methylation editing. Here, we describe the development and comparative analysis of dCas9-based epigenome editing systems that recruit the catalytic domain of DNMT3A, with a particular focus on comprehensive assessment and minimization of off-target binding and DNA methylation induction, and utilization of optimized systems for modulating TF binding.
Results
Direct fusion of dCas9 to the DNMT3A catalytic domain results in high off-target DNA methylation
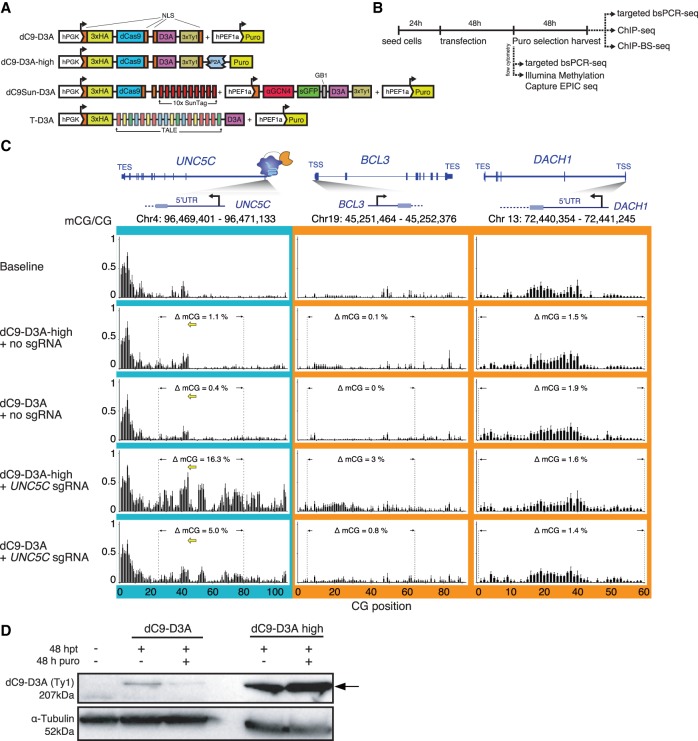
The direct fusion of dCas9 to the catalytic domain of DNMT3A was previously reported and used to induce cytosine methylation at targeted loci (Qi et al. 2013; Amabile et al. 2016; Liu et al. 2016; Vojta et al. 2016; Stepper et al. 2017). However, the potential off-target binding and methylation induced by these constructs has not been comprehensively assessed. In order to better understand the on-target and off-target effects of this system, we generated two dCas9-DNMT3A constructs, dC9-D3A and dC9-D3A-high (Fig. 1A), where the sole difference was in their puromycin selection marker. While identical in their structural design, dC9-D3A-high had the puromycin N-acetyltransferase fused to a self-cleavable peptide (P2A) (Kim et al. 2011), whereas the dC9-D3A construct had the puromycin-selectable gene expressed via its own constitutive promoter. MCF-7 cells or HeLa cells were transiently transfected with these two constructs and a gRNA targeting the UNC5C promoter and incubated for 48 h to allow for adequate protein expression followed by 48 h puromycin selection. Cells were then harvested, and DNA or chromatin was extracted for targeted bisulfite sequencing (targeted bsPCR-seq), chromatin immunoprecipitation (ChIP), or ChIP bisulfite sequencing (ChIP-bs-seq) (Fig. 1B; Brinkman et al. 2012). Initially we measured induced on-target DNA methylation at the UNC5C promoter and a proxy for potential off-target DNA methylation at the promoters of the BCL3 and DACH1 genes (Fig. 1C) in HeLa cells with dC9-D3A or dC9-D3A-high. Surprisingly, dC9-D3A exhibited poor on-target DNA methylation induction at the UNC5C promoter, with the highest observed increase in methylation at any single CG site within the region of 21%, and an average ΔmCG increase of 5% over all 62 CGs in the region. In contrast, dC9-D3A-high induced DNA methylation up to 52% at single CG sites in the UNC5C promoter and an average ΔmCG increase of 16% over the promoter region. However, dC9-D3A-high displayed strong off-target DNA methylation activity compared to dC9-D3A at the BCL3 promoter (average increase of 3% and 0.8%, respectively, over 46 CGs) but less so at the DACH1 promoter (average increase of 1.6% and 1.4%, respectively, over 48 CGs) (Fig. 1C). We speculated that the different on- and off-target DNA methylation rate was the result of sgRNA design as well as differing protein levels of dC9-D3A and dC9-D3A-high. Indeed, western blot on protein extracts from cells before and after puromycin selection demonstrated that protein levels were substantially higher in cells transfected with dC9-D3A-high compared to dC9-D3A, regardless of selection (Fig. 1D). We consequently reasoned that modulating the on- and off-target efficiency for targeted DNA methylation could not be accomplished by merely changing expression levels of the direct fusion constructs and that there was an inherent tradeoff in the on-target methylation efficacy versus off-target methylation with the direct fusion system. A redesigned, modular system for independent recruitment of the DNMT3A effector may be effective for overcoming this shortcoming (Fig. 1A).
Figure 1.
Characterizing on-target and off-target mCG deposition efficiency by dC9-D3A direct fusion system. (A) Schematics of the dCas9 (dC9) and TALE (T) constructs used, indicating positioning of nuclear localization sequences (NLS), protein tags (human influenza hemagglutinin [3xHA, 3xTy1]), promoter choice (glycerol kinase promoter [hPGK], human elongation factor-1 alpha promoter [hPEF1a]), solubility tag (protein G B1 domain [GB1]), selectable marker (puromycin resistance [puro]), single-chain Fv antibody against GCN4 domain (αGCN4), and human DNTM3A catalytic domain (D3A). (B) Timeline and outline for experimental design for measuring DNA methylation and TF occupancy (CTCF and NRF1). (C) Targeted DNA methylation deposition to the UNC5C promoter in HeLa cells, measured by bsPCR-seq. sgRNA placement is shown with yellow arrows; dotted lines indicate interval for CGs included in quantitation. (D) Western blot of relative dC9-D3A protein abundance (anti-Ty1) per 50 µg of total cell lysate: (lane 1) untransfected HeLa cells, (lanes 2,4) 48 h post transfection (hpt), (lanes 3,5) 48 hpt and 48 h puromycin (puro) selection, loading control anti-Tubulin. Arrow indicates dC9-D3A.
Adapting a modular dCas9 system for achieving high and specific DNA methylation
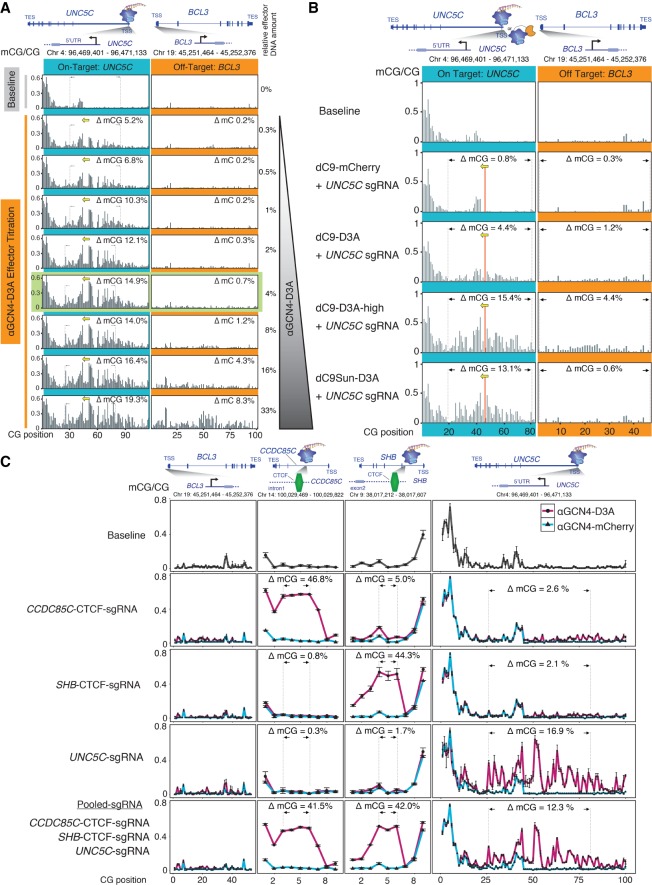
A modular dCas9 system has previously been reported utilizing the SunTag array (Tanenbaum et al. 2014). This repeating array of short repeat peptide sequences is fused to dCas9, thus acting as an epitope docking station that allows multiple proteins (fused to the counterpart single-chain antibody, scFv-GCN4, subsequently referred to as αGCN4) to be recruited to a desired target site. We hypothesized that this system would allow us to independently modulate the expression of the DNMT3A catalytic domain and the dCas9-SunTag, with the aim of limiting dCas9-SunTag abundance to favor its binding to its highest affinity sites in the genome while restricting the presence of excess DNMT3A in order to avoid its nonspecific activity. Such a system should reduce spurious off-target DNA methylation while maintaining high on-target mCG induction, thus improving on the design of the dCas9 molecule directly fused to a single DNMT3A. We adapted the previously reported SunTag system by fusing 10 GCN4 peptides with a flexible linker to dCas9 (dC9Sun) and the counterpart antibody to DNMT3A (αGCN4-D3A; dC9Sun-D3A) (Fig. 1A). The resulting dual constructs were tested in a titration series (Fig. 2A) that aimed to maximize on-target DNA methylation deposition at the UNC5C promoter, while minimizing off-target DNA methylation induction. The BCL3 promoter was used as an initial reporter locus for detecting the induction of excessive off-target DNA methylation. The entire set of transfections was carried out in HeLa cells that were transfected with a fixed amount of pGK-dCas9-SunTag plasmid and varying amounts of effector (αGCN4-D3A). The latter was titrated from 0% to 33% of the total amount of DNA transfected. The optimum amount of pEF1a-αGCN4-D3A effector that yielded the highest amount of on-target DNA methylation and the least amount of off-target DNA methylation was determined to be 4% of the total amount of DNA transfected. The change in DNA methylation (ΔmCG) was calculated by subtracting the average of all mCG/CG ratios in a defined region (e.g., 62 CGs at the UNC5C promoter) from the same CG sites in control-treated cells (baseline). Our adapted system was able to achieve high on-target mCG deposition of up to 49% at individual CpG dinucleotides and an average ΔmCG of 12.6% over all 62 CG dinucleotides, while maintaining off-target DNA methylation at only 0.7% (Fig. 2A, green highlight). A complementary experiment that titrated the amount of pGK-dCas9-SunTag plasmid while maintaining a fixed αGCN4-D3A effector quantity did not substantially improve on-target DNA methylation in HeLa cells, but altering the dC9Sun to effector ratio might be beneficial for other cell types (Supplemental Fig. S1). Hereafter, the optimized ratios of pGK-dCas9-SunTag plasmid to pEF1a-αGCN4-D3A effector plasmid from Figure 2A were used for the remainder of the study and will be collectively referred to as dC9Sun-D3A. The use of the SunTag system to recruit DNMT3A via TALEs to the DACH1 and UNC5C promoters also substantially improved on-target mCG deposition compared to a TALE-DNMT3A single fusion (Supplemental Figs. S2, S3). However, while TALEs are known for their high specificity, they are time consuming to adapt for multiple target sites and challenging to multiplex; hence, we focused on the more rapidly configurable dCas9 system for the remainder of the study.
Figure 2.
Modular dC9Sun-D3A system outperforms dC9-D3A direct fusion. (A) Titration of αGCN4-D3A effector (D3A [human DNMT3A catalytic domain]). Fraction of mCG is shown in black bars; dotted lines and black arrows indicate region used to calculate mCG change. (B) Comparison of dC9-D3A high, dC9-D3A, dC9Sun-D3A, and dC9Sun-mCherry (CRISPRi control) at the UNC5C promoter (on-target) versus the BCL3 promoter (off-target) by targeted bsPCR-seq (average mCG/CG, n = 3 replicates; error bars, SD). (C) mCG deposition efficiency by dC9Sun-D3A at three different loci (CCDC85C, SHB, and UNC5C promoters) measured by targeted bsPCR-seq (average mCG/CG, n = 3 replicates; error bars, SD).
To control for changes in methylation due to the presence of dCas9 binding in the vicinity of our targeted region, we initially used a catalytic mutant of the DNMT3A catalytic domain (D3AMut), which harbored four alanine substitutions in its catalytic center (F39A, E63A, E155A, R284A). However, when mCG deposition was tested at three target sites (CCDC85C intron, SHB intron, and MIR152) (Supplemental Fig. S4A–C), we detected residual mCG deposition of up to 22%, 16%, and 8%, respectively. We speculate that the DNMT3A catalytic domain might be able to recruit endogenous functional WT DNMT3A by forming oligomeric complexes as previously described (Holz-Schietinger et al. 2011). Accordingly, to eliminate any potential functional impacts based on residual induction of cytosine methylation, we determined that an αGCN4-mCherry effector would be a more appropriate control for the effect of dCas9 binding alone, compared to the αGCN4-D3AMut construct.
Targeting of the UNC5C promoter was repeated with dC9Sun-D3A, dC9-D3A, and dC9-D3A-high to compare the performance of each system, measuring the methylation level at CpG dinucleotides surrounding the UNC5C target region as well as at the BCL3 promoter, which served as a proxy for widespread off-target DNA methylation (Fig. 2B). dC9Sun-D3A induced mCG at the UNC5C promoter to comparable levels as dC9-D3A-high (ΔmCG over 62 CG sites at UNC5C was 4%, 15%, and 13% for dC9-D3A, dC9-D3A-high, and dC9Sun-D3A, respectively), without the shortcomings of promiscuous off-target methylation (ΔmCG over 46 CG sites at the off-target BCL3 region was 1.2%, 4.4%, and 0.6% for dC9-D3A, dC9-D3A-high, and dC9Sun-D3A, respectively). Notably, the off-target mCG induction by dC9Sun-D3A was reduced by >80% compared to dC9-D3A-high (Fig. 2B, BCL3 promoter off-target methylation).
To determine whether this improved DNA methylation induction and reduced off-target effects are achievable at multiple different loci, the set of target sites was expanded both independently and via multiplexed targeting. This was achieved by selecting a series of CTCF sites that were previously shown to be mCG sensitive (Maurano et al. 2015), hypomethylated, and occupied by CTCF in HeLa cells. Notably, high mCG induction was consistently achieved across both CTCF core binding sites (CCDC85C intron [47%] and SHB intron [44%]) and the UNC5C promoter (17%) (Fig. 2C, row 2–4). Multiplexing all three single guide RNAs (sgRNAs) resulted in a loss of on-target mCG deposition of <5% at all three targets (Fig. 2C, highlighted regions, cf. rows 2–4 and row 5), strongly suggesting that the dC9Sun-D3A system is a viable multiplexing option for mCG deposition. The maximum ΔmCG observed was at the UNC5C promoter, with a 57% increase. dC9Sun-D3A was able to induce DNA methylation at distances ≥300 bp; however, the SunTag system did not appear to add any additional distance compared to the dCas9 direct fusion (Supplemental Fig. S5). The same was observed when the T-D3A direct fusion is compared to TSun-D3A; however, adding the SunTag system to a TALE domain targeting the UNC5C promoter improves on-target mCG deposition from 30% to 64% (Supplemental Fig. S5).
Furthermore, we investigated whether a similar dC9Sun system was able to facilitate DNA demethylation when coupled to human TET1 (tet methylcytosine dioxygenase 1) catalytic domain (Morita et al. 2016) at the GAD1 intron 3. We found that the efficacy of the system was highly dependent on the sgRNA placement, with dC9Sun-TET1 achieving up to 60.0% mCG reduction at the GAD1 intron 3 (Supplemental Fig. S6). Taken together, the system of dC9Sun in combination with epigenetic effectors coupled to αGCN4 is highly effective in targeted deposition or removal of DNA methylation, without the limitations of high off-target effects.
Global assessment of binding and methylation specificity
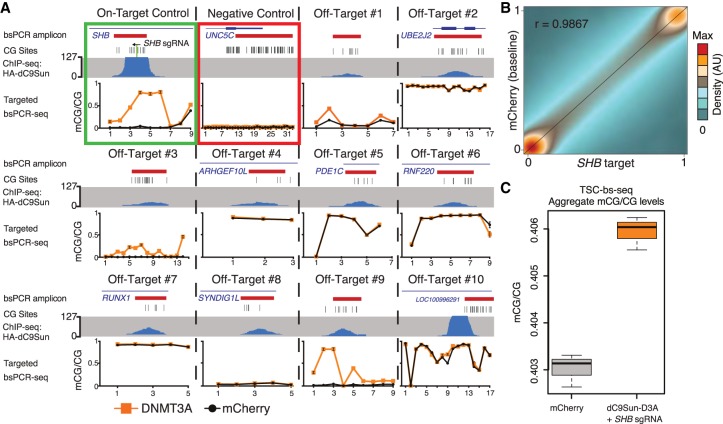
In order to more comprehensively assess the specificity of the dC9Sun-D3A system, we performed ChIP-seq upon dC9Sun using the 3xHA epitopes present at the N terminus of the protein, after targeting the construct to intron 1 of SHB (Fig. 3A). ChIP-seq peak calling identified 13 significant peaks throughout the entire genome (q-value < 0.01), with the top 10 most significant peaks shown in Figure 3A. All but one off-target peak were found to have <10% of the ChIP-seq normalized read density compared to the on-target binding site peak in the SHB intron (Supplemental Table S1). The reciprocal αGCN4-D3A ChIP-seq experiment using the C-terminal 3xTy1 epitope yielded only one significant peak at the on-target SHB site (Supplemental Table S1).
Figure 3.
Genome-wide off-target DNA methylation assessment. (A) Compilation of dC9Sun-D3A ChIP-seq (blue peaks) and targeted bsPCR-seq for CGs covered by ChIP-seq (bsPCR amplicon location in red), bsPCR-seq mCherry-only expressing cells (black line), and SHB sgRNA + dC9Sun-D3A (orange line) (sorted cells, n = 3 biological replicates; error bars, SD). (B) Correlation of mCG values for each CG site (>2.6 × 106, ≥5× coverage) of combined replicates (n = 3) from Illumina TruSeq methyl capture EPIC pulldown experiment. mCherry (baseline) HeLa cells shown on y-axis; SHB target dC9Sun-D3A HeLa cells shown on x-axis. Pearson correlation coefficient shown in upper left corner. (C) Boxplot of mCG/CG from all covered CGs from mCherry only HeLa cells (gray) and SHB sgRNA + dC9Sun-D3A (orange) (n = 3 biological replicates; thick black line indicates median; error bars, SD).
To further explore the extent of off-target mCG deposition and dC9Sun-D3A occupancy, we designed all three components of the system (dC9Sun, αGCN4-D3A, and SHB sgRNA) to express different fluorescent proteins (BFP, GFP, and mCherry, respectively), so that cells transfected with all three constructs could be enriched using flow cytometry (Supplemental Fig. S7A). The cell sorting approach, enriching for cells that have an intermediate level of expression of dC9Sun (BFP coexpression) and αGCN4-D3A (GFP fusion), was chosen to first guarantee expression of all three components (dC9-Sun, αGCN4-D3A, and sgRNA) in a single cell, and second, to achieve the highest possible on-targeted mCG deposition at SHB promoter while keeping off-target mCG change to a minimum (Supplemental Fig. S7B). Next, targeted bsPCR-seq in these sorted cells was performed for off-target sites and the on-target site SHB intron. Of all the off-target ChIP peaks, only three exhibited an increase in DNA methylation of ΔmCG >10% at one or more underlying CG dinucleotides (off-target peak regions 1, 3, and 9) (Fig. 3A), possibly driven by the hypomethylated state of these loci. A plausible explanation for the off-target binding of dC9Sun-D3A to these sites is that they contain partial matches to the SHB sgRNA, with the matching bases ranging from 11–15 nt out of the 17 nt of the SHB sgRNA target sequence (Supplemental Fig. S8). Therefore, the specificity of this system is likely only restricted by the uniqueness of the genomic sequence to which dCas9 is targeted and could be improved by alternative sgRNA design. Overall, these results demonstrate that the dC9Sun-D3A system can exhibit very high specificity of binding in the genome.
While binding of dC9Sun-D3A appeared to be highly specific, there is the potential that expression of the components of the system could still induce off-target methylation without detectable binding of dC9Sun-D3A. Therefore, to identify any mCG changes more broadly throughout the genome, the Illumina TruSeq methyl capture EPIC kit was used for targeted capture and detection of the DNA methylation state at single-base resolution (target solution capture bisulfite sequencing [TSC-bs-seq]) for 2.6 million CpG sites located in genomic regulatory regions in the DNA isolated from the previously fluorescently sorted cells (Supplemental Fig. S7). This approach allows quantitation of the DNA methylation state of ∼12% of all CGs present in the human genome, which are specifically targeted to cover the majority of known constitutively or conditionally unmethylated and lowly methylated regions in the genome (97% of CpG islands, 95% GENCODE promoters, 66% open chromatin regions, 98% FANTOM5 enhancers, 78% TF binding sites). Therefore, this provides an effective approach for high coverage (≥5 reads per CG and replicate) base resolution detection and quantitation of potential off-target methylation in the regions of the genome that could potentially become methylated. These probes captured regions including the on-target SHB intron and off-target site 9, where up to 80.5% and 82.1% methylation was observed, respectively, as observed in the targeted bsPCR-seq analysis (Supplemental Fig. S9). Notably, on-target mCG deposition was measured independently to be 80.5% and 78.0% for the CGs in the CTCF binding site (SHB intron) by either TSC-bs-seq or targeted bsPCR-seq, respectively. The methylation levels quantitated at CG sites by both targeted bsPCR-seq and TSC-bs-seq methods were very similar to each other, with Spearman correlation coefficient for controls and dC9Sun-D3A treatment of r = 0.976 and r = 0.967, respectively (total number of CGs = 94, n = 3 replicates). Furthermore, both sets of measurements suggest that sorting cells for optimal expression of all three components (dC9Sun, αGCN4-D3A, and sgRNA) reduces off-target mCG deposition by 2.1-fold (UNC5C promoter) and improves on-target mCG deposition in the cell population by 17.3% compared to puromycin selection (Supplemental Fig. S10). The high specificity of targeted DNA methylation induction by the dC9Sun-D3A system was further demonstrated by the very similar mCG levels (Pearson correlation coefficient r = 0.9867) at the 2,629,232 CGs covered (depth ≥5 reads; median CG coverage ≥16) throughout the targeted regions of the genome between cells expressing the control construct (dC9Sun-mCherry) and those expressing dC9Sun-D3A and the SHB intron 1 sgRNA (Fig. 3B). This was also observed for all pairwise correlations of each replicate (Supplemental Fig. S11). Moreover, the difference in average fraction mCG/CG for all CGs covered (≥5 reads coverage, 2,629,232 CGs, n = 3 replicates) between cells expressing the control construct (dC9Sun-mCherry) and those expressing dC9Sun-D3A and the SHB intron sgRNA was <0.3% (Fig. 3C). Overall, this demonstrates that the dC9Sun-D3A system exhibits very high specificity in both binding sites and induction of DNA methylation, in stark contrast to the high off-target activity of dC9-D3A (Figs. 1C, 2B), the system that has been most commonly used to date (Amabile et al. 2016; Liu et al. 2016; Vojta et al. 2016).
Direct assessment of the effect of DNA methylation on DNA–protein interactions
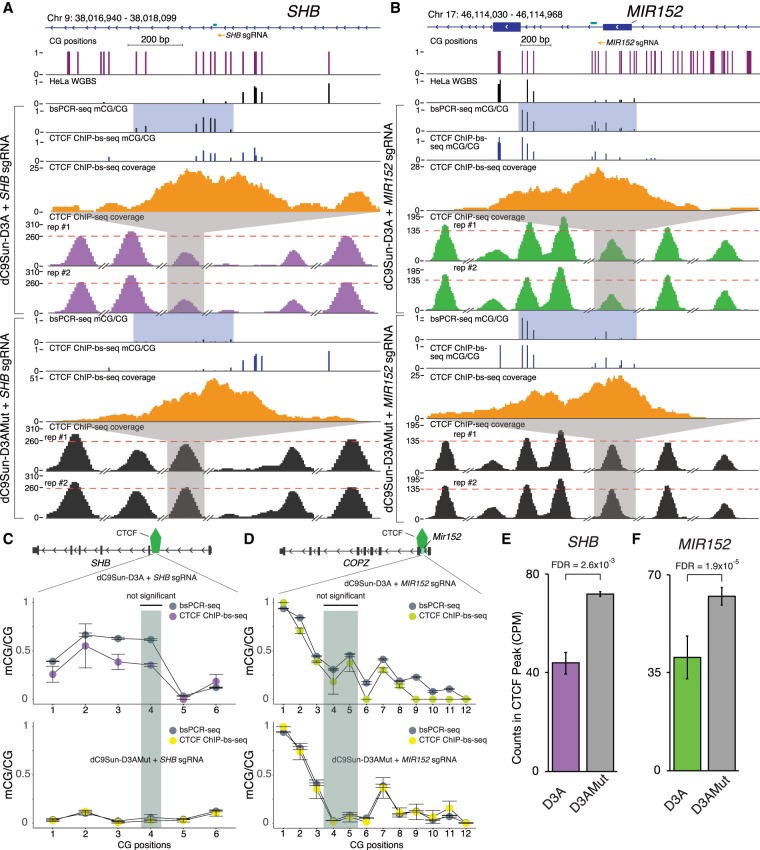
Having established the specificity and efficiency of the dC9Sun-D3A system, we sought to examine the consequences of targeted mCG deposition upon binding of methylation sensitive TFs. Previously, Maurano et al. (2015) reported that a subset of CTCF binding sites are sensitive to mCG reduction, induced by either DNMT triple knockout in HCT116 cells or 5-azacytidine treatment in K562 cells. Based on these findings, we speculated that CTCF binding sites sensitive to mCG loss would exhibit a reciprocal phenotype when mCG was specifically deposited in CTCF core binding sites in HeLa cells. To that end, we selected target regions for testing by intersecting the CTCF binding sites that were mCG sensitive in both K562 and HCT116 sites (FDR < 0.01) with CTCF sites occupied in HeLa cells. These sites were further required to each contain at least one hypomethylated CG in their core binding site. Three resulting CTCF binding sites were selected, in (1) a SHB intron, (2) a region upstream of MIR152, and (3) a CCDC85C intron. CTCF binding sites for MIR152, SHB intron, and CCDC85C intron are completely unmethylated in HeLa cells, as determined by whole-genome bisulfite sequencing (WGBS) (Fig. 4A,B; Supplemental Fig. S12, HeLa WGBS track). This observation was confirmed by targeted bsPCR-seq, where the amplicon covered the entire CTCF core binding site (Fig. 2C, baseline panel, CCDC85D and SHB). Next, we targeted mCG to the CTCF core binding sites by recruiting either dC9Sun-D3A or dC9Sun-D3AMut with sgRNAs binding within 100 bp of the CTCF core binding site. dC9Sun-D3AMut was used to control for possible steric hindrance effects on CTCF binding; however, we subsequently utilized dC9Sun-mCherry since dC9Sun-D3AMut exhibited residual DNA methylation activity (Supplemental Fig. S4). After 48 h of puromycin selection, the on-target mCG deposited at the CTCF core binding sites was 62% for the single CG in the CTCF binding site in SHB (Fig. 4C), 31% and 46% for the two CGs in the CTCF binding site upstream of MIR152 (Fig. 4D), and 52% and 35% for the two CGs in the CTCF binding site in CCDC85C (Supplemental Fig. S12B). We then investigated whether CTCF binding was impacted by the targeted DNA methylation deposited in its core binding site by performing CTCF ChIP-seq in duplicates for each condition. Importantly, all three CTCF binding sites, which were independently targeted by dC9Sun-D3A, showed a significant decrease in CTCF occupancy compared to dC9Sun-D3AMut (0.39-fold, 0.35-fold, and 0.67-fold reduction in normalized ChIP-seq read density for SHB, MIR152, and CCDC85C, respectively (edgeR, FDR = 2.6 × 10−3, 1.9 × 10−5, 5.2 × 10−12, respectively). Notably, the reduction in CTCF binding at the targeted loci (MIR152, SHB, and CCDC85C) were ranked either the most or second most decreased CTCF peaks compared to control, as determined by edgeR analysis of normalized peak counts (for SHB, see Fig. 4E; for MIR152, see Fig. 4F; for CCDC85C, see Supplemental Fig. S12C). Further, we performed CTCF ChIP-bisulfite sequencing (ChIP-bs-seq) (Brinkman et al. 2012; Hon et al. 2012; Statham et al. 2012) to assess the DNA methylation status of the DNA that was directly bound by CTCF (Fig. 4A,B; Supplemental Fig. S12A). This confirmed that the DNA bound by the remaining CTCF was methylated and comparable to the initial on-target DNA methylation measured by targeted bsPCR-seq (Fig. 4C,D, top panel comparing gray to colored circles), suggesting that CTCF occupancy is reduced by >30% for all three CTCF target sites (SHB, MIR152, and CCDC85C) due to mCG deposition. The remaining chromatin-bound CTCF was found to have mCG in its core binding, indicating a possible transition state where CTCF is either poised to leave or mCG is targeted for active DNA demethylation.
Figure 4.
Impact of targeted DNA methylation induction on CTCF binding. (A) Genome Browser display of targeted CTCF binding site in an SHB intron. Sets of experiments include (from top to bottom): targeted bsPCR-seq (fraction mCG/CG), CTCF-ChIP-bs-seq (fraction mCG/CG), CTCF-ChIP-bs-seq coverage, and CTCF ChIP-seq coverage. CTCF core binding site is highlighted in shaded green. CTCF ChIP-seq coverage (TMM normalized counts) is shown with adjacent peaks for reference (broken x-axis). Red dotted line is set to maximum targeted CTCF peak in the control samples. (B) Genome Browser snapshot of targeted CTCF binding site upstream of MIR152. (C,D) Quantitation of mCG/CG in CTCF core binding site (green shaded region) and adjacent to core binding site comparing targeted bsPCR-seq (gray circles) to ChIP-bs-seq (purple or green circles for αGCN4-D3A and yellow circles for αGCN4-D3AMut) for SHB and MIR152, respectively (replicates n = 2; error bars, SD; Fisher's exact test). (E,F) Quantitation of CTCF CPM normalized ChIP-seq peak at SHB and MIR152, respectively (replicates n = 2; error bars, SD; statistic edgeR, Benjamini-Hochberg multiple test corrected P-values).
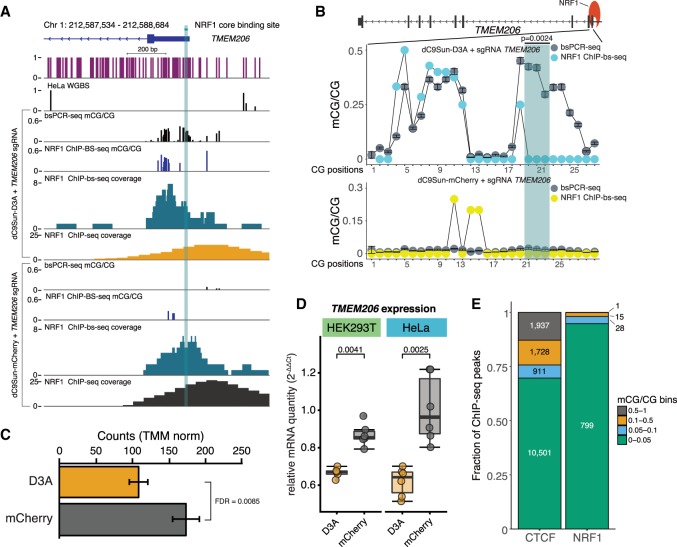
To extend the findings of targeted mCG deposition on CTCF binding to an additional TF, we next investigated the impact of mCG deposition on the binding of NRF1. This TF was selected as previous studies have found that NRF1 binding appears to be DNA methylation sensitive in vitro, as determined by a differential array binding assay (Hu et al. 2013), as well as in cell culture, where NRF1 occupies binding sites that lose mCG in DNMT triple knockout embryonic stem cells (Domcke et al. 2015). Similarly to our CTCF analyses, we set out to target four NRF1 occupied binding sites in HeLa cells, with each having at least one hypomethylated CG in the core binding site (TMEM206 promoter, downstream from TRAPPC3, upstream of MSANTD3, and downstream from TEF) with six different sgRNAs each. These targets were chosen based on both strong NRF1 occupancy as well as having narrow hypomethylated binding sites, where methylation was found within a 1-kb window both upstream of and downstream from the NRF1 binding site. As noted before (Hinz et al. 2015; Moreno-Mateos et al. 2015; Horlbeck et al. 2016) and seen in the resulting on-target DNA methylation patterns, sgRNA placement greatly affected the extent of mCG induced by dC9Sun-D3A (Supplemental Fig. S13A–D). Consequently, we focused solely on the TMEM206 promoter as it showed the greatest susceptibility to targeted mCG induction based on the presence of three CGs in its 15-bp core NRF1 binding site and appeared to be strongly occupied by NRF1 (Fig. 5A). To maximize on-target DNA methylation deposition for all three CGs, we chose sgRNA 2 for all further experiments (Supplemental Fig. S13A). Next we performed targeted bsPCR-seq to measure the average DNA methylation induction for all puromycin selected cells and found a 30%, 32%, and 32% increase in methylation level for each of the three CGs in the TMEM206 core binding site, respectively (Fig. 5B). As with our CTCF analyses, we then tested whether NRF1 could also bind to methylated DNA at the TMEM206 promoter by performing NRF1 ChIP-bs-seq. Due to the very low quantity of NRF1-immunoprecipitated DNA as the result of fewer NRF1 binding sites in the genome compared to other DNA-binding proteins such as CTCF, we merged data from multiple replicates (n = 3) to gain sufficient coverage to accurately quantitate mCG levels in the NRF1 core binding site. The NRF1 core binding site and CGs upstream of and downstream from it are completely hypomethylated as judged by bsPCR-seq and NRF1 ChIP-bs-seq when dC9Sun-mCherry is recruited to the TMEM206 promoter (Fig. 5B, lower panel). When TMEM206 promoter was targeted by dC9Sun-D3A, no mCG was detectable in the ChIP-bs-seq data of the NRF1 core binding site (Fig. 5B); however, upstream of this binding site, mCG was still detectable to the same level as seen previously in the targeted bsPCR-seq (Fig. 5B). This observation suggested that mCG was only tolerated upstream of the NRF1 binding site. Consequently, NRF1 either blocked mCG deposition in its core binding site or lost its binding capacity once mCG was deposited by dC9Sun-D3A. To understand the impact on NRF1 binding, we performed NRF1 ChIP-seq and found that NRF1 binding is reduced by 30% (edgeR, FDR = 0.0085, ranked 63 out of 607) when targeted by dC9Sun-D3A compared to cells targeted by dC9Sun-mCherry (Fig. 5C). Furthermore, while dC9Sun-D3A may have the potential to recruit other factors that cause the observed reduction in NRF1 binding, this would not be consistent with the observed DNA methylation pattern of NRF1-bound DNA as measured by ChIP-bs-seq (Fig. 5B). Notably, when DNA methylation was targeted to the NRF1 core binding site in the TMEM206 promoter, in both HeLa and HEK293T cells, TMEM206 transcript abundance was decreased, suggesting that the DNA methylation–dependent loss of NRF1 binding to the TMEM206 promoter appears to be causative for a change in transcription (Fig. 5D).
Figure 5.
Impact of targeted DNA methylation induction on NRF1 binding. (A) Genome Browser display of the targeted NRF1 binding site in the TMEM206 promoter. Sets of experiments include (from top to bottom): bsPCR-seq (mCG/CG), NRF1-ChIP-bs-seq (mCG/CG), NRF1-ChIP-bs-seq coverage, and NRF1 ChIP-seq-coverage. The NRF1 core binding site is highlighted in shaded green. (B) Quantitation of mCG/CG in the NRF1 core binding site (green shaded region) and adjacent to core binding site, comparing targeted bsPCR-seq (gray circles) to ChIP-bs-seq (cyan circles for αGCN4-DNMT3A and yellow circles for αGCN4-mCherry) at the TMEM206 promoter (NRF1 ChIP-bs samples combined n = 3 for coverage; bsPCR-seq, n = 3; error bars, SD; statistic: Fisher's exact test). (C) Quantitation of the TMEM206 promoter NRF1 ChIP-seq peak counts (TMM normalized) in samples treated with αGCN4-D3A (orange) compared to αGCN4-mCherry (gray) (mCherry n = 2; D3A n = 3; statistic edgeR, Benjamini-Hochberg multiple test corrected P-values). (D) qRT-PCR analysis of TMEM206 expression (normalized to geometric mean of the housekeeping genes RPS18, GAPDH, and HSPC3) comparing dC9Sun-D3A and dC9Sun-mCherry targeted to the NRF1 binding site in the TMEM206 promoter. Cells tested are HEK293T and HeLa cells, respectively (biological replicates n = 6; reference GFP-Puro transfected, puromycin-treated HeLa or HEK293T cells n = 4; statistic: Wilcoxon/Mann-Whitney U test, one-tailed). (E) Comparison of average mCG/CG in the CTCF and NRF1 core binding sites, respectively, by binning them into intervals of no (≥0 and ≤0.05), low (>0.05 and ≤0.1), intermediate (>0.1 and ≤0.5), or high (>0.5) levels of DNA methylation.
To compare the different levels of CTCF and NRF1 DNA methylation tolerance, core binding sites in the genome were interrogated by extracting the position of CGs in each core binding site (CTCF and NRF1 positional weight matrix) followed by averaging mCG/CG levels at all CG sites within each core binding site of CTCF and NRF1 ChIP-bs-seq data. Further, we parsed the average level of mCG/CG per core binding site for CTCF and NRF1 into bins based on the average mCG level (TF binding motif mCG level <5%, 5%–10%, 10%–50%, and >50%) (Fig. 5E), where 94.8% (799 out of 843) of NRF1 binding sites genome-wide have DNA methylation levels <5%. In contrast, only 69.6% (10,501 out of 15,077) of CTCF binding sites have DNA methylation levels <5%, which may explain why we detected that CTCF could bind to its core binding site even after targeted DNA methylation induction by dC9Sun-D3A (Fig. 4C,D). Taken together, we show that targeted DNA methylation, but not dCas9 interference (Figs. 4C,D, 5B), displaced both CTCF and NRF1, with the latter showing the higher DNA methylation sensitivity. Future work could leverage precise mCG deposition of the dC9Sun-D3A system to test direct mCG sensitivity for a variety of TFs in vivo.
Discussion
Previous work has highlighted the ability of dCas9 to be used to induce DNA methylation at desired regions in the genome (Amabile et al. 2016; Liu et al. 2016; Vojta et al. 2016; Lei et al. 2017; Stepper et al. 2017). However, a key issue that to date has not been sufficiently addressed is the potential for off-target DNA methylation induction, which could lead to nonspecific genomic responses and limits the utility of the systems for unambiguous assessment of the effect of DNA methylation at a locus. Single fusion constructs, such as dC9-D3A, lack the ability to fine-tune on- versus off-target DNA methylation deposition, and exhibit extensive off-target activity. When dC9-D3A expression is increased to attain high on-target methylation, the off-target methylation rate reached unacceptably high levels. Conversely, limiting the expression of the single fusion to reduce off-target methylation levels resulted in poor induction of on-target DNA methylation. Therefore, the single fusion constructs present multiple significant deficiencies. Here, we present a highly specific and tunable system to perform targeted alteration of DNA methylation, based on the modular SunTag system (Tanenbaum et al. 2014), allowing independent variation of the expression of the DNA targeting module (dC9Sun) and the effector module (αGCN4-D3A). While no significant difference in the distance at which DNA methylation deposition was observed between the dC9Sun-D3A system and the direct fusion systems, dC9Sun-D3A exhibited greatly improved specificity for depositing targeted methylation compared to dC9-D3A and achieved effective targeting of multiple loci simultaneously with on-target DNA methylation induction levels comparable to single-guide experiments. Recently, a similar dC9Sun system employing full-length DNMT3A1 was described by Huang et al. (2017), where lentiviral delivery was used. Huang et al. (2017) investigated off-target mCG deposition by reduced representation bisulfite sequencing (RRBS) at a limited subset of CpG islands (about 6100, 21.3% of all possible CpG islands), with a single sample per condition and reported off-target mCG deposition to be lower than dC9-D3A direct fusion systems. In contrast, we employed the more comprehensive approach of using TSC-bs-seq, covering 2.1 million CGs at ≥5 reads per sample (∼11× the number of CGs surveyed in Huang et al. [2017] at equivalent coverage). The Illumina EPIC TCS-bs-seq system is designed to cover a large number of putative regulatory regions in the genome that are frequently in an unmethylated state, here covering 12.7% of all hypomethylated CGs (at ≥5 reads per condition) in the HeLa genome (∼5.5 × 106 hypomethylated CGs in HeLa genome, mCG/CG ≤0.2). In contrast, the method used by Huang et al. (2017) would only cover 1.8% of the hypomethylated CG sites in the HeLa genome. We found that our system was about 40-fold less susceptible to gain or loss of mCG at CG islands compared to the system described by Huang et al. (2017) (Supplemental Fig. S14). Thus, through well-replicated use of this TCS-bs-seq system, we have performed one of the most extensive surveys of on- and off-target methylation induced by dCas9-based DNA methylation modifying systems to date, demonstrating that our dC9Sun-D3A system is highly specific.
Having established the specificity and fidelity of our dC9Sun-D3A system, we subsequently directly determined the impact of targeted DNA methylation on the binding of two DNA-binding proteins, CTCF and NRF1. Although CTCF sensitivity to DNA methylation had previously been reported (Liu et al. 2016; Lei et al. 2017), our work is the first to directly test the impact of targeted DNA methylation deposition upon NRF1 binding. Our results indicate that both CTCF and NRF1 are impacted by DNA methylation deposition. CTCF was still able to tolerate and bind methylated DNA in its core binding site for three independent sites. A possible explanation for this is that the binding of CTCF is in a transitional state, where its core binding site has been methylated but CTCF has yet to be evicted. Alternatively, CTCF may be binding 5-hydroxymethylcytosine (5hmC), the byproduct of Tet-mediated oxidation of 5-methylcytosine [5mC]), as previously described (Feldmann et al. 2013; Teif et al. 2014; Marina et al. 2016) since CTCF ChIP-bs-seq cannot distinguish between 5mC and 5hmC. In contrast to CTCF, we did not observe NRF1 binding to any intermediate 5mC or possible 5hmC states, as there was a loss in NRF1 occupancy upon DNA methylation deposition. Taken together, the dC9Sun-D3A technology opens up avenues to perform highly precise DNA methylation deposition and could be adapted to effectively implement alternative epigenetic editing such as altering histone modifications or DNA demethylation with the same advantages as described above. It is also conceivable to leverage the array of αGCN4 binding sites on the SunTag for multiplexed epigenetic editing, employing more than one epigenetic effector simultaneously.
Conclusion
The influence of DNA methylation changes on gene regulation is still a highly debated topic, with broad implications for interpretation of the potential effect of differential methylation states evident in a variety of contexts, such as development and cancer. However, it is paramount to study the effects of targeted DNA methylation changes at key regions with a high degree of confidence, such that off-target effects by spurious DNA methylation deposition are minimized. To that end, the dC9Sun-D3A system described here is highly adaptable and tunable and capable of high on-target mCG deposition while not suffering from widespread spurious off-target DNA methylation induction as observed with some other previously described dCas9 systems. Therefore, the dC9Sun-D3A system could be employed in a variety of different cell types and systems to test the direct effect of mCG deposition on TF binding, splicing, or transcription in general.
Methods
Cell lines
MCF-7 and HeLa cells were cultured in a humidified cell culture incubator at 37°C with 5% (v/v) CO2. MCF-7 cells were maintained using MEM (minimum essential medium) alpha (Life Technologies, catalog no. 12571071) supplemented with 10% (v/v) fetal bovine serum (FBS) (Integrated Sciences, catalog no. HYCSV3017603), 1× (v/v) 5.5% sodium bicarbonate (Life Technologies, catalog no. 25080094), and 1× (v/v) Glutamax (Life Technologies, catalog no. 35050061) while HeLa cells were cultured with DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS and 1× (v/v) Glutamax.
Cell transfections
Transfections were carried out using a 1:3 ratio of DNA to FuGENE HD (Promega, catalog no. E2311) following manufacturer's instructions. Briefly, MCF-7 or HeLa cells were seeded at 80% confluency in either six-well culture plate (In-vitro Technologies, catalog no. FAL353046) or 15-cm culture dishes (In-vitro Technologies, catalog no. COR430599). Twenty-four hours post seeding, 500 ng of TALE-DNMT3A or dCas9-DNMT3A constructs and 500 ng of individual sgRNA plasmids were transfected into cells. For the TALE and dCas9-SunTag constructs, 330 ng was used, along with 330 ng of individual or pooled sgRNAs, 40 ng of αGCN4-DNMT3A, and 300–320 ng of pEF1a-GFP-Puro (transfection control plasmid) to make the final amount to 1 µg. This was scaled up for 15-cm plate transfections. Cells were screened after 48 h for expression of reporter gene (GFP or mCherry) by fluorescent microscopy followed by either FACS sorting or 48 h of puromycin selection (2 µg/mL, Life Technologies, catalog no. A1113803) to enrich for positively transfected cells. After puromycin selection, surviving cells were subsequently either used for ChIPs, ChIP followed by bisulfite sequencing, or targeted bisulfite sequencing where DNA was extracted by the ISOLATE II DNA extraction kit (Bioline, catalog no. BIO-52067). Triple positive cells containing the tagBFP2 (dCas9-Suntag vector), mCherry (sgRNA vector), and sfGFP (αGCN4-DNMT3A vector) were FACS sorted and subjected to DNA extraction using the ISOLATE II DNA extraction kit (Bioline, catalog no. BIO-52067), which was further used in determining genome-wide off-target DNA methylation deposition using the TruSeq methyl capture EPIC library prep kit (Illumina, catalog no. FC-151-1002) as well as for targeted bisulfite sequencing.
Plasmid construction
The human codon optimized Cas9 was amplified using PCR from the hCas9 D10A plasmid (Addgene, 41816). Subsequently, PCR mutagenesis was conducted to introduce the H840A mutation to generate the catalytically dead Cas9 (dCas9). The DNMT3A sequence was PCR amplified from the ZF-598-DNMT3A plasmid (gift from Pilar Blancafort) and the N-terminal 3xHA tag, linker peptide between dCas9 and DNMT3A, and C-terminal 3xTy1 tag were ordered as separate gBlocks (IDT). Finally, the dCas9 cassette was inserted into a plasmid backbone containing a puromycin selectable marker. The single fusion TALE expression vector backbone was based on Sanjana et al. (2012), and Gibson assembly (Gibson et al. 2009) was used to replace the nuclease with DNMT3A. The linker peptide for the TALE-DNMT3A expression vector was identical to the dCas9 counterpart.
Gibson assembly was used to clone the empty TALE-Sun Tag backbone using plasmids pHRdSV40-dCas9-10XGCN4_ v4_P2A-BFP (Addgene, 60903) and pEF1a-TALE-DNMT3A-WT (Addgene, 100937). Similarly, the dCas9-SunTag construct was assembled from pGK-dCas9-DNMT3A-WT (V3; Addgene, 100938), pHRdSV40-dCas9-10XGCN4_v4_P2A-BFP (Addgene, 60903), and a poly(A) gBlock sequence (IDT). Following this, pHRdSV40-scFv-GCN4-sfGFP-VP64-GB1-NLS (Addgene, 60904), pEF1a-TALE-DNMT3A-WT (Addgene, 100937), and a gBlock linker sequence (IDT) was used to assemble αGCN4-DNMT3A, which was also used to create the DNMT3A FEER > AAAA mutation. This same backbone was used to Gibson assemble the αGCN4-mCherry control plasmid, where the DNMT3A domain was replaced with a mCherry fluorophore. Furthermore, restriction ligation was used to insert TALE targeting sequences within the empty TALE-SunTag backbone using MluI and AfeI (NEB). TALE DNA-binding domain inserts were synthesized in-house using iterative capped assembly as previously described (Briggs et al. 2012). Finally, gRNA plasmids were synthesized according to the protocol by Mali et al. (2013), and target sequences for TALEs and gRNA are listed in Supplemental Table S2. All plasmids constructed for this study can be found at https://www.addgene.org/browse/article/28191639/.
Targeted BS-PCR primer design and sequencing
For each bisulfite PCR amplicon, primers were designed with MethPrimer tool (Li and Dahiya 2002) (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) with all parameters set to default except the following: product size 200–400 bp; primer Tm min: 55°C, opt: 60°C and max: 64°C; primer size min: 20 nt, opt: 25 nt and max: 30 nt. Each primer pair was tested in a 15 µL PCR reaction with either NEB EpiMark (NEB, M0490L) or 2× MyTaq (Bioline, BIO-25041) for a range of annealing temperatures ranging from 50°C–64°C for 40 cycles. Primers that amplified specific products of the expected size were used, and these amplicons were confirmed as our regions of interest via Sanger sequencing. Five-hundred nanograms of extracted genomic DNA from transfected HeLa or MCF-7 cells and 0.5% spiked in lambda DNA (nonconversion control) was bisulfite converted using the EZ DNA-methylation-gold kit (Zymo Research, catalog no. D5006) following the manufacturer's instructions. We used 20 ng of bisulfite converted DNA for generating PCR products, which were subsequently pooled in an equimolar ratio and purified by Agencourt AMPure XP beads (Beckman Coulter, catalog no. A63880). Libraries were prepared by A-tailing and ligating Illumina-compatible Y-shaped adapters to amplicons that were amplified by three PCR cycles and subsequently purified by Agencourt AMPure XP beads (Beckman Coulter, catalog no. A63880). Single-end 300-bp sequencing was performed on an Illumina MiSeq platform.
qRT-PCR
Transfected HeLa and HEK293T cells were harvested with trypsin, washed with Dulbecco's phosphate-buffered saline (dPBS), and centrifuged at 300g for 3 min, and then RNA was extracted using RNeasy Mini Kit (Qiagen, #74104). A cDNA library was made using SuperScript IV Reverse Transcriptase (ThermoFisher, #18090050) supplemented with poly d(T) and random hexamer reverse transcription primer mix with 500 ng of RNA. qPCR primers (listed in Supplemental Table S2) were designed using Primer 3 for GAPDH, RPS18, and HSPC3 as housekeeping controls and TMEM206 as the target. Luna qPCR universal master mix (Genesearch, #M3003E) was used for qPCR, along with 2 µL of 1:5 diluted cDNA for each reaction. Activation was analyzed with the Applied Biosystems ViiA7 instrument with the following program: 95°C for 3 min, followed by 40 cycles at 95°C for 10 sec, 63°C for 20 sec, and then 72°C for 5 sec. Gene expression levels were normalized to the geometric mean of the three control genes with ΔΔCt calculated by comparing to the untransfected samples.
ChIP-sequencing
Transfected HeLa cells were used for performing ChIPs for CTCF and NRF1 as described previously (Wang et al. 2012; Domcke et al. 2015) with some modifications. Briefly, cells were crosslinked for 10 min in 1% formaldehyde and quenched in 125 mM glycine. Chromatin was sheared (Covaris, S220) for 5 min, 5% duty cycle, 200 cycles per burst, and 140 watts peak output and incubated with either a CTCF (Cell Signaling, 2899B), NRF1 (Abcam, ab55744), HA (Biolegend, 901502), or Ty1 (Sigma, SAB4800032) antibody and subsequently conjugated to a 50:50 mix of protein A/G Dynabeads (Invitrogen, M-280). After wash steps, DNA was eluted, crosslinks were reversed, and immunoprecipitated DNA was purified by Agencourt AMPure XP beads (Beckman Coulter, catalog no. A63880). Libraries were prepared from the entire ChIP eluate volume containing either 1 ng or 0.1 ng input material per replicate for CTCF or NRF1, respectively, using the ThruPLEX DNA-seq 12S kit (Rubicon Genomics, R400429). After limited PCR amplification, libraries were purified using Agencourt AMPure XP beads (Beckman Coulter, catalog no. A63880), and eluted in a final volume of 20 µL. Single-end 100 bp sequencing was performed on an Illumina HiSeq 1500.
ChIP-bs-seq
ChIP was performed using CTCF and NRF1 antibodies as stated above. Either 1 ng or 10 ng per replicate of CTCF and NRF1 ChIP eluate was used and subjected to bisulfite conversion using Zymo methylation direct kit (Zymo Research, D5020). Bisulfite converted DNA was immediately handled for library preparation using the Accel-NGS Methyl-Seq DNA library kit (Swift Biosciences, catalog no. 30024) following the manufacturer's instructions. Libraries were amplified for either nine or 13 cycles for CTCF and NRF1, respectively. Single-end 100-bp sequencing was performed on an Illumina HiSeq 1500.
Targeted enrichment for determining off-target DNA methylation deposition
HeLa cells were transfected with pEF1a-GFP-Puro and pEF1a-mCherry for control baseline or pGK-dCas9-Suntag-tagBFP2, pEF1a-mCherry-SHB1 sgRNA, and pEF1a-αGCN4-DNMT3A for targeted methylation at SHB locus in triplicate. Transfected cells were sorted for GFP and mCherry positive (control baseline) or GFP, mCherry and BFP positive (targeted methylation at SHB locus) and subjected to DNA extraction using the ISOLATE II DNA extraction kit (Bioline, catalog no. BIO-52067). We used 250 ng of genomic DNA from each replicate for library preparation and subsequently pooled for target enrichment using the TruSeq methyl capture EPIC library prep kit (Illumina, catalog no. FC-151-1002) following manufacturer's instructions. Libraries were bisulfite converted using EZ DNA methylation Lightning kit (Zymo Research, D5030) and amplified for 10 cycles. Single-end 100 bp sequencing was performed on an Illumina HiSeq 1500.
Western blot
HeLa cells were seeded at 80% confluency and transfected with equal amounts of sterilized plasmids dC9-D3A high (or dC9-D3A) and SHB-1-sgRNA to 15.8 µg, along with three times the amount of FuGENE HD transfection reagent (Promega, catalog no. E2311) following the manufacturer's instructions. Samples were either collected 48 h post-transfection or following an additional 48 h after puromycin selection (2 µg/mL, Life Technologies, catalog no. A1113803) to replicate the conditions employed while generating cells used for DNA methylation analysis. Cells were harvested and spun down at 300g for 5 min and then lysed in RIPA buffer with 4% SDS and 1 µL Benzonase (Sigma, catalog no. E1014). Samples were further lysed by sonication (Covaris, S220) and gently pipetted with a 21G needle (Livingston, catalog no. DN21Gx1.0LV) to reduce viscosity. Standard immunoblotting was performed on 50 µg of cell lysate for each sample. Mouse anti-Ty1 (1:1000, Sigma, catalog no. AB4800032) antibody was used, along with mouse anti-α-Tubulin (1:1000, GenScript, catalog no. A01410-40) as the loading control.
Data processing
ChIP-seq and ChIP-bs-seq data were quality checked and hard trimmed (Accel-NGS Methyl-Seq DNA library kit requirements) using Trimmomatic (Bolger et al. 2014) with the following parameters: ILLUMINACLIP:2:30:10, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:20, MINLEN:25. Trimmed ChIP-seq data were mapped to the human genome (GRCh37/hg19) using Bowtie1.1.1 (Langmead et al. 2009) with the options “--wrapper basic-0 -m 1 -S -p 4 -n 1” allowing up to one mismatch. Reads mapping to multiple locations were then excluded, and reads with identical 5′ ends and strand were presumed to be PCR duplicates and were excluded using Picard MarkDuplicates. Bigwig coverage tracks and differential peak calling were produced by THOR (Allhoff et al. 2016) using TMM normalization. Normalized peak counts were analyzed by edgeR (Robinson et al. 2010) for fold-change and filtered by multiple-test-corrected false-discovery rate (FDR < 0.05). Targeted bs-PCR-seq data and ChIP-bs-seq data were mapped to the GRCh37/hg19 genome and processed using BS-Seeker2 (Guo et al. 2013) with default parameters, Bowtie 1, and allowing for one mismatch. Data processing pipelines leveraged sambamba (Tarasov et al. 2015) and Gnu parallel (Tange 2011) to speed up sample analysis. This work used regions and sites that are well established in hg19 and did not improve in human genome assembly hg38. Further, previous work that described specific CTCF binding sites (Maurano et al. 2015), as well as NRF1 binding sites (Domcke et al. 2015), that were reported to be DNA methylation sensitive were also aligned to hg19. Consequently, conclusions drawn from our manuscript would not change based on aligning to hg38.
Data access
All HeLa-S3 raw and processed data generated in this study (WGBS-seq, ChIP-seq, ChIP-bs-seq, bsPCR-seq, and TSC-bs-seq) have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107607.
Supplementary Material
Acknowledgments
We thank O. Bogdanović, P. Blancafort, and the members of the Lister and Polo laboratories for helpful discussions and feedback. This work was funded by the Australian National Health and Medical Research Council (GNT1069830), the Australian Research Council (ARC) Centre of Excellence program in Plant Energy Biology (CE140100008), the National Institutes of Health (5R01DA036906), and the Raine Medical Research Foundation. Support was provided by an Australian Research Council Future Fellowship to R.L. (FT120100862), Sylvia and Charles Viertel Senior Medical Research Fellowships (to R.L. and J.M.P.), and a Howard Hughes Medical Institute International Research Scholarship to R.L.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.233049.117.
References
- Allhoff M, Seré K, F Pires J, Zenke M, G Costa I. 2016. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res 44: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, Lombardo A. 2016. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 167: 219–232.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. 2007. The mammalian epigenome. Cell 128: 669–681. [DOI] [PubMed] [Google Scholar]
- Bernstein DL, Le Lay JE, Ruano EG, Kaestner KH. 2015. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J Clin Invest 125: 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. 2012. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res 40: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman AB, Gu H, Bartels SJJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, et al. 2012. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 22: 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. 2015. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528: 575–579. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463. [DOI] [PubMed] [Google Scholar]
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schübeler D. 2013. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet 9: e1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford EE, Grimmer MR, Stolzenburg S, Bogdanovic O, de Mendoza A, Farnham PJ, Blancafort P, Lister R. 2017. Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. bioRxiv 10.1101/170506. [DOI]
- Galonska C, Charlton J, Mattei AL, Donaghey J, Clement K, Gu H, Mohammad AW, Stamenova EK, Cacchiarelli D, Klages S, et al. 2018. Genome-wide tracking of dCas9-methyltransferase footprints. Nat Commun 9: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Teschendorff AE. 2017. Epigenetic and genetic deregulation in cancer target distinct signaling pathway domains. Nucleic Acids Res 45: 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M. 2013. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X. 2017. Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol Cell 66: 711–720.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM, Laughery MF, Wyrick JJ. 2015. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry 54: 7063–7066. [DOI] [PubMed] [Google Scholar]
- Holz-Schietinger C, Matje DM, Harrison MF, Reich NO. 2011. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J Biol Chem 286: 41479–41488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. 2012. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Witkowsky LB, Guglielmi B, Replogle JM, Gilbert LA, Villalta JE, Torigoe SE, Tjian R, Weissman JS. 2016. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife 5: e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, Shin J, Cox E, Rho HS, Woodard C, et al. 2013. DNA methylation presents distinct binding sites for human transcription factors. eLife 2: e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Su J, Lei Y, Brunetti L, Gundry MC, Zhang X, Jeong M, Li W, Goodell MA. 2017. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol 18: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31–39. [DOI] [PubMed] [Google Scholar]
- Jurkowski T, Ravichandran M, Stepper P. 2015. Synthetic epigenetics—towards intelligent control of epigenetic states and cell identity. Clin Epigenetics 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köferle A, Stricker SH, Beck S. 2015. Brave new epigenomes: the dawn of epigenetic engineering. Genome Med 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhang X, Su J, Jeong M, Gundry MC, Huang YH, Zhou Y, Li W, Goodell MA. 2017. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat Commun 8: 16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. 2016. Editing DNA methylation in the mammalian genome. Cell 167: 233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, et al. 2018. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172: 979–992.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marina RJ, Sturgill D, Bailly MA, Thenoz M, Varma G, Prigge MF, Nanan KK, Shukla S, Haque N, Oberdoerffer S. 2016. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J 35: 335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, Stamatoyannopoulos JA. 2015. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep 12: 1184–1195. [DOI] [PubMed] [Google Scholar]
- McDonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, Challen GA. 2016. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open 5: 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12: 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, et al. 2016. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol 34: 1060–1065. [DOI] [PubMed] [Google Scholar]
- Nunna S, Reinhardt R, Ragozin S, Jeltsch A. 2014. Targeted methylation of the epithelial cell adhesion molecule (EpCAM) promoter to silence its expression in ovarian cancer cells. PLoS One 9: e87703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, Mackay JP, Farnham PJ, Segal DJ. 2017. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res 45: 9901–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- Perino M, Veenstra GJC. 2016. Chromatin control of developmental dynamics and plasticity. Dev Cell 38: 610–620. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. 2007. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem 282: 33336–33345. [DOI] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. 2012. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7: 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. 2012. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 7: 171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. 2013. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 425: 479–491. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. 2012. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res 22: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, Reik W, Jeltsch A, Jurkowski TP. 2017. Efficient targeted DNA methylation with chimeric dCas9–Dnmt3a–Dnmt3L methyltransferase. Nucleic Acids Res 45: 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenburg S, Beltran AS, Swift-Scanlan T, Rivenbark AG, Rashwan R, Blancafort P. 2015. Stable oncogenic silencing in vivo by programmable and targeted de novo DNA methylation in breast cancer. Oncogene 34: 5427–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O. 2011. GNU parallel: the command-line power tool. The USENIX Mag 36: 42–47. [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31: 2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Beshnova DA, Vainshtein Y, Marth C, Mallm JP, Höfer T, Rippe K. 2014. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res 24: 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. 2016. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 44: 5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22: 1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong T, Meister GE, Workman RE, Kato NC, Spellberg MJ, Turker F, Timp W, Ostermeier M, Novina CD. 2017. Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci Rep 7: 6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356: eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.