Abstract
Molecular maps of the human brain alone do not inform us of the features unique to humans. Yet, the identification of these features is important for understanding both the evolution and nature of human cognition. Here, we approached this question by analyzing gene expression and H3K27ac chromatin modification data collected in eight brain regions of humans, chimpanzees, gorillas, a gibbon, and macaques. An analysis of spatial transcriptome trajectories across eight brain regions in four primate species revealed 1851 genes showing human-specific transcriptome differences in one or multiple brain regions, in contrast to 240 chimpanzee-specific differences. More than half of these human-specific differences represented elevated expression of genes enriched in neuronal and astrocytic markers in the human hippocampus, whereas the rest were enriched in microglial markers and displayed human-specific expression in several frontal cortical regions and the cerebellum. An analysis of the predicted regulatory interactions driving these differences revealed the role of transcription factors in species-specific transcriptome changes, and epigenetic modifications were linked to spatial expression differences conserved across species.
Changes in gene expression were long thought to underlie the rapid evolution of phenotypic traits, including the evolution of human cognition. To date, a number of studies have investigated this topic and have reported scores of genes showing expression unique to the human brain in comparison to closely related living primate relatives: apes and old-world monkeys (Enard et al. 2002; Cáceres et al. 2003; Brawand et al. 2011; Konopka et al. 2012). Early studies conducted in up to seven brain regions using microarray suggested the complex picture of gene expression evolution (Cáceres et al. 2003; Khaitovich et al. 2004; Oldham et al. 2006). Subsequent studies, however, mainly focused on the investigation of temporal gene expression changes during human, ape, and macaque brain development in one or several brain regions (Somel et al. 2009, 2010; Liu et al. 2012; Bakken et al. 2016).
In parallel, several studies investigated the regulatory mechanisms driving human-specific changes in gene expression. Analyses of transcription factors (TFs) revealed predicted TF regulatory networks and individual TFs driving gene expression differences between human and chimpanzee brains (Nowick et al. 2009; Liu et al. 2012). Analyses of epigenetic modifications identified promoters and enhancers potentially involved in human corticogenesis (Reilly et al. 2015), as well as active human-specific cis-regulatory elements in adult brains (Vermunt et al. 2016).
Although no further steps have been taken toward the understanding of human-specific expression across multiple brain regions using comparisons to other primates, several studies have focused on the human brain itself (Roth et al. 2006; Oldham et al. 2008; Johnson et al. 2009; Kang et al. 2011; Hawrylycz et al. 2012). The analyses conducted across as many as 170 brain regions and subregions (Hawrylycz et al. 2012), 16 neuronal subtypes (Lake et al. 2016), and 466 individual cells (Darmanis et al. 2015) revealed the complex transcriptional landscape. Hence, integrated analyses of multiple brain regions in the context of human evolution may uncover alterations in spatial profiles of expression levels, revealing changes that collectively contribute to human brain functionality.
Here, we determined uniquely human transcriptome features based on gene expression profiles collected across eight anatomically and functionally distinct brain regions of humans, chimpanzees, gorillas, and a gibbon. We further utilized published epigenome data (Vermunt et al. 2016) collected in eight brain regions of humans, chimpanzees, and macaques to assess and compare putative regulatory mechanisms that drive gene expression differences among species.
Results
Data description
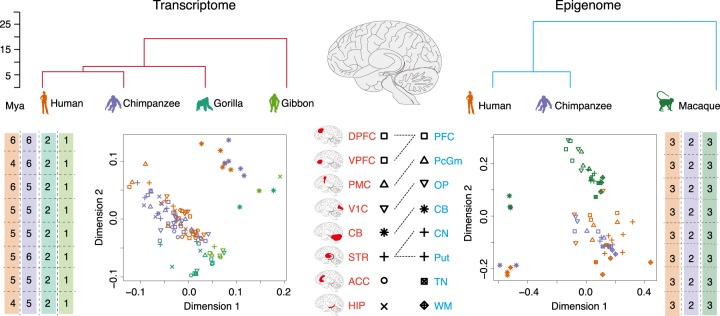
We assessed gene expression differences among humans, chimpanzees, gorillas and gibbons by measuring the poly(A)+ RNA fraction in eight brain regions of each species (Fig. 1). In each brain region, we measured RNA expression in four to six humans, five to six chimpanzees, two gorillas and one gibbon. The brain regions included five neocortical areas, as well as parts of the hippocampus (HIP), striatum (STR) and cerebellum (CB) (Fig. 1; Supplemental Tables S1, S2). The neocortical areas included dorsolateral prefrontal cortex (DPFC), ventrolateral prefrontal cortex (VPFC), premotor cortex (PMC), anterior cingulate cortex (ACC), and primary visual cortex (V1C). The paired-end sequencing of these samples on the Illumina platform (RNA-seq) resulted in 36 million read pairs per sample on average (Supplemental Table S3), yielding a total of 27,991 expressed genes.
Figure 1.
Primate brain transcriptome and epigenome data sets. (Top) The trees show phylogenetic relationship among species for transcriptome (red) and epigenome (blue) data sets. The silhouette figure colors mark species identity throughout the figure. (Mya) Million years ago. (Bottom center) Locations of brain regions, corresponding symbols, and labels included in the transcriptome (red) and epigenome (blue: [PFC] prefrontal cortex; [PcGm] precentral gyrus; [OP] occipital pole; [CN] caudate nucleus; [Put] putamen; [CB] cerebellum; [WM] white matter; [TN] thalamic nuclei) data sets. The dashed lines show brain regions shared between the data sets. (Bottom, sides) The numbers of biological replicates for each of the brain regions and plots showing two dimensions resulting from the multidimensional scaling (MDS) analyses of the transcriptome (left) and epigenome (right) data. The colors and shapes of the symbols on the MDS plots indicate the species and brain region identities of the samples.
To investigate regulatory mechanisms associated with gene expression level differences among species, we further reanalyzed published H3K27ac chromatin immunoprecipitation sequencing data (ChIP-seq) collected in eight brain regions of three humans, two chimpanzees, and three rhesus monkeys (Vermunt et al. 2016). Six of the eight regions overlapped with brain regions used for the RNA-seq measurements (Fig. 1). The reanalysis of the ChIP-seq data yielded 51,283 genomic regions that were significantly enriched in H3K27ac modification signal, termed predicted cis-regulatory elements (CREs) (Supplemental Table S4).
The multidimensional scaling (MDS) and t-distributed stochastic neighbor embedding (t-SNE) (van der Maaten and Hinton 2008) performed based on gene or CRE coverage showed a difference between the cerebellum and the other brain regions, as well as separation or a separation trend among species (Fig. 1; Supplemental Figs. S1A,B, S2).
Human–chimpanzee divergence across brain regions
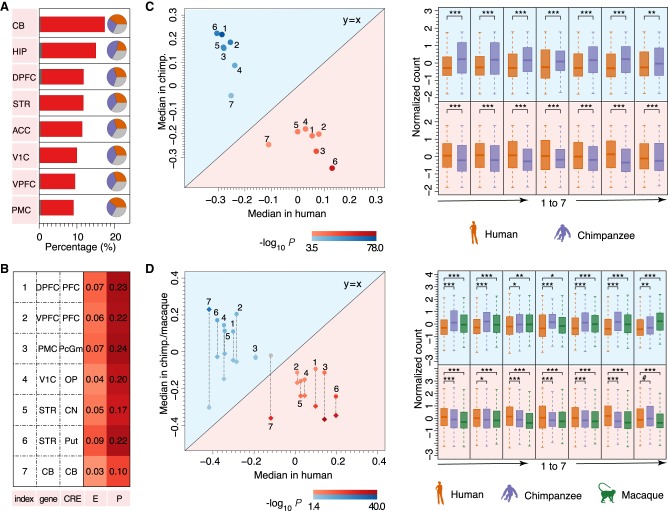
A comparison between humans and chimpanzees revealed substantial differences in gene expression and CRE coverage: On average, 12% of genes and 8% of CREs differed significantly between species, with more differences found in subcortical regions compared to the neocortex (Benjamini and Hochberg [BH] FDR-corrected P < 0.05, fold change >2) (Fig. 2A; Supplemental Fig. S1C). This extent of differential expression was not affected by PCR duplicates generated during the RNA-seq procedure (Supplemental Fig. S3). Similarly, the confounding variables, such as the samples’ RNA quality and experimental batch, as well as the sex, age, and postmortem interval of each individual, did not contribute substantially to gene expression differences between humans and chimpanzees (Supplemental Figs. S4–S8).
Figure 2.
Gene expression differences between humans and chimpanzees and their relationship with epigenetic differences. (A, left) The percentage of genes that show significant gene expression differences between humans and chimpanzees (red bars) and expected by chance (gray bars); (right) pie charts showing the percentages of human-specific (orange), chimpanzee-specific (purple), and unassigned (gray) gene expression differences sorted using combined gorilla and gibbon outgroup information. (B) The percentages of gene expression variance between humans and chimpanzees explained by promoter CREs (P) and enhancer CREs (E) coverage differences in each brain region shared between transcriptome (gene) and epigenome (CRE) data sets. (C) Normalized H3K27ac read counts for CREs located next to genes differentially expressed between humans and chimpanzees: (Left) The color of the dots shows the direction of the expression difference: (red) higher in humans; (blue) lower in humans. (Right) The box plots show distributions of CRE coverage in humans (orange) and chimpanzees (purple) for the corresponding dots. The significance of the coverage differences between humans and chimpanzees in a one-sided Wilcoxon test was shown by each dot's color gradient (left) and the asterisks (right): (***) P < 0.0005; (**) P < 0.005. The numbers indicate brain regions in accordance with the index shown in B. (D) Normalized H3K27ac read counts for CREs located next to genes showing human-specific gene expression changes. (Left) The color of the dots shows the direction of the expression difference, as in C. Each dot's shape represents nonhuman species: (circle) chimpanzees; (diamond) macaques. (Right) The box plots show the distributions of CRE coverage in humans (orange), chimpanzees (purple), and macaques (green) for the corresponding dots. The significance of the coverage differences between humans and nonhuman primates in a one-sided Wilcoxon test was shown by each dot's color gradient (left) and the asterisks (right): (***) P < 0.0005; (**) P < 0.005; (*) P < 0.05; (#) P > 0.05 as the gray dot shows. The numbers indicate brain regions in accordance with the index shown in B.
Using gorilla and gibbon transcriptome data or macaque epigenetic data as outgroup information, we assigned differences in gene expression and CRE coverage to the human and chimpanzee evolutionary lineages. Overall, the percentages of genes and CREs assigned to the two lineages were balanced across brain regions (Fig. 2A; Supplemental Fig. S1C). This result was robust at different lineage assignment thresholds and remained stable using only gorilla or gibbon data as outgroup information (Supplemental Fig. S1D).
We then tested the relationship between gene expression and epigenetic differences between humans and chimpanzees in each brain region by coupling CREs to the nearest gene. This resulted in 8120 putative promoter CREs located within 1000 bp (1 kb) from the genes’ transcription start site (TSS) and 43,163 putative enhancer CREs located beyond this distance. We found a significant positive correlation between epigenetic and gene expression differences between species: In linear regression analysis, the promoter and enhancer CREs explained on average 20% and 6% of gene expression differences, respectively (permutations, P < 0.001) (Fig. 2B). Correspondingly, CREs located next to genes differentially expressed between humans and chimpanzees or genes showing species-specific expression had significantly more corresponding coverage differences (hypergeometric test, BH-corrected P < 0.05) (Supplemental Tables S5, S6). Furthermore, the direction of the differences was consistent with the known role of H3K27ac modification as a marker of chromatin regions associated with active transcription (Fig. 2C,D; Supplemental Fig. S1E; Supplemental Table S7; Wang et al. 2008).
Human-specific coexpression modules
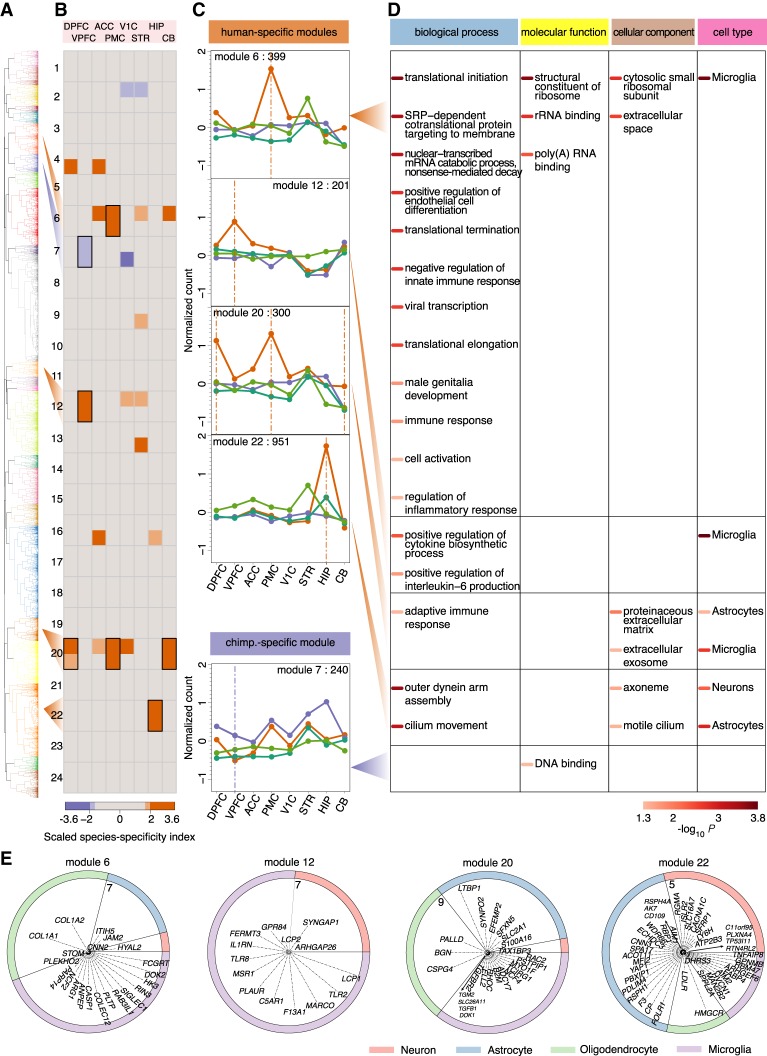
To assess the spatial distribution of human-specific gene expression differences among brain regions in a more comprehensive manner, we constructed a coexpression network based on 9726 genes showing significant expression differences between humans and chimpanzees in at least one brain region. Unsupervised clustering of their expression profiles yielded 24 coexpression modules, each characterized by its representative pattern (Fig. 3A; Supplemental Table S8; Supplemental Fig. S9). Most of these patterns were observed using alternative clustering procedures, such as k-means clustering or signed WGCNA, detected at different module definition cutoffs, and robust to jackknife resampling (Supplemental Fig. S10).
Figure 3.
Characteristics of species-specific coexpression modules. (A) Dendrogram based on unsupervised hierarchical clustering of expression profiles of 9726 genes with significant expression differences between humans and chimpanzees in at least one brain region. The colors indicate 24 identified modules. (B) The heat map shows scaled species-specificity indices calculated using gorilla (upper) and gibbon (lower) data for each module in each brain region: (top) each of the eight brain regions; (left) module labels. The colors indicate species-specificity based on Z-score thresholds corresponding to 5% and 10% cutoffs as indicated by the bar below: (orange) human-specific; (purple) chimpanzee-specific. The black rectangles indicate species-specific modules supported by both outgroup species. (C) Gene expression profiles of human-specific and chimpanzee-specific modules. The dots show normalized read counts averaged across genes within each module for respective species: (orange) humans; (purple) chimpanzees; (dark green) gorillas; (light green) gibbon. The label at the top of each panel shows the number of genes within the module. The vertical lines show brain regions showing significant species-specificity within the module. (D) The enrichment of genes in species-specific modules in three GO term categories and cell-type markers. The shade of the color bars beside the term and cell-type names indicates P-values of a hypergeometric test after BH correction, as shown by the bar below. (E) The distribution of cell-type marker genes within the four human-specific modules. Each ring represents one human-specific module, showing the relative proportions of neuronal, astrocytic, oligodendrocytic, and microglial marker genes within the module with expression human-specificity ratio greater than one, normalized to the overall number of marker genes for each cell type. The corresponding marker gene names are shown within the rings, with the distance between each gene and the center representing the log2-transformed human-specificity ratio. The numbers show the maximum of the log2-transformed human-specificity ratio range within the modules. The human-specificity ratio was calculated as the ratio of the absolute expression difference between humans and the outgroup species to the absolute difference between chimpanzees and the outgroup species.
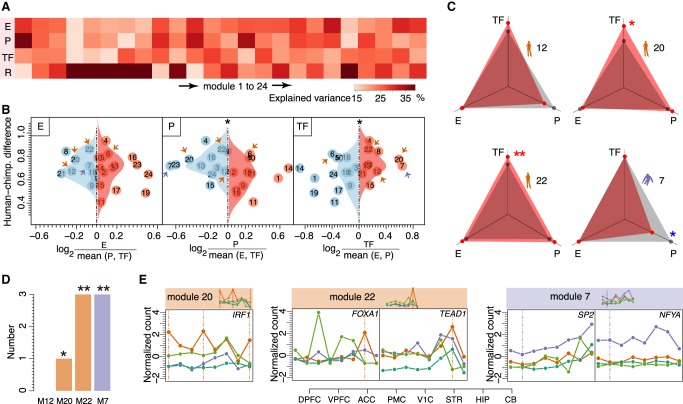
For each module, we calculated a species-specificity index defined as the ratio of the number of genes with human-specific expression to that with chimpanzee-specific expression in each brain region. Notably, despite an overall similarity of human-specific and chimpanzee-specific gene numbers in individual brain regions, human-specific expression differences clustered in particular coexpression modules far more frequently than the chimpanzee-specific expression differences. Using a 5% cutoff (|Z| > 1.96), seven modules were enriched in human-specific expression differences and only one in chimpanzee-specific differences (Fig. 3B). A more stringent criterion, requiring at least one brain region to be classified as species-specific using both gorilla and gibbon brain samples as outgroups, yielded four human-specific modules (modules 6, 12, 20, and 22) and one chimpanzee-specific module (module 7) (Fig. 3B,C).
In modules 6, 12, and 20, human-specific expression differences mainly localized in neocortical regions (PMC, VPFC, and DPFC) as well as CB for module 20 genes, whereas module 22 represented human-specific expression differences particular only to the hippocampus (Fig. 3C). An analysis based on 585 published cell-type marker genes contained in the modules (Sharma et al. 2015) showed the strong enrichment of modules 6, 12, and 20 in microglial markers (hypergeometric test, BH-corrected P-value <0.05) (Fig. 3D,E). In contrast, module 22 was enriched in neuronal and astrocytic markers (hypergeometric test, BH-corrected P-value <0.05) (Fig. 3D,E). The cell-type enrichment results were robust to the use of alternative cell-type marker sets (Supplemental Table S9; Supplemental Fig. S11; Darmanis et al. 2015; Zeisel et al. 2015; Zhang et al. 2016). Accordingly, genes in modules 6, 12, and 20 were enriched in Gene Ontology (GO) biological function terms associated with immune response, the primary task of microglia (Fig. 3D; Supplemental Table S10; Filiano et al. 2015). In contrast, module 22 genes were enriched in outer dynein arm assembly and cilium movement functions (Fig. 3D; Supplemental Table S10). This might be noteworthy, because motile cilia are located on ependymal cells that have been shown to be present in the mouse hippocampus (Zeisel et al. 2015).
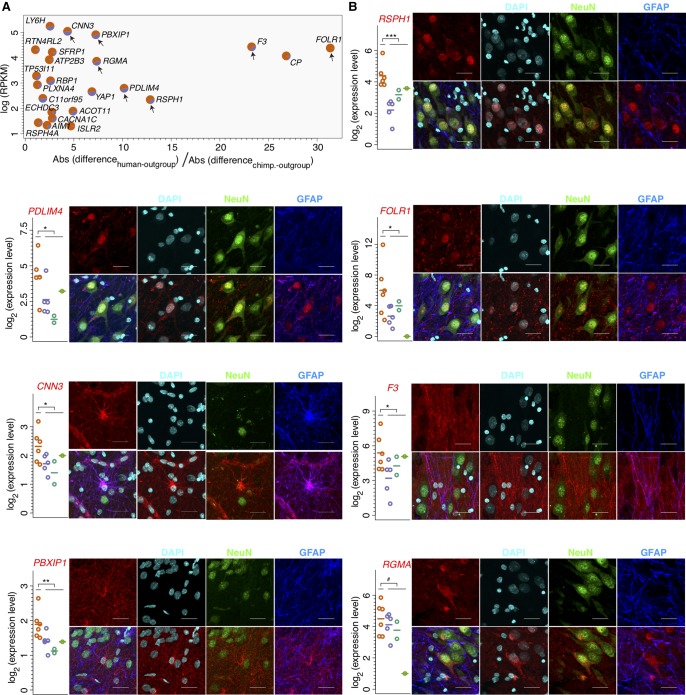
To test the authenticity of gene expression differences particular to the human hippocampus and to assess their histological localizations, we conducted quantitative PCR (RT-qPCR) and immunohistochemistry (IHC) experiments for a subset of module 22 genes. Among the 63 module 22 genes reported to be expressed in mouse neurons and astrocytes (Sharma et al. 2015), we selected seven genes highly expressed in the human hippocampus (average RPKM >50) and covering most of the human-specificity ratio range: RSPH1, PDLIM4, FOLR1, CNN3, F3, PBXIP1, and RGMA (Fig. 4A).
Figure 4.
Human-specificity of module 22 genes in the hippocampus. (A) Scatter plot showing expression levels and human-specificity ratio calculated using combined outgroup data for a subset of 23 module 22 genes selected based on expression and cell-type–specificity information (Methods). Each gene is represented by a pie chart showing relative specificity of expression to the hippocampus, compared to the average expression across the rest of the brain regions, in humans (orange) and chimpanzees (purple). The arrows indicate seven genes measured in RT-qPCR and IHC experiments. (B) The expression and cellular localization of the seven selected genes with human-specific expression in the hippocampus. Each panel shows expression levels measured using RT-qPCR (left) and cellular localization in the human hippocampus determined using IHC (right). (Left) The log2-transformed expression levels are shown by colored symbols. Each symbol represents an individual (n = 6 for humans, n = 5 for chimpanzees, n = 2 for gorillas, and n = 1 for gibbon). The colors represent species: (orange) humans; (purple) chimpanzees; (dark green) gorillas; (light green) gibbon. The horizontal lines show the mean expression across biological replicates. The asterisks indicate the significance of expression differences between humans and the other three primate species in a one-sided Wilcoxon test: (***) P < 0.0005; (**) P < 0.005; (*) P < 0.05; (#) P = 0.1. (Right) The IHC images show the signals from the investigated protein (red), the general nuclear marker DAPI (light blue), the neuronal marker NeuN (green), the astrocytic marker GFAP (blue), as well as merged images (scale bar, 25 µm). The RSPH1, PDLIM4, FOLR1, and RGMA images were taken in the CA3, CNN3, and F3 images—in the CA1, PBXIP1 images—in the dentate gyrus.
Six of the seven genes had significantly higher expression in the human hippocampus compared to chimpanzees, gorillas, and gibbon in RT-qPCR experiments, consistent with RNA-seq measurements (one-sided Wilcoxon rank-sum test, P < 0.05) (Fig. 4B). The remaining gene, RGMA, showed the same expression trend (one-sided Wilcoxon rank-sum test, P = 0.1). The RT-qPCR result, as well as higher expression of tested genes in the human hippocampus in the original RNA-seq data, was not caused by a particular human sample (Supplemental Figs. S12, S13).
IHC experiments showed that RSPH1, PDLIM4, FOLR1, and RGMA proteins localize in neurons, and CNN3 in astrocytes (Fig. 4B). PBXIP1 and F3 staining revealed morphology compatible with astrocyte projections, and F3 signal partially colocalized with GFAP staining marking the majority of astrocytes and their projections. Notably, the localization of CNN3, PBXIP1, and F3 in astrocytes and their projections was reported in both mice and humans (Darmanis et al. 2015; Sharma et al. 2015; Zhang et al. 2016), whereas the localization of RSPH1 and RGMA in neurons was reported in humans (Zhang et al. 2016) and mice (Sharma et al. 2015), respectively (Supplemental Table S9).
Regulation of coexpression modules
A combination of gene expression and H3K27ac modification data allows us to assess the influence of various regulatory mechanisms on the species-specific alterations of spatial expression profiles represented by the 24 gene modules. Specifically, we assessed the influence of (1) H3K27ac modification levels at promoter regions, (2) H3K27ac modification levels at putative enhancer regions, and (3) expression levels of TFs with binding sites present in promoter regions identified based on TF binding profiles from the JASPAR CORE database (Sandelin et al. 2004), on the spatial profile of each gene in each of the 24 modules (Supplemental Table S11).
Each regulatory mechanism explained 12%–38% of the total expression variation across brain regions within the modules, with a mean of 24% for epigenetic modifications and 22% for TFs (Fig. 5A). Notably, modules predominantly regulated by TFs showed more spatial expression differences between humans and chimpanzees, whereas modules predominantly regulated by promoter CREs were more conserved (one-sided Wilcoxon rank-sum test, P < 0.05) (Fig. 5B). The greater extent of spatial expression differences for modules preferentially regulated by TFs was further reproduced using TFs with binding sites located within distal enhancer CREs (Supplemental Fig. S14). The modules predicted to be regulated by enhancer CREs showed intermediate conservation. Consistently, four of the five species-specific modules (modules 7, 12, 20, and 22) showed a greater predicted regulatory impact of TFs and a lesser impact of promoter CREs (Fig. 5B,C). Furthermore, two of these modules (modules 7 and 20) were predominantly regulated both by TFs with binding sites located within proximal promoter regions and by TFs with binding sites located within distal enhancer CREs (Fig. 5B; Supplemental Fig. S14).
Figure 5.
Putative spatial expression regulators. (A) The mean percentage of gene expression variation explained by epigenetic modification levels of enhancer CREs (E), promoter CREs (P), TF expression levels for TFs identified using binding sites present in proximal promoter regions (TF), and the residual variance (R) for each module. (B) Influence of the three regulatory mechanisms on spatial gene expression differences between humans and chimpanzees. The dots represent modules. The horizontal axis shows the relative influence of a regulatory mechanism in each module. The vertical axis and the distributions show human–chimpanzee expression differences for modules with the major (red) and minor (blue) contribution of a given regulatory mechanism. The asterisks indicate the significance of the difference between two distributions (one-sided Wilcoxon test, here and further: [*] P < 0.05; [**] P < 0.005). The arrows mark species-specific modules: (orange) human; (purple) chimpanzee. (C) The influence of the three regulatory mechanisms on spatial expression in species-specific modules. Red dots denote the normalized median of absolute Pearson correlation coefficients based on spatial profiles of genes and corresponding regulators. The gray dots denote values expected by chance calculated by randomly assigning regulators to genes (Methods). The asterisks represent the significance of the difference between observed and expected values (one-sided Wilcoxon test: [red] greater; [blue] smaller). (D) Numbers of TFs showing correlated expression with targets in species-specific modules. The asterisks indicate the probabilities of finding the observed or greater number of correlated TFs by chance (1000 permutations of module labels). (E) Spatial expression patterns of five TFs (IRF1, FOXA1, TEAD1, SP2, and NFYA) predicted to regulate target genes in modules 20, 22, and 7, respectively: (orange) human; (purple) chimpanzee; (dark green) gorilla; (light green) gibbon. The module expression patterns are shown above the panels. The vertical lines indicate brain regions responsible for species-specificity of the modules.
The expression of seven TFs significantly positively or negatively correlated with the expression of their predicted target genes within modules: one TF (IRF1) in module 20, three TFs (FOXA1, TEAD1, PRDM1) in module 22, and three TFs (SP2, NFYA, PAX5) in module 7 (one-sided Wilcoxon rank-sum test, BH-corrected P < 0.05, Pearson correlation coefficient, |r| > 0.6) (Fig. 5D,E; Supplemental Fig. S15).
The previously reported functions of these TFs matched their roles in the regulation of spatial expression in brains. FOXA1 has been reported to play a role in the development and maintenance of the dopaminergic neuron system affected in Alzheimer's (Wruck et al. 2016) and Parkinson's diseases (Domanskyi et al. 2014). Mutations in the TEAD1 gene were shown to cause Aicardi syndrome, which is characterized by severe developmental brain defects (Schrauwen et al. 2015). The expression of PRDM1, previously characterized as a transcriptional repressor (Keller and Maniatis 1991), negatively correlated with the expression of its predicted targets in module 22. Moreover, in the brain, PRDM1 inhibition was linked to gliomas’ progression (Wang et al. 2013). Another gene IRF1, correlating positively with the expression of its predicted targets in module 20, was shown to be involved in immune response and reactive microglia activation (Masuda et al. 2015; Langlais et al. 2016), which matches functional enrichment results for this module.
Discussion
Different regions are tightly integrated within anatomical and functional architecture of the brain. In this study, we took advantage of this phenomenon to identify uniquely human gene expression features representing eight different brain regions.
The identification of lineage-specific gene expression changes is based on a comparison to closely related species, analogously to a DNA sequence–level analysis. Unlike DNA sequence, gene expression represents a continuous spectrum and may evolve rapidly on a given lineage (Pollard et al. 2006). Accordingly, comparisons to distant species could lead to incorrect lineage placement due to multiple gene expression changes. Measurements of gene expression in the brains of humans, chimpanzees, orangutans, and old-world monkeys showed that although expression in the brain is more conserved than in the other tissues, the differences still accumulate rapidly (Khaitovich et al. 2006). Thus, the use of two outgroup species proximal to humans and chimpanzees, gorillas and gibbons, enables us to get a more reliable placement of expression differences to the evolutionary lineages in our study.
An analysis based on individual brain regions showed that the percentages of gene expression changes assigned to the human and the chimpanzee lineages deviated little from the 50:50 ratio. This balanced ratio reflects the equal length of the two lineages after the separation from the most recent common ancestor. In contrast, an analysis based on expression profiles across brain regions revealed eight human-specific coexpression modules (four of them supported by both outgroup species) and one chimpanzee-specific module. The four human-specific modules representing expression changes in neocortical areas, cerebellum, and hippocampus contained 1851 genes, cumulatively. The chimpanzee-specific module contained 240 genes. Thus, the differences in the shape of gene expression trajectories across brain regions were heavily biased toward the human lineage.
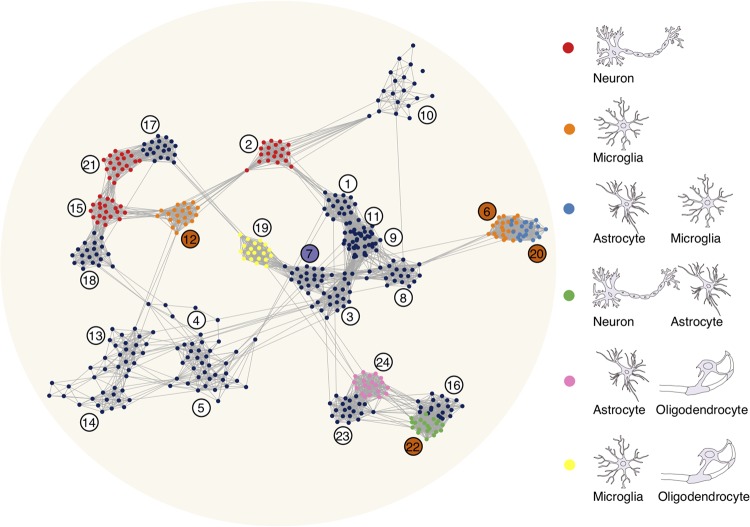
More than half of all human-specific gene expression profiles formed a single module, module 22, characterized by elevated expression in the human hippocampus. Furthermore, an analysis of the coexpression network revealed an additional module (module 16) closely linked to module 22 and showing human-specific hippocampal overexpression of a smaller magnitude (Fig. 6; Supplemental Fig. S9). This result is notable, because the hippocampus is central to learning and memory functions believed to be particularly enhanced in humans (Premack 2007), and aligns with the reported acceleration of transcriptome evolution and an increase in transcriptional connectivity in the human hippocampus compared to other primates (Konopka et al. 2012). A functional analysis of genes in modules 16 and 22, as well as in module 22 alone, revealed enrichment in neuronal and astrocytic markers, as well as functions related to axoneme and cilium movements (Fig. 3D; Supplemental Fig. S16), indicating the complex character of human-specific changes. Previous studies indicated the particularity and interconnected functionality of hippocampal neurons and astrocytes, suggesting the possibility of region-specific alterations involving the two cell types. Both neurons and astrocytes, as well as their interactions, were implicated in learning and memory processes localized in the hippocampus (Henneberger et al. 2010; Suzuki et al. 2011; Ota et al. 2013; Hassanpoor et al. 2014; Tadi et al. 2015; Gao et al. 2016; Pabst et al. 2016). Astrocytes were further shown to contribute to the integration of adult-born neurons in the hippocampus, although adult hippocampal neurogenesis in humans remains controversial (Krzisch et al. 2015; Sultan et al. 2015; Sorrells et al. 2018). Moreover, the involvement of hippocampal astrocytes and astrocyte–neuron interactions in Huntington's disease, major depressive disorder, and schizophrenia have been reported (Kolomeets and Uranova 2010; Cobb et al. 2016; L'Episcopo et al. 2016).
Figure 6.
Visualization of the 24 modules in the coexpression network. The nodes represent genes. For each module, the 20 genes best correlated within a module are drawn. The edges represent Pearson correlation coefficients >0.8. The colors of the nodes denote significant enrichment of genes from the corresponding module in cell-type markers, as illustrated by the graphical legend on the right. The colors of the module labels indicate species-specific modules: human (orange); chimpanzee (purple).
The remaining three modules containing human-specific changes involving neocortical areas of the frontal lobe (DPFC, VPFC, and PMC), as well as cerebellar cortex (CB), formed two clusters within the coexpression network, but showed similar functionality (Fig. 6). This shared functionality included microglia as the main cell type, extracellular matrix as the main cellular component, and immune response as the main biological process. Microglia, constituting 5%–12% of all cells in the mouse and human brain (Lawson et al. 1990; Lyck et al. 2009; Olah et al. 2018) and primarily known for their role in immune response, have recently been implicated in multiple brain functions including the regulation of learning-related synapse formation, synaptic plasticity, and cognition (Parkhurst et al. 2013; Hong et al. 2016; Tay et al. 2017) and are linked with neurodegenerative and psychiatric disorders (Wolf et al. 2017). Furthermore, microglial cells were shown to have different transcriptional identities in different brain regions (Grabert et al. 2016). Although microglial expression has been shown to be affected by environmental transitions (Gosselin et al. 2017), there was no overlap between these reported changes and the human-specific microglial expression features detected in our study (hypergeometric test, P-value = 0.702) (Supplemental Fig. S17). Taken together, these observations indicate the possibility of regional changes in microglial expression potentially linked to the evolution of human cognition.
Our analysis of regulatory mechanisms that drive evolutionary changes of spatial gene expression patterns revealed a clear and significant trend. We show that between TFs and H3K27ac epigenetic modifications, which are associated with active promoters and enhancers, TFs represent the main driving force underlying recent evolutionary changes. This observation matches the studies linking particular TFs with rapidly evolving human-specific expression features (Gilad et al. 2006; Blekhman et al. 2008; Nowick et al. 2009; Liu et al. 2012). Similarly, we identified the TFs potentially responsible for some of the detected human-specific expression differences, including FOXA1, TEAD1, and PRDM1 for module 22 genes and IRF1 for module 20 genes. In contrast, we found that module genes predominantly controlled by epigenetic modifications associated with active promoters were the most conserved in their expression among species. The regulation by enhancer epigenetic modifications resulted in the intermediate expression conservation.
Taken together, our results show that an investigation of the spatial profiles of expression levels across multiple brain regions provides novel insights into the evolution of expression features unique to the human brain transcriptome. Although functional interpretation of these results is limited by the absence of experimental transcriptome manipulation and perturbation tools, our study provides important insights into the possible nature of the molecular mechanisms underlying human cognitive evolution. Specifically, our study singles out the potential roles of neurons and astrocytes in the human hippocampus, as well as the diverse functions of microglia in the frontal cortex and cerebellum, as potential focal points for further research of the molecular mechanisms underlying the particularity of the human brain.
Methods
Sample preparations
The autopsied brains of eight Japanese patients (six males and two females, aged 51–76 years) with no neuropsychiatric disorders were obtained from the Brain Research Institute, Niigata University, Japan. Written informed consent for autopsy, collection of samples, and subsequent use for research purposes was obtained from the next of kin of the deceased. A total RNA from the hippocampus was purchased from Clontech Laboratories Inc. (Catalog no. 636593). The use of postmortem human brain tissues was authorized by the Niigata University Medical Ethics Committee (No. 468), the National Institutes of Natural Sciences Bioethics Committee (No. 24-02, 25-03, 25-04), and the Kyoto University Medical Ethics Committee (No. G421). All ape brain samples were obtained from institutes or zoos after their natural deaths under the collaborative research project of the Great Ape Information Network (GAIN) (https://shigen.nig.ac.jp/gain/). These included six chimpanzees: Genki (ID: 0135), Iyo (ID: 0305), Kenny (ID: 0363), Mayumi (ID: 0459), Reiko (ID: 0432), Yoshio (ID: 0461); two gorillas: Gon (ID: 0056), Sakura (ID: 0065); and a gibbon Daichi (ID: 0039).
Brain dissection was performed from either fresh specimens or frozen tissue slabs by the same person to ensure sampling consistency between specimens. The areas dissected for each brain region included Brodmann areas (BA) 9/46 for DPFC, BA44/45 for VPFC, BA6 for PMC, BA17 for V1C, BA24 for ACC, caudate and putamen for STR, CA1/2/3 and dentate gyrus for the hippocampus, and posterior lobe for CB. About 100 mg of dissected tissues were used from each individual. Total RNA was extracted using RNeasy Plus Universal Kit (Qiagen). Quality and quantity measurements of extracted RNA were performed using NanoDrop (Thermo Fisher Scientific) and Qubit Fluorometer (Thermo Fisher Scientific), respectively, and the RNA Integrity Numbers (RIN) were determined using Bioanalyzer RNA 6000 Nano Kit (Agilent).
RNA-sequencing
Sequencing libraries were prepared using the NEBNext mRNA Library Prep Kit for nondirectional libraries and the NEBNext Ultra Directional RNA Library Prep Kit for directional libraries (New England BioLabs) according to the manufacturer's instructions. The RNA-seq libraries were each sequenced (101 cycles) using the Illumina HiSeq 1500, HiSeq 2000, and HiSeq 4000 platforms.
Gene expression quantification
Raw reads of four species were aligned to the reference genomes (human, hg19; chimpanzee, panTro4; gorilla, gorGor3; gibbon, nomLeu1) by “STAR” (Dobin et al. 2013) (parameters: outFilterMultimapNmax = 1, outFilterMatchNminOverLread = 0.95, outFilterMismatchNoverLmax = 0.05, outFilterIntronMotifs = RemoveNoncanonical). Due to the fact that the human hg38 reference genome was not available at the start of the project, the hg19 genome version was used here. PCR duplicate removal was then conducted using SAMtools (Li et al. 2009). Only uniquely mapped reads were considered as valid to avoid estimation ambiguity. We further conducted a parallel analysis skipping the removal of PCR duplicates and obtained consistent results (Supplemental Fig. S3).
To locate read abundance on gene level, we obtained chimpanzee, gorilla, and gibbon orthologs of human genes (GENCODE v19) by reciprocal liftOver, requiring the same exon orders and no less than 50% of exons preserved. Based on these annotations, numbers of reads were counted per gene per sample using htseq-count within HTSeq (Anders et al. 2015). Only genes with read counts >0 in all four species in at least one brain region were used in the following analysis.
We took advantage of the median ratio method implemented in DESeq2 (Love et al. 2014) to normalize the gene abundance to get normalized read count for further comparisons across species and brain regions. We further calculated the reads per kilobase per million mapped reads (RPKM) as the expression level of each gene for comparable expression levels among different genes in some explorations.
CREs’ H3K27ac coverage quantification
Primary CREs were directly obtained from published data (Vermunt et al. 2016) (Gene Expression Omnibus accession no. GSE67978). Unlike published work, our study mainly focused on a comparison between humans and chimpanzees. Thus we made some changes with respect to CRE ortholog detection in the referred paper described as follows: Candidate CREs were obtained by merging the primary CREs of humans and chimpanzees, and then reciprocal liftOver was utilized to get corresponding regions in chimpanzees and rhesus monkeys from humans based on these candidate CREs, which require <50% changes in length.
We used Bowtie (Langmead et al. 2009) to map raw reads to reference genomes (human, hg38; chimpanzee, panTro4; rhesus monkeys, rheMac3) with the same parameters as the referred paper. The integrated analysis of transcriptome and epigenome data in humans was based on Ensembl ID matching between the hg19 and hg38 versions of the human genome. Mapping RNA-seq reads to each of the versions resulted in nearly identical gene expression level estimates (Supplemental Fig. S18). Other preprocessing was similar with those in the gene level.
Global pattern exploration
Two means were included for detecting the overall patterns of gene expression or CRE coverage across species and brain regions—MDS and t-SNE. MDS was performed on the basis of sample dissimilarities, defined as one minus Pearson correlation coefficient between pairwise samples using the “cmdscale” function in R (R Core Team 2016). t-SNE was conducted in a similar way by the “tsne” package in R. To assess the species’ and brain region identity of the gibbon hippocampus sample, we examined the third and the fourth dimensions of the MDS analysis, as well as the correlations between gene expression in the gibbon hippocampus sample and expression in each of gorilla brain regions calculated using genes showing hippocampus-specific expression. Genes showing hippocampus-specific expression were defined as genes with at least twofold higher expression in the hippocampus compared to the other brain regions in human and chimpanzee data.
Detection of differences between humans and chimpanzees
Genes/CREs showing differential expression/coverage between humans and chimpanzees in each brain region were identified using the Bioconductor package “DESeq2” (Love et al. 2014) in R. Differentially expressed genes or differential coverage CREs were determined by the criteria of false discovery rate (FDR) <0.05 and fold change >2. To find out whether confounding factors such as individual sexes, RIN, experimental batch, age, and postmortem interval influenced our analysis, we compared the fold changes calculated between humans and chimpanzees based on all human and chimpanzee samples and those based on the samples within the same condition in each brain region. The number of genes/CREs showing differential expression/coverage expected by chance was estimated by shuffling the human and chimpanzee sample labels 100 times for genes or exhaustive permutations for CREs.
Lineage assignment of differences between humans and chimpanzees
We assigned significant differences in gene expression or CRE coverage between humans and chimpanzees in each brain region to human or chimpanzee evolutionary lineage based on evolutionary outgroups. Specifically, genes/CREs with absolute differences between humans and outgroups more than a certain fold (here 2, 2.5, and 3) as great as those between chimpanzees and outgroups were determined as human-specific genes/CREs and vice versa for chimpanzee-specific genes/CREs. For differentially expressed genes, lineage assignment was conducted based on outgroups of gorillas, a gibbon, and the combination of the two. For differential coverage CREs, rhesus monkeys were used as the outgroup information. Figure 2A and Supplemental Figure S1C show the lineage assignment results under the threshold of 2.5-fold change.
Relationship between transcriptome and epigenome regarding human–chimpanzee differences
The classification of CREs into promoter CREs and enhancer CREs resembled the method mentioned in Vermunt et al. (2016), that is, every CRE was coupled to its closest gene based on transcription start site (TSS) of each gene. A distance cutoff of 1000 bp (1 kb) finally resulted in 8120 promoter CREs located within 1 kb from TSS and 43,163 enhancer CREs located beyond this distance.
The enrichment of CREs with the nearest gene differentially expressed between humans and chimpanzees or showing species-specific expression in CREs with differential coverage between species or species-specific CREs was evaluated by a hypergeometric test with a statistical background of CREs coupled to all expressed genes or all differentially expressed genes between humans and chimpanzees. The enrichment with a BH-corrected P-value of <0.05 was considered to be significant. Moreover, H3K27ac coverage dissimilarity across species for CREs located next to differentially expressed genes between humans and chimpanzees or species-specific genes was assessed using a one-sided Wilcoxon rank-sum test.
The association between human–chimpanzee gene expression differences and CREs’ H3K27ac coverage differences in individual brain regions was assessed using a linear regression analysis of log2 fold changes between humans and chimpanzees of differentially expressed genes and coupled promoter or enhancer CREs. Permutations were done via random coupling between genes and CREs 1000 times to estimate the significance of these associations.
Coexpression module construction
Genes’ spatial profiles across brain regions were obtained by averaging the biological replicates. We then used the unsupervised hierarchical clustering (function “hclust” in R) to detect coexpression modules based on 9726 genes differentially expressed between humans and chimpanzees in at least one brain region. Dissimilarity between pairwise genes was defined as one minus Pearson correlation coefficient calculated between the spatial profiles of the genes in humans. To test the reproducibility of the detected 24 coexpression modules, we also used cutoffs resulting in 16 and 36 modules and determined gene set overlap using hypergeometric test implemented in “overlapTable” function in the Bioconductor package “WGCNA” (Langfelder and Horvath 2008) in R.
Other methods for detecting coexpression modules, like k-means clustering and weighted gene coexpression network analysis (WGCNA), were meanwhile performed. Twenty-seven modules were derived from WGCNA (tag-value pairs: corType = “bicor”, power = 19, networkType = “signed”, TOMType = “unsigned”, minModuleSize = 100, deepSplit = 4, reassignThreshold = 0, pamRespectsDendro = F, mergeCutHeight = 0.1) and 24 were detected by k-means. Gene set overlap between modules detected by hierarchical clustering and those by k-means and WGCNA was assessed in the same manner described above.
To further check the influence of particular samples on module detection, we performed a fivefold jackknife procedure: Samples were randomly separated into five equal-sized groups and module construction was repeated by leaving one group out every cycle using hierarchical clustering. Robustness was assessed in the same manner described above.
Species-specificity of coexpression modules and functionality enrichment analysis
On the basis of species-specificity of each gene in each brain region defined above in Methods, we evaluated the species-specificity index, defined as the ratio of the number of genes with human-specific differences to that with chimpanzee-specific differences, for each brain region in each of the 24 identified modules. The species-specificity indices across all 24 modules per brain region were standardized to assess the relative species-specificity of each module compared to other modules. We used a Z-score of ±1.64 (two-sided P-value = 0.1) and ±1.96 (two-sided P-value = 0.05) as species-specificity cutoffs to detect human- and chimpanzee-specific modules. Only modules with stable species-specificity regardless of different outgroups were considered as species-specific.
Gene Ontology (GO) terms enriched in genes within each module were uncovered by the Bioconductor package “topGO” (http://bioconductor.org/packages/release/bioc/html/topGO.html) using Fisher's exact test for GO enrichment. GO term categories including biological processes, molecular functions, and cellular components with P-values after BH correction of <0.05 were considered to be significantly enriched.
Cell-type marker genes from a previously published paper (Sharma et al. 2015) were used to determine the enriched cell-type markers in genes of each module by means of a hypergeometric test. The BH-corrected hypergeometric test P-value of <0.05 was used as the judging boundary of enrichment. At the same time, three additional cell-type data sets were analyzed to confirm our results (Darmanis et al. 2015; Zeisel et al. 2015; Zhang et al. 2016). We further assessed the relative contribution of each cell type to the module composition for each human-specific module. The relative contributions were calculated as the proportions of the marker genes for a specific cell type falling into a module normalized by the overall number of detected marker genes for this type. As expression in microglia was shown to be affected by the environmental transition (Gosselin et al. 2017), we intersected the microglial marker genes used to specify the three human-specific modules enriched in microglial cell types with reported environmental transition-sensitive genes and assessed the overlap significance using the function “phyper” in R. We further examined the expression level fold change after environmental transition for the corresponding microglial markers using data from Gosselin et al. (2017).
RT-qPCR validation
Figure 4A illustrated a subset of 23 module 22 genes with expression levels (RPKM) in the human hippocampus of >3, specificity of expression in humans defined as the ratio of absolute difference between humans and combined gorilla and gibbon outgroup to that between chimpanzees and combined outgroup >1, and expression in mouse neuron and astrocytes reported previously (Sharma et al. 2015). To confirm the observed human-specific differences of module 22 genes in the hippocampus, quantitative reverse transcription PCR (RT-qPCR) experiments were conducted for seven genes (RSPH1, PDLIM4, FOLR1, CNN3, F3, PBXIP1, RGMA) selected from the 23 genes, using a RNA-direct SYBR Green Realtime PCR Master Mix (TOYOBO), in accordance with the manufacturer's instructions. The same brain-derived RNAs used for the transcriptome analysis were also used for this experiment. For humans, two more individuals were used in addition to those in the RNA-seq measurements. The RT-qPCR primers were made in perfect conserved regions (100% identity) in the four species (Supplemental Table S12). The quantification of expression level for each gene was determined by comparative calculation using the internal marker GAPDH. To assess the influence of particular human samples on the RT-qPCR result, for each tested gene we removed the human sample with the highest expression level in the RT-qPCR data and recalculated the significance of expression differences between humans and nonhuman primates using a one-sided Wilcoxon rank-sum test.
Immunohistochemistry
Postmortem brain samples from six humans (males and females aged 45–59) were provided by city clinical hospital #12, Moscow, Russia. Written informed consent for autopsy, collection of samples, and subsequent use for research purposes was obtained from the next of kin of the deceased. The individuals died from natural causes, having no history of neurological or psychiatric illness. After brain extraction, each hippocampal region was dissected from the left hemisphere, sliced into 10-mm-thick sections, and immediately placed in freshly prepared 4% paraformaldehyde solution for 5 d at 4°C, then rinsed several times in 0.01M PBS pH 7.4 containing 0.01% sodium azide and stored in the latter solution at 4°C until further use. The postmortem interval was no more than 12 h. Fixed brain samples were cut on Leica VT1200S vibratome in 30-µm-thick serial sections and processed for multiple fluorescent immunohistochemistry, as detailed below. All staining steps were performed in 24-well cell culture plates using individual inserts with permeable bottoms.
For immunofluorescence reaction, the free-floating sections were subjected to heat-induced epitope retrieval with antigen retrieval acidic reagent (R&D CTS014), then washed in phosphate-buffered saline 0.01M PBS pH 7.4 three times for 10 min each. Then sections were permeabilized for 1 h at room temperature in 1% PBST (0.01M PBS with 0.5% Triton X-100 detergent) with 5% normal horse serum (NHS) and 5% normal donkey serum (NDS) and washed again. Reactions with mixture of primary antibodies (1:250–1:1000) (Supplemental Table S13), typically for one human-specific antigen with two markers (glial and neuronal), were performed in the blocking buffer (5% NHS, 5%NDS in 0.5% PBST) overnight at 4°C. Following washing in 0.5% PBST three times for 10 min each and incubation with biotinylated horse anti-rabbit immunoglobulin IgG (1:250) (Supplemental Table S14), corresponding to human-specific antigen antibody, in the blocking buffer for 2 h at room temperature, washed sections were processed with a mixture of donkey anti-mouse-AlexaFluor488 and donkey anti-chicken-AlexaFluor647, corresponding to neuronal and glial marker antibodies, and streptavidin-AlexaFluor568 for detection of human-specific antigens, 2 h at room temperature (1:500) (Supplemental Table S14). After washing, to block lipofuscin autofluorescence, sections were incubated in 1% sudan black B solution in 70% ethanol for 10 min each. Then sections were washed in PBS, attached to the glass slides, mounted with Fluoromount aqueous mounting medium (Sigma) with an addition of blue fluorescent nuclear counterstain DAPI, coverslipped, and sealed with nail polish. No signal was seen in control sections processed without the primary antibody, except autofluorescence in the blood vessels.
Images were obtained using an Olympus FluoView 10i confocal laser scanning microscope with UPLSAPO 10X/0.40 and UPLSAPO 60X/1.20 W objectives. Image analyses were performed using Imaris (Bitplane).
Regulation of spatial patterns in coexpression modules
To find the associated transcription factors (TFs) of each gene, we utilized the nonredundant sets of transcription factor binding sites (TFBSs) in humans from the JASPAR CORE database (Sandelin et al. 2004). We aligned the position weight matrix (PWM) of each TF to the adjacent 2-kb area (TSS ± 1 kb) of each gene or the distal enhancer region identified as described in Methods using the Bioconductor package “TFBSTools” (Tan and Lenhard 2016) in R. The top three significant TFs in terms of alignment scores were considered to be the corresponding TFs of each gene.
To explore the percentage of variance explained by epigenetic modification levels of promoter CREs, enhancer CREs, and TF expression levels in each coexpression module, we performed linear regression with gene spatial patterns in humans as response variables and those of coupled regulators as explanatory variables by the package “relaimpo” (Gromping 2006) in R. Spatial patterns across brain regions shared between transcriptome and epigenome data sets were used for genes, as well as coupled regulators. The first principal component (PC1) of spatial patterns for each kind of coupled regulators that was sufficient to represent the whole patterns was used as an unbiased estimation with regard to different numbers of members in each kind of coupled regulators (Supplemental Fig. S19). The “lmg” metric was used to decompose total R2 by averaging along orders of variables. The mean explained percentage across genes containing all three kinds of coupled regulators was calculated for each module.
Association between regulatory mechanisms and human–chimpanzee differences
We defined the relative influence of each regulatory mechanism (promoter CREs, enhancer CREs, and TFs) in each module as the explained variance by a given regulator relative to the average of the other two regulators. The spatial gene expression differences between humans and chimpanzees were calculated as the average of one minus the Pearson correlation coefficient calculated between their spatial patterns. For each regulatory mechanism, the dissimilarities between human–chimpanzee differences of regulator-driven modules (relative influence >1) and the other modules (relative influence <1) were assessed by a one-sided Wilcoxon rank-sum test.
Moreover, absolute Pearson correlation coefficients were calculated between spatial patterns of genes and each kind of coupled regulators for four species-specific modules (modules 7, 12, 20, 22). The first principal component (PC1) of spatial patterns for each kind of coupled regulators was used as mentioned above. Only genes containing all three kinds of coupled regulators were considered in each module. To estimate the correlation expected by chance, we randomly chose genes from all expressed genes as TFs or CREs from all detected CREs as promoter/enhancer CREs and calculated the correlation between genes and randomly chosen regulators 1000 times (Fig. 5C, gray dots). A one-sided Wilcoxon rank-sum test was used to assess the significance of greater correlation for TFs or smaller correlation for promoter CREs compared to expected values.
Putative TFs regulating species-specific modules
We inspected the TFs showing significant correlation with their predicted target genes in four species-specific modules (modules 7, 12, 20, 22) by checking the absolute Pearson correlation coefficients between the spatial patterns of TF and corresponding predicted target genes in the module. Significance was assessed by comparing the absolute Pearson correlation coefficients calculated within the module to that calculated between TF and its corresponding predicted target genes located outside of the module using the one-sided Wilcoxon rank-sum test. TFs with BH-corrected P < 0.05 and absolute Pearson correlation coefficients >0.6 were determined to be correlated. To estimate the significance of the observed number of correlated TFs in each module, we permuted module labels and re-performed the above procedure 1000 times.
Visualization of modules in a coexpression network
A coexpression network of all 24 modules in a graphopt layout was achieved by the package “igraph” (Csárdi and Nepusz 2006) in R using the force-directed graphopt algorithm based on Pearson correlation coefficients calculated between spatial patterns of pairwise genes. The top 20 representative genes in each module were chosen according to each gene's intramodular connectivity, defined as the sum of Pearson correlation coefficient calculated between this gene and remaining genes in the module.
Data access
All raw sequencing reads from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE100796.
Supplementary Material
Acknowledgments
We thank the following facilities for providing the chimpanzee, gorilla, and gibbon samples: the Primate Research Institute of Kyoto University, the Kumamoto Sanctuary, the Wildlife Research Center of Kyoto University, the Kyoto City Zoo in Kyoto, and the Kobe Oji Zoo in Kobe. These samples were provided through the Great Ape Information Network. We thank Dr. Masayuki Tanaka (Kyoto City Zoo), Dr. Hiroo Imai (Primate Research Institute, Kyoto University), the Functional Genomics Facility, the National Institute for Basic Biology (NIBB) Core Research Facilities for technical support, and I. Burke and A. Shmelkova for their helpful comments on the manuscript. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDB13010200) to P.K., the National Natural Science Foundation of China (grant numbers 31420103920, 91331203) to P.K., the National One Thousand Foreign Experts Plan (grant number WQ20123100078) to P.K., the Russian Science Foundation (grant numbers 16-14-00220) to P.K., the NIBB Collaborative Research Program (12-311, 13-725, 14-714, 15-815, 16-460) to Y.G., the Collaborative Research Project (2011-2310, 2012-2414, 2014-2625) of the Brain Research Institute, Niigata University to Y.G., and the Cooperation Research Program (2013-E-27, 2014-B-57, 2015-B-60, 2016-B-93, 2017-B-49) of the Primate Research Institute, Kyoto University to Y.G. This work was funded by Japan Society for the Promotion of Science (JSPS) KAKENHI grants, numbered JP22770240, JP25711027, JP26640065, JP16H04849 to Y.G.
Author contributions: Y.G. conceived the project and designed the experiments. C.X., Q.L., L.H., S.T., and Y.G. analyzed the data. O.E. and V.S. performed the IHC experiments. T.O., T.U., and Y.G. prepared the nonhuman ape brains. A.K. and H.N. prepared the human brains. K.Y., S.S., and Y.G. performed RNA-seq experiments. C.X., Q.L., P.K., and Y.G. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231357.117.
Freely available online through the Genome Research Open Access option.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. 2016. A comprehensive transcriptional map of primate brain development. Nature 535: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. 2008. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet 4: e1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. 2003. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci 100: 13030–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, O'Neill K, Milner J, Mahajan GJ, Lawrence TJ, May WL, Miguel-Hidalgo J, Rajkowska G, Stockmeier CA. 2016. Density of GFAP-immunoreactive astrocytes is decreased in left hippocampi in major depressive disorder. Neuroscience 316: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csárdi G, Nepusz T. 2006. The igraph software package for complex network research. InterJournal, Complex Systems 1695: 1–9. [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. 2015. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci 112: 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanskyi A, Alter H, Vogt MA, Gass P, Vinnikov IA. 2014. Transcription factors Foxa1 and Foxa2 are required for adult dopamine neurons maintenance. Front Cell Neurosci 8: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340–343. [DOI] [PubMed] [Google Scholar]
- Filiano AJ, Gadani SP, Kipnis J. 2015. Interactions of innate and adaptive immunity in brain development and function. Brain Res 1617: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao V, Suzuki A, Magistretti PJ, Lengacher S, Pollonini G, Steinman MQ, Alberini CM. 2016. Astrocytic β2-adrenergic receptors mediate hippocampal long-term memory consolidation. Proc Natl Acad Sci 113: 8526–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245. [DOI] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O'Connor C, Fitzpatrick C, Pasillas MP, et al. 2017. An environment-dependent transcriptional network specifies human microglia identity. Science 356: eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, McColl BW. 2016. Microglial brain region–dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromping U. 2006. Relative importance for linear regression in R: the package relaimpo. J Stat Softw 17 10.18637/jss.v017.i01. [DOI] [Google Scholar]
- Hassanpoor H, Fallah A, Raza M. 2014. Mechanisms of hippocampal astrocytes mediation of spatial memory and θ rhythm by gliotransmitters and growth factors. Cell Biol Int 38: 1355–1366. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger C, Papouin T, Oliet SH, Rusakov DA. 2010. Long-term potentiation depends on release of D-serine from astrocytes. Nature 463: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Dissing-Olesen L, Stevens B. 2016. New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol 36: 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, Geschwind DH, Mane SM, State MW, Šestan N. 2009. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62: 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. 1991. Identification and characterization of a novel repressor of β-interferon gene expression. Genes Dev 5: 868–879. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. 2004. Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14: 1462–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Pääbo S. 2006. Evolution of primate gene expression. Nat Rev Genet 7: 693–702. [DOI] [PubMed] [Google Scholar]
- Kolomeets NS, Uranova N. 2010. Ultrastructural abnormalities of astrocytes in the hippocampus in schizophrenia and duration of illness: a postortem morphometric study. World J Biol Psychiatry 11: 282–292. [DOI] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM, et al. 2012. Human-specific transcriptional networks in the brain. Neuron 75: 601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzisch M, Temprana SG, Mongiat LA, Armida J, Schmutz V, Virtanen MA, Kocher-Braissant J, Kraftsik R, Vutskits L, Conzelmann KK, et al. 2015. Pre-existing astrocytes form functional perisynaptic processes on neurons generated in the adult hippocampus. Brain Struct Funct 220: 2027–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. 2016. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352: 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D, Barreiro LB, Gros P. 2016. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med 213: 585–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P, Gordon S. 1990. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39: 151–170. [DOI] [PubMed] [Google Scholar]
- L'Episcopo F, Drouin-Ouellet J, Tirolo C, Pulvirenti A, Giugno R, Testa N, Caniglia S, Serapide MF, Cisbani G, Barker RA, et al. 2016. GSK-3β-induced Tau pathology drives hippocampal neuronal cell death in Huntington's disease: involvement of astrocyte–neuron interactions. Cell Death Dis 7: e2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, et al. 2012. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res 22: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyck L, Santamaria ID, Pakkenberg B, Chemnitz J, Schrøder HD, Finsen B, Gundersen HJ. 2009. An empirical analysis of the precision of estimating the numbers of neurons and glia in human neocortex using a fractionator-design with sub-sampling. J Neurosci Methods 182: 143–156. [DOI] [PubMed] [Google Scholar]
- Masuda T, Iwamoto S, Mikuriya S, Tozaki-Saitoh H, Tamura T, Tsuda M, Inoue K. 2015. Transcription factor IRF1 is responsible for IRF8-mediated IL-1β expression in reactive microglia. J Pharmacol Sci 128: 216–220. [DOI] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. 2009. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci 106: 22358–22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah M, Patrick E, Villani AC, Xu J, White CC, Ryan KJ, Piehowski P, Kapasi A, Nejad P, Cimpean M, et al. 2018. A transcriptomic atlas of aged human microglia. Nat Commun 9: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. 2006. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci 103: 17973–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. 2008. Functional organization of the transcriptome in human brain. Nat Neurosci 11: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota Y, Zanetti AT, Hallock RM. 2013. The role of astrocytes in the regulation of synaptic plasticity and memory formation. Neural Plast 2013: 185463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M, Braganza O, Dannenberg H, Hu W, Pothmann L, Rosen J, Mody I, van Loo K, Deisseroth K, Becker AJ, et al. 2016. Astrocyte intermediaries of septal cholinergic modulation in the hippocampus. Neuron 90: 853–865. [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR III, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. 2013. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155: 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443: 167–172. [DOI] [PubMed] [Google Scholar]
- Premack D. 2007. Human and animal cognition: continuity and discontinuity. Proc Natl Acad Sci 104: 13861–13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. 2015. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347: 1155–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A. 2006. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics 7: 67–80. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Szelinger S, Siniard AL, Corneveaux JJ, Kurdoglu A, Richholt R, De Both M, Malenica I, Swaminathan S, Rangasamy S, et al. 2015. A de novo mutation in TEAD1 causes non–X-linked Aicardi syndrome. Invest Ophthalmol Vis Sci 56: 3896–3904. [DOI] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, et al. 2015. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci 18: 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. 2009. Transcriptional neoteny in the human brain. Proc Natl Acad Sci 106: 5743–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, et al. 2010. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res 20: 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, et al. 2018. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555: 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S, Li L, Moss J, Petrelli F, Cassé F, Gebara E, Lopatar J, Pfrieger FW, Bezzi P, Bischofberger J, et al. 2015. Synaptic integration of adult-born hippocampal neurons is locally controlled by astrocytes. Neuron 88: 957–972. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Stern SA, Bozdagi O, Huntley GW, Walker RH, Magistretti PJ, Alberini CM. 2011. Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 144: 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadi M, Allaman I, Lengacher S, Grenningloh G, Magistretti PJ. 2015. Learning-induced gene expression in the hippocampus reveals a role of neuron-astrocyte metabolic coupling in long term memory. PLoS One 10: e0141568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Lenhard B. 2016. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32: 1555–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay TL, Savage JC, Hui CW, Bisht K, Tremblay ME. 2017. Microglia across the lifespan: from origin to function in brain development, plasticity and cognition. J Physiol 595: 1929–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J Mach Learn Res 9: 2579–2605. [Google Scholar]
- Vermunt MW, Tan SC, Castelijns B, Geeven G, Reinink P, de Bruijn E, Kondova I, Persengiev S, Netherlands Brain Bank, Bontrop R, et al. 2016. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci 19: 494–503. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang K, Han L, Zhang A, Shi Z, Zhang K, Zhang H, Yang S, Pu P, Shen C, et al. 2013. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/β-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett 331: 211–219. [DOI] [PubMed] [Google Scholar]
- Wolf SA, Boddeke HW, Kettenmann H. 2017. Microglia in physiology and disease. Annu Rev Physiol 79: 619–643. [DOI] [PubMed] [Google Scholar]
- Wruck W, Schröter F, Adjaye J. 2016. Meta-analysis of transcriptome data related to hippocampus biopsies and iPSC-derived neuronal cells from Alzheimer's disease patients reveals an association with FOXA1 and FOXA2 gene regulatory networks. J Alzheimers Dis 50: 1065–1082. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347: 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. 2016. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89: 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.