Abstract
Background
The cancer-testis specific gene Opa interacting protein 5 (OIP5) is reactivated in many human cancers, but its functions in glioblastoma remain unclear. Here, we assessed the significance of OIP5 in the tumorigenesis and metastasis of glioblastoma for the first time.
Methods
An immunohistochemistry assay was performed to detect OIP5 expression changes in glioblastoma patients. Overall survival analysis was performed to evaluate the prognostic significance of OIP5. Growth curve, colony formation, and transwell assays were used to analyze cell proliferation and metastasis. Tumorigenicity potential was investigated in orthotopic tumor models, and immunoprecipitation, chromatin immunoprecipitation, and luciferase assays were employed to explore the mechanisms underlying the activation of OIP5 expression by E2F transcription factor 1 (E2F1) to stabilize and maintain E2F1 signaling.
Results
OIP5 was found to be upregulated in glioblastoma patients and to impair patient survival, and the increased expression of OIP5 was positively correlated with tumor stage. Compared with short hairpin green fluorescent protein cells, cells in which OIP5 was knocked down exhibited significantly reduced proliferation, metastasis, colony formation, and tumorigenicity abilities, whereas OIP5 recovery enhanced these abilities. OIP5 was highly correlated with cell cycle progression but had no obvious effects on apoptosis. Notably, we demonstrated a feedback loop in which E2F1 activates the expression of OIP5 to stabilize and maintain E2F1 signaling and promote the E2F1-regulated gene expression that is required for aggressive tumor biology.
Conclusions
Collectively, our findings demonstrate that OIP5 promotes glioblastoma progression and metastasis, suggesting that OIP5 is a potential target for anticancer therapy.
Keywords: E2F1, glioblastoma, metastasis, OIP5, tumorigenesis
Importance of the study
Cancer-testis specific genes have been pursued as potential targets for anticancer therapy. The mechanisms of these genes in tumor tissue development and whether their reactivation plays roles in supporting tumorigenic features have not been well described. We confirmed universal OIP5 upregulation in glioblastoma and demonstrated that increased OIP5 expression impairs patient survival. Our orthotopic mice model showed that OIP5 depletion significantly inhibited tumor growth in vivo and improved mice survivability. Notably, we demonstrated a novel feedback loop in which E2F1 activates the expression of OIP5 to stabilize and maintain E2F1 signaling, persistently augment pathway activity, and promote the E2F1-regulated gene expression that is required for aggressive tumor biology. Our findings elucidate the importance of OIP5 in glioblastoma as a therapeutic target and show the potential for using OIP5 as a clinically relevant biomarker to predict tumor progression and survival.
Cancer-testis specific genes have been pursued as potential targets of anticancer therapy for more than 20 years,1 and they are named as such because original results indicated that these genes are only expressed in testes and multiple tumor tissues. The first cancer-testis specific gene, now named MAGE-A1, was identified in melanoma patients by Professor Van Der Bruggen and his colleagues in Belgium,2 which was followed by the identification of a series of genes utilizing a similar strategy.3–7 Subsequently, gene expression profiling and other high-throughput methods have identified more than 200 genes that are classified as being cancer-testis specific.8–10 The mechanisms of these genes in tumor tissue development and whether their reactivation plays roles in supporting tumorigenic features have been studied to a lesser extent.
Opa interacting protein 5 (OIP5), also called LINT-2511 and hMis18beta12 in previous studies, was first identified by a yeast 2-hybrid system.13 While recent research has shown that OIP5 is a cancer-testis specific gene that is upregulated in many human cancers,14–17 the functional role and tumor-promoting mechanisms of OIP5 in cancer cells remain unknown.
E2F transcription factor 1 (E2F1) is a key transcription factor that participates in various biological processes.18 Aberrant E2F1 upregulation, which is associated with malignant processes and poor prognosis, is frequently observed in many kinds of human cancers.19–22 Moreover, E2F1 is also associated with radiosensitivity23 and chemosensitivity24,25 in cancer cells. While some findings showing that E2F1 is involved in cell senescence and apoptosis may suggest its dual role in tumor formation,26 several events accelerating the development of cancer may neutralize the tumor-suppressive functions of E2F1. For example, the apoptosis and cell senescence processes mediated by E2F1 may lose their efficacy in p53- or p14-defective cells,27,28 and oncogenic epidermal growth factor receptor and phosphatidylinositol-3 kinase/Akt signals also inhibit the apoptotic functionality of E2F1.29,30 Additionally, E2F1 has been demonstrated to promote tumor formation and angiogenesis as well as epithelial-mesenchymal transition in several cases.31–34 In brief, the adjustable oncogenic activity of E2F1 is modulated by other signals, suggesting that OIP5 may have a role in these processes.
In this study, we show for the first time that OIP5 is upregulated in glioblastoma patients and correlated with poor prognosis. In addition, OIP5 is critical for the proliferation, migration, and invasion of glioblastoma cells. Specifically, we found OIP5 to be an undiscovered downstream gene of E2F1 and demonstrated that OIP5 interacts with E2F1 to stabilize and maintain E2F1 signaling.
Materials and Methods
Reagents and Antibodies
The OIP5 (12142-1-AP) and cyclin-dependent kinase 1 (CDK1) (19532-1-AP) antibodies were purchased from Proteintech Group, and antibodies against CDK2 (#2546), CDK4 (#12790), and ubiquitin (#3936) were purchased from Cell Signaling Technology. The E2F1 (sc-251) antibody was purchased from Santa Cruz Technology, and the bromodeoxyuridine (BrdU) antibody (ab6326) was purchased from Abcam. The tubulin (AT819) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; AG019) antibodies were purchased from Beyotime, and the Ki-67 (550609) antibody was purchased from BD Pharmingen. Cycloheximide (C7698), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; M5655), BrdU (B5002), dimethyl sulfoxide (D5879), and MG132 (M7449) were purchased from Sigma-Aldrich.
Cell Culture
All cell lines were originally obtained from American Type Culture Collection and cultured as previously described.35
Plasmids, Transfection, and Infection
Human OIP5 short hairpin (sh)RNA (#1, TRCN0000074087; #2, TRCN0000074083) and E2F1 shRNA (TRCN0000039659) were purchased from Sigma-Aldrich. A vector expressing OIP5 was constructed by PCR amplification, and the PCR product was subsequently inserted into the pCDH-CMV-MCS-EF1-copGFP vector. Transfection and infection were performed as previously described.35
Patient Data Analysis
Patient and gene expression data were obtained from the R2 Genomics Analysis and Visualization Platform database (http://hgserver1.amc.nl/cgi -bin/r2/main.cgi). Overall survival analysis was performed using high and low OIP5 expression cutoffs based on the R2 algorithm, and P-values were also obtained from the database.
Immunohistochemistry Assay
For immunohistochemistry (IHC) analysis, paraffin-embedded tissues were cut into sections 5 mm thick. After deparaffinization, hydration, and antigen retrieval, the tissue sections were exposed to antibodies against OIP5 (1:100) or E2F1 (1:100) at 4°C overnight. The sections were then treated with a horseradish peroxidase detection system (GBI Labs). Staining was visualized using a 3,3ʹ-diaminobenzidine kit, and hematoxylin was used as the counterstain. All images were acquired using a light microscope.
Quantitative Real-Time PCR
Quantitative real-time (qRT) PCR was performed as previously described.36
Western Blot and Immunoprecipitation Analysis
Western blot analyses were performed as previously described.35 For immunoprecipitation analysis, cells were plated in 100-mm plates and then lysed in western blot and immunoprecipitation lysis buffer. Whole-cell lysates were incubated with the indicated antibodies at 4°C overnight and then incubated with Protein A+G agarose. Following extensive washing in phosphate buffered saline (PBS), bound proteins were recovered by boiling the agarose in 1× sodium dodecyl sulfate–polyacrylamide gel electrophoresis sample loading buffer.
Cell Viability and Proliferation Assays
Cell viability and proliferation assays were performed as previously described.36
Colony Formation Assay and Orthotopic Model
Colony formation assays were performed as previously described.35 Nude mice were used for the orthotopic model. Briefly, 1 × 105 cells were suspended in 10 μL PBS and injected into the brains at a position of 2 mm lateral, 1 mm anterior to the bregma, and 4 mm deep. When all the mice of the short hairpin green fluorescent protein (shGFP) group died due to cancer progression, their brains were collected for hematoxylin and eosin (H&E) and IHC staining.
All animal work was conducted in accordance with the 2006 Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China) and approved by the Animal Ethics Committee of Southwest University.
Flow Cytometry Analysis
For cell cycle analysis, cells were harvested and fixed in 70% ethanol at 4°C overnight. Next, the cells were incubated with 40 µg/mL RNase A and 1% propidium iodide for 30 minutes. DNA content was measured using a BD Accuri C6 flow cytometer and analyzed with FlowJo software. Apoptosis analysis was performed as previously described.35
Migration, Invasion, and Wound Healing Assays
In brief, migration and invasion were assessed using a 24-well Millicell Hanging Cell Culture Insert (Merck Millipore) with a pore size of 8.0 µm. The insert was covered by Matrigel (Corning) for the invasion assays according to the manufacturer’s protocol. Cells were cultured for 12 hours in the migration assays and 48 hours in the invasion assays at 37°C in a cell culture incubator. Cells in the upper compartment were removed by wiping the upper membrane with an alcohol tampon, and cells on the bottom of the microporous membrane were stained with crystal violet. For the wound healing assays, cells were seeded into 6-well plates and wounded in a line across the wells with a 10 µL plastic pipette tip. Cellular debris was washed with PBS, and the distances migrated during the indicated times were measured using a microscope.
Chromatin Immunoprecipitation
A chromatin immunoprecipitation (ChIP) assay kit (Beyotime) was used according to the manufacturer’s protocol.
Luciferase Reporter Assay
The pGME2F-luc plasmid was purchased from Genomeditech. The OIP5 wild-type and OIP5 mutant promoter sequences were synthesized by the Beijing Genomics Institute and subsequently ligated into the pGL3-basic vector (Promega). The pGL3, pRL-TK (Promega), and pCMV-E2F1 (Addgene) vectors were co-transfected into cells, which were then incubated for 48 more hours. The luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Statistical Analysis
Statistical analyses were performed using Microsoft Excel. Data were presented as mean ± SEM and analyzed using unpaired 2-tailed t-tests. Graphics were generated using GraphPad and R software. P-values of <0.05 (*) and <0.01 (**) were considered statistically significant.
Results
OIP5 Is Upregulated in Glioblastoma Patients and Correlated with Poor Prognosis
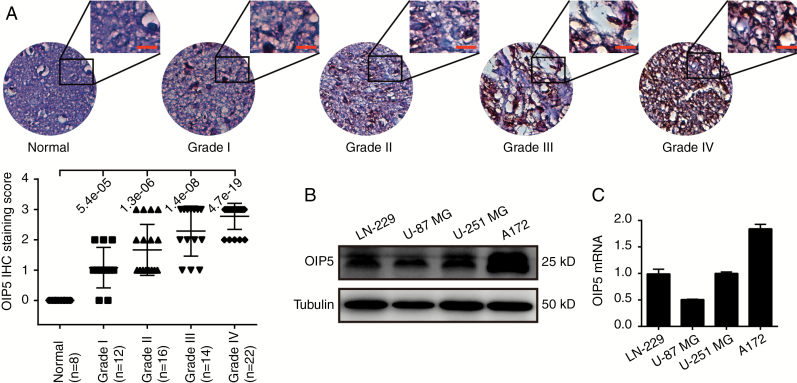
The IHC assay was performed to detect OIP5 expression changes in glioblastoma patients. OIP5 expression was silenced in the tissues of 8 normal human brains but highly expressed in glioblastoma tissues in a grade-dependent manner (Fig. 1A). We next examined OIP5 expression in several glioblastoma cell lines, including LN-229, U-87 MG, U-251 MG, and A172. We found that OIP5 was commonly expressed in each line (Fig. 1B and C). Furthermore, we examined OIP5 expression in tumor tissue and peritumoral tissue. We found that OIP5 was upregulated in the tumor tissues (Supplementary Fig. S1A).
Fig. 1 .
OIP5 is commonly expressed in glioblastoma patients and cell lines. (A) Immunohistochemical assays (top) and statistical analyses (bottom) of OIP5 expression in normal brain and glioblastoma tissue. The data were analyzed using 2-tailed Student t-tests with P-values indicated. (B) Western blot and (C) qRT-PCR analysis of OIP5 expression in 4 glioblastoma cell lines. Scale bars 50 μm.
We further validated the upregulation of OIP5 expression in 10 databases using the R2 Genomics Analysis and Visualization Platform. The expression of OIP5 was low and not significantly different in 2 normal brain databases (P = 0.59). However, OIP5 was upregulated in the tumor tissues and cell lines of 8 glioma and glioblastoma databases, with significant differences from samples from the normal brain database (Supplementary Fig. S1B). Furthermore, OIP5 expression was also correlated with tumor stage in French’s database (Supplementary Fig. S1C), and OIP5 expression was observed to be lower in living patients than in dead patients according to Paugh’s data (Supplementary Fig. S1D). We also checked the OIP5 expression in the new World Health Organization classification of tumors of the CNS using GlioVis datasets.37 Results suggested that OIP5 was highly expressed in the isocitrate dehydrogenase wild-type group (Supplementary Fig. S1E) according to The Cancer Genome Atlas low-grade glioma dataset.38 Survival probability was analyzed using 4 glioblastoma and glioma databases, revealing high OIP5 expression to be closely related to poor survival and low OIP5 expression to be associated with good overall survival (Supplementary Fig. S1F). Together, these results definitively demonstrate that OIP5 is upregulated in glioblastoma patients, suggesting that OIP5 may play a role in glioblastoma development and progression.
OIP5 Is Required for Glioblastoma Cell Proliferation In Vitro
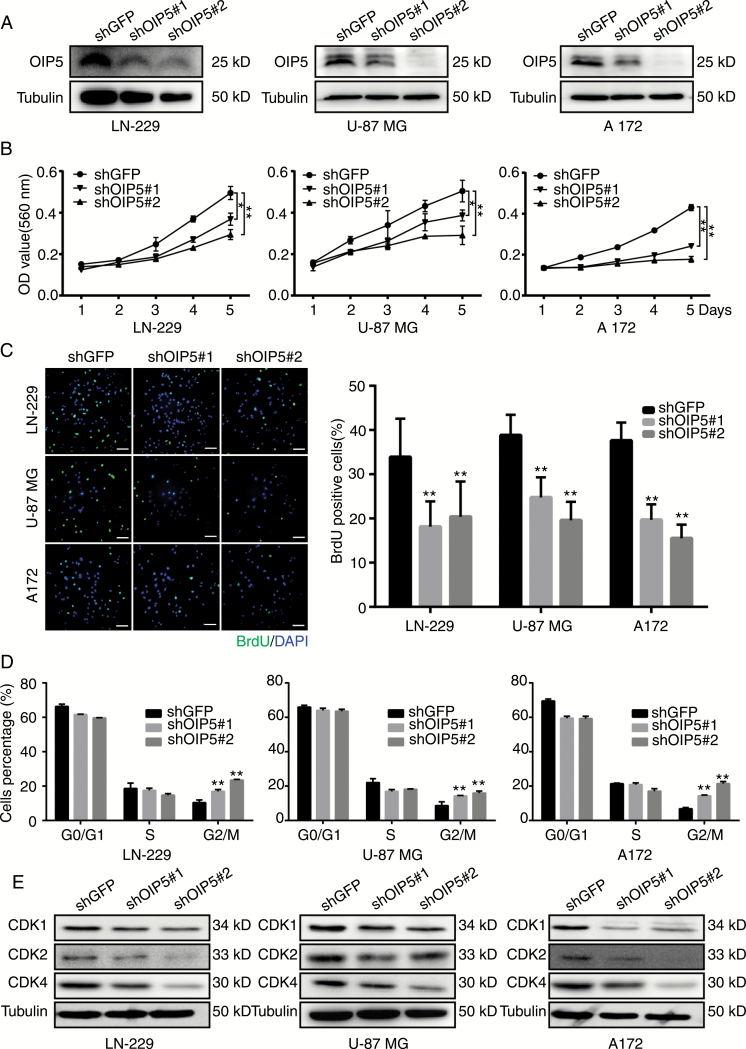
Universal OIP5 upregulation exposes an attractive possibility that OIP5 may act as a cancer promoter in glioblastoma. To evaluate the role of OIP5 in glioblastoma development and progression, OIP5 was stably knocked down using 2 different lentiviral shRNA sequences (shOIP5#1 and shOIP5#2) in LN-229, U-87 MG, and A172 cells; shGFP was used as a negative control. Western blotting (Fig. 2A) and qRT-PCR (Supplementary Fig. S2A) analysis showed that both shRNA sequences were successfully downregulated OIP5 expression, although optimal efficiency was detected with shOIP5#2. Three days after lentiviral shRNA infection, evident morphological changes were observed with a microscope in cells in which OIP5 was knocked down. The number of shOIP5#1 and shOIP5#2 cells was visibly lower than the number of shGFP cells (Supplementary Fig. S2B). MTT assays showed that OIP5 knockdown significantly suppressed the proliferation of glioblastoma cells in vitro (Fig. 2B), and BrdU incorporation was reduced in the shOIP5 groups compared with that in shGFP cells (Fig. 2C). These results suggested that OIP5 silencing suppresses glioblastoma cell proliferation.
Fig. 2 .
OIP5 is required for proliferation of glioblastoma cells and regulates cell cycle progression. (A) Western blot analysis of OIP5 expression in LN-229, U-87 MG, and A172 cell lines expressing shGFP, shOIP5#1, or shOIP5#2. (B) Growth curves of LN-229, U-87 MG, and A172 cells expressing shGFP, shOIP5#1, or shOIP5#2. (C) Representative fluorescent micrographs and quantification for BrdU staining of LN-229, U-87 MG, and A172 cells expressing shGFP, shOIP5#1, or shOIP5#2. (D) Quantification of cell population in each phase of LN-229, U-87 MG, and A172 cells expressing shGFP, shOIP5#1, or shOIP5#2. (E) Protein expression of the indicated genes in LN-229, U-87 MG, and A172 cells expressing shGFP, shOIP5#1, or shOIP5#2. Data were analyzed using 2-tailed Student’s t-tests, *P < 0.05, **P < 0.01. Scale bars 20 μm.
To further confirm the involvement of OIP5 in glioblastoma proliferation, we recovered OIP5 expression in OIP5 knockdown cells, and OIP5 was successfully overexpressed (Supplementary Fig. S2C). Predictably, cell growth curves (Supplementary Fig. S2D) and BrdU-positive rates (Supplementary Fig. S2E) were obviously increased. These results indicate that OIP5 is essential for glioblastoma cell proliferation.
OIP5 Regulates Glioblastoma Cell Cycle Progression but Not Apoptosis
To determine which cellular processes are associated with OIP5 expression, we performed mini-ontology analysis in French’s database using gene sets related to OIP5, which were identified by a positive correlation coefficient of R > 0.7. We found that OIP5 was significantly correlated with cell cycle progression (Supplementary Fig. S3A). Flow cytometry (Supplementary Fig. S3B) and statistical analyses (Fig. 2D) showed that OIP5 knockdown in LN-229, U-87 MG, and A172 cells led to G2/M cell cycle arrest, and the expression levels of the cell cycle–related proteins CDK1, CDK2, and CDK4 were reduced in shOIP5 cells (Fig. 2E).
We also used flow cytometry techniques to detect the apoptosis of shOIP5 cells. However, OIP5 knockdown did not significantly increase apoptosis in these cells compared with that observed after GFP knockdown (Supplementary Fig. S3C). Additionally, the expression levels of apoptosis-related proteins were not markedly changed (Supplementary Fig. S3D). These data demonstrate that OIP5 is a cell cycle regulator of glioblastoma.
OIP5 Is Essential for the Migration and Invasion of Glioblastoma Cells In Vitro
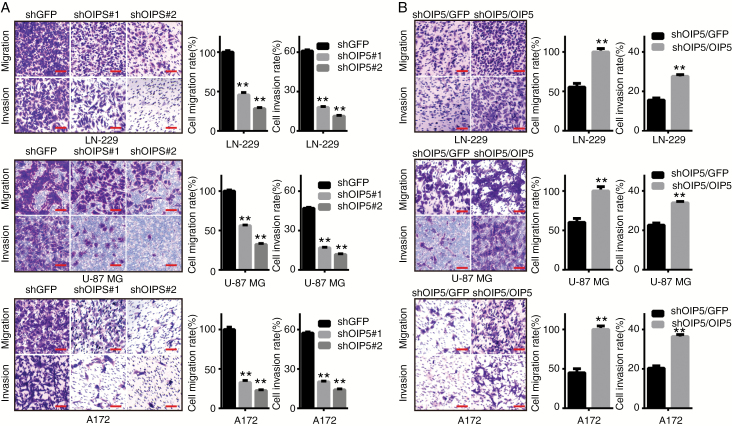
A key characteristic of cancer cells is neoplastic metastasis, a main challenge affecting cancer-related mortality in oncotherapy. To determine whether OIP5 is essential for glioblastoma cell migration and invasion, we performed wound healing assays and transwell assays. OIP5 knockdown significantly reduced the ability of glioblastoma cell wound healing (Supplementary Fig. S4A). Similarly, transwell assays showed a lower migration and invasion ability of shOIP5 cells than shGFP cells (Fig. 3A). Additionally, restoration of OIP5 expression in OIP5 knockdown cells accelerated their wound healing (Supplementary Fig. S4B). Comparable observations were also observed in the migration and invasion assays (Fig. 3B). Collectively, these data demonstrated that OIP5 can promote glioblastoma cell migration and invasion in vitro.
Fig. 3 .
OIP5 is essential for the migration and invasion of glioblastoma cells. (A) Migration and invasion assays were performed in LN-229, U-87 MG, and A172 cells expressing shGFP, shOIP5#1, or shOIP5#2 (left), and the relative percentage of migratory or invasive cells was calculated (right). (B) Migration and invasion assays were performed in OIP5-rescued OIP5 knockdown cells (left), and the relative percentage of migratory or invasive cells was calculated (right). Data were analyzed using 2-tailed Student’s t-tests, **P < 0.01. Scale bars 20 μm.
OIP5 Is Required for Glioblastoma Colony Formation In Vitro and Tumor Formation In Vivo
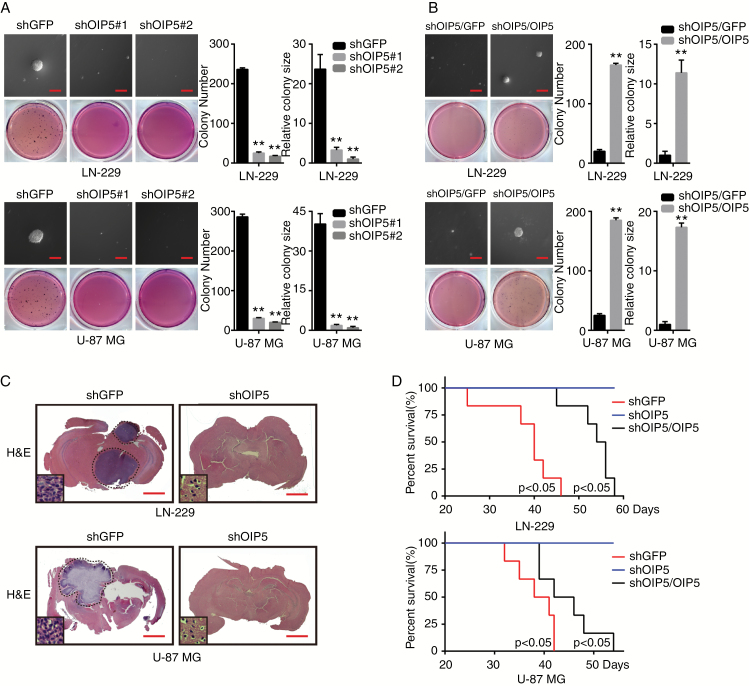
We next aimed to delineate whether OIP5 could modulate cellular properties that are frequently associated with increased tumor initiation potential and aggressiveness, such as clonogenic and self-renewal abilities. While OIP5 knockdown significantly decreased colony formation in LN-229 and U-87 MG cells (Fig. 4A), OIP5 overexpression in OIP5 knockdown cells enhanced colony formation (Fig. 4B).
Fig. 4 .
OIP5 is required for glioblastoma colony formation in vitro and tumor formation in vivo. (A) Colony formation ability of LN-229 and U-87 MG cells expressing shGFP, shOIP5#1, or shOIP5#2. (B) Colony formation ability of the OIP5-rescued OIP5 knockdown cells. (C) Orthotopic implantation was performed after OIP5 knockdown in LN-229 and U-87 MG cells. Representative images of the hematoxylin and eosin staining are presented. (D) Survival rates of mice were analyzed after orthotopic implantation. Data were analyzed using 2-tailed Student’s t-tests, **P < 0.01. Scale bars Panels A and B: 50 μm; Panel C: 3 mm.
To further examine the effects of OIP5 on the tumor formation potential of glioblastoma cells, we performed in vivo orthotopic implantation in nude mice. In the orthotopic transplantation tumor model, OIP5 depletion significantly inhibited tumor growth in vivo (Fig. 4C). In addition, improved survivability was observed in shOIP5 mice compared with shGFP mice. Additionally, restoration of OIP5 expression in OIP5 knockdown cells impaired mice survivability (Fig. 4D). These results show that OIP5 is necessary for the colony formation and tumorigenesis of glioblastoma cells.
OIP5 Regulates and Stabilizes E2F1 Signaling
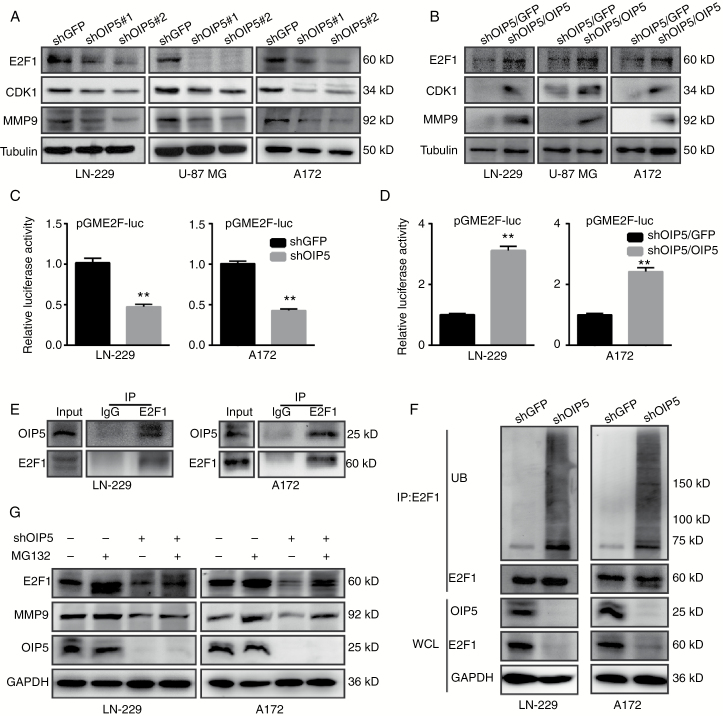
We next investigated the effects of OIP5 inhibition on E2F1 and its regulated genes, including CDK1 and matrix metalloproteinase 9 (MMP9). OIP5 knockdown cells exhibited decreased E2F1, CDK1, and MMP9 expression levels compared with those in shGFP cells (Fig. 5A). Conversely, OIP5 overexpression in OIP5 knockdown cells elevated E2F1, CDK1, and MMP9 levels (Fig. 5B). OIP5 depletion inhibited E2F1 activity (Fig. 5C), as evaluated using an E2F1-binding element luciferase reporter, whereas OIP5 recovery yielded the opposite effect (Fig. 5D).
Fig. 5.
OIP5 regulates and stabilizes E2F1 signaling. (A) Protein expression levels of E2F1, CDK1, and MMP9 were analyzed by western blot in LN-229, U-87 MG, and A172 cells expressing shGFP, shOIP5#1, or shOIP5#2. (B) E2F1, CDK1, and MMP9 expression levels of the OIP5-rescued OIP5 knockdown cells. (C) Luciferase assay in LN-229 and A172 cells co-transfected with the reporter plasmids combined with the indicated shRNA. (D) Luciferase assay in OIP5-rescued OIP5 knockdown cells. (E) Interaction of endogenous OIP5 with endogenous E2F1. Equal amounts of LN-229 and A172 cell lysates were used for each immunoprecipitation (IP) reaction. (F) LN-229 and A172 cells expressing shGFP or shOIP5 were treated with MG132 for 6 h before harvesting. Whole cell lysates (WCL) were incubated with anti-E2F1 antibody, and the immunocomplexes were immunoblotted with antibodies against ubiquitin (UB) and E2F1. (G) LN-229 and A172 cells expressing shGFP or shOIP5 were treated with or without MG132 for 6 h before harvesting. Equal amounts of cell lysates were immunoblotted with the indicated antibodies. Data were analyzed using 2-tailed Student’s t-tests, **P < 0.01.
We hypothesized that OIP5 may interact with and modulate the activity of E2F1. Indeed, immunoprecipitation assays confirmed an interaction between OIP5 and E2F1 in LN-229 and A172 cells (Fig. 5E), and OIP5 knockdown significantly enhanced the ubiquitination of E2F1 (Fig. 5F). Furthermore, OIP5-mediated E2F1 degradation could be rescued by the proteasome inhibitor MG132 (Fig. 5G). To test whether OIP5 stabilizes E2F1 protein, we measured the effects of both OIP5 overexpression and depletion on degradation of E2F1 protein using the cycloheximide chase assay. As expected, OIP5 overexpression promoted E2F1 protein stability (Supplementary Fig. S5A), while OIP5 depletion significantly decreased the protein half-life of E2F1 compared with that in shGFP cells (Supplementary Fig. S5B). These results collectively indicate that OIP5 interacts with and stabilizes E2F1 in glioblastoma cells.
OIP5 Is a Downstream Gene of E2F1
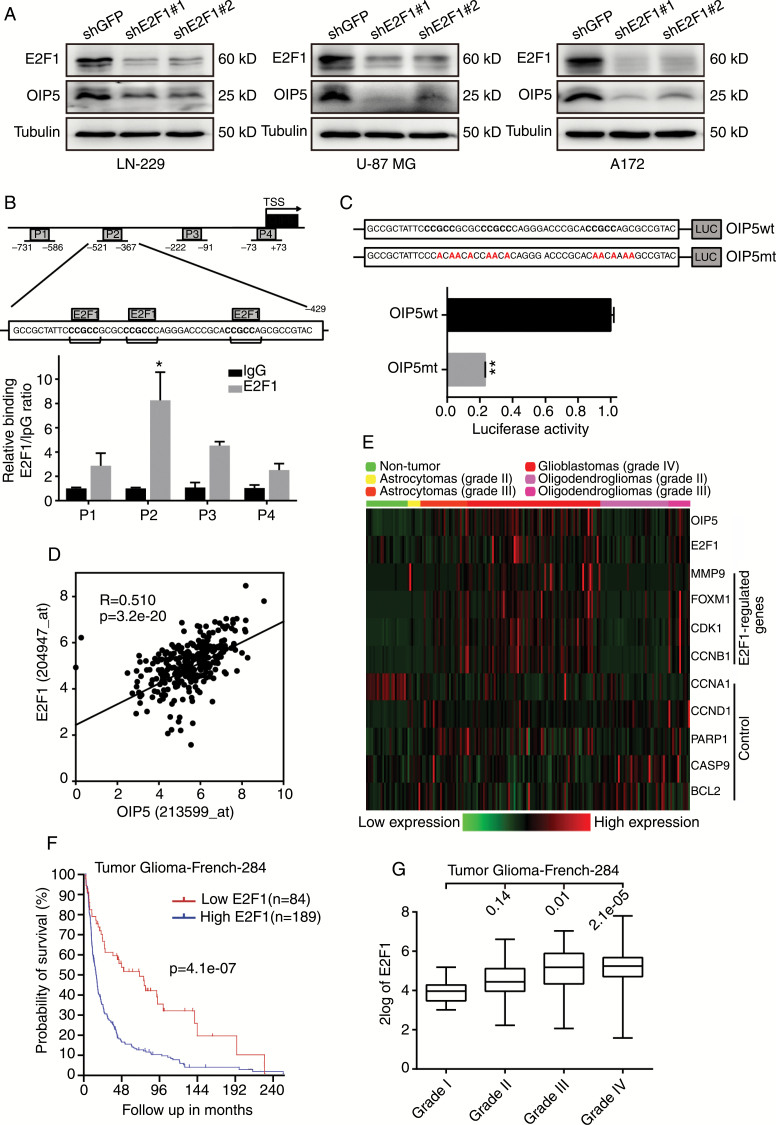
Interestingly, OIP5 expression was also significantly reduced when E2F1 was stably knocked down (Fig. 6A). Therefore, we further explored the relationship between OIP5 and E2F1. As OIP5 was determined to be highly correlated with cell cycle progression, E2F1 also participates in cell cycle regulation as a master transcription factor. Thus, we wanted to determine whether E2F1 is a transcriptional regulator of OIP5. The ChIP assay showed that OIP5 promoter sequences were robustly recruited by E2F1 relative to immunoglobulin G controls, and 3 E2F1-binding elements were detected in the P2 region of the OIP5 promoter (Fig. 6B). The luciferase assay demonstrated that mutating the E2F1-binding elements could significantly inhibit the transcriptional activity of this promoter region (Fig. 6C).
Fig. 6.
OIP5 is a downstream gene of E2F1. (A) Immunoblot analysis of E2F1 and OIP5 in LN-229, U-87 MG, and A172 cells expressing shGFP, shE2F1#1, or shE2F1#2. (B) A total of 4 sets of primers were designed within the human OIP5 promoter, and a ChIP assay was performed using E2F1 antibodies. Immunoglobulin G was used as the negative control. Three E2F1 conserved recognition sequences were identified in the OIP5 promoter in the P2 region. (C) The OIP5 wild-type (wt) and OIP5 mutant (mt) promoter sequences were ligated into the pGL3 plasmid and co-transfected with pRL-TK. Luciferase activity was examined 48 h after transfection. (D) The expression of OIP5 correlates with E2F1 levels. (E) Gene expression profile analysis on the GDS1962 dataset using E2F1-regulated genes and other control genes. (F) Kaplan–Meier analysis of overall survival in French’s database. (G) E2F1 expression was correlated with tumor stage in French’s database. The log-rank test P-values are shown. Data were analyzed using 2-tailed Student’s t-tests, *P < 0.05, **P < 0.01.
Moreover, OIP5 expression was positively correlated with E2F1 expression in French’s database (Fig. 6D). To further study the correlation between OIP5 and E2F1, we performed gene expression profile analysis on the GDS1962 dataset using E2F1-regulated genes and other control genes. OIP5 was highly correlated with E2F1 and its regulated genes, including MMP9, FOXM1, CDK1, and CCNB1, but the control genes CCNA1, CCND1, PARP1, CASP9, and BCL2 were not significantly correlated with OIP5 expression (Fig. 6E). These correlations support a role for OIP5 in E2F1 signaling. Survival probability was also analyzed using French’s database, revealing high E2F1 expression to be closely related to poor survivability and low E2F1 expression to be associated with good overall survival (Fig. 6F). Similar to OIP5, E2F1 expression was correlated with tumor stage in French’s database (Fig. 6G).
These data indicate that E2F1 binds the OIP5 promoter and regulates OIP5 expression, suggesting that OIP5 is a downstream gene of E2F1 in glioblastoma. Moreover, these findings reveal the cancer-promoting activity of E2F1 signaling in glioblastoma.
To investigate whether OIP5 is important in clinical therapy, we downregulated the expression of OIP5 in the primary human glioma cell 080611T (Supplementary Fig. S6A), and results show that OIP5 depletion significantly suppressed the proliferation of primary human glioma cell 080611T (Supplementary Fig. S6B). In addition, OIP5 depletion significantly decreased colony formation in 080611T cells (Supplementary Fig. S6C). Furthermore, we also wanted to know whether OIP5 contributes to resistance against anticancer treatment—we knocked down OIP5 and then treated cells with 1 μM doxorubicin or 50 μM of the E2F inhibitor HLM006474. Results indicated that downregulation of OIP5 increased the sensitivity against anticancer treatment in glioblastoma cells (Supplementary Fig. S7). These data indicate that OIP5 is a potential target for clinical therapy.
Discussion
Anticancer therapy remains unoptimized because of a lack of effective targets. Because of their testis/tumor-biased expression patterns, cancer-testis specific genes have been pursued as potential targets for nearly 20 years. Accumulating evidence has revealed cancer-testis specific genes as having critical roles in cancer progression.1 Currently, no uniform conclusions have been reached as to why cancer-testis specific genes are reactivated in cancer cells. However, these reactivations are undeniably associated with poor prognosis, indicating that cancer-testis specific genes can play functional roles in supporting tumorigenesis. Here, we combined database analysis and experimental studies on human glioblastoma samples to demonstrate that OIP5 is upregulated in glioblastoma patients and impairs patient survival (Fig. 1 and Supplementary Fig. S1). We also utilized gain- and loss-of-function analyses to demonstrate a critical role of OIP5 in the growth (Fig. 2A–C and Supplementary Fig. S2) and metastatic ability (Fig. 3 and Supplementary Fig. S4) of glioblastoma cells. OIP5 activation was negatively correlated with patient survival, indicating that OIP5 activation is a prognostic marker. These findings can enlarge the scope of cancer-testis specific genes in tumor-supporting functions.
OIP5 has been reported as a potential cancer-promoting candidate gene in other screens, including screens for gastric cancer,14 lung cancer,15 clear cell renal cell carcinoma,16 and breast cancer,17 raising the possibility that OIP5 is involved in the progression of a wide variety of cancers. Although OIP5 upregulation has been reported in many human cancers,14–17 to our knowledge we are the first to describe OIP5 as a proliferation and metastasis promoter in glioblastoma patients. A recent study reported that OIP5 inhibition can impair breast cancer cell proliferation,39 and another research group found OIP5 to be required for hepatocellular carcinoma growth and metastasis.40 These studies are identical to our findings in that decreased OIP5 levels suppressed the proliferation and metastasis of glioblastoma cells. However, the mechanism underlying how OIP5 supports tumor proliferation and metastasis is still not fully understood.
Not surprisingly, OIP5 was associated with cell cycle progression (Supplementary Fig. S3A). OIP5 was first identified as a centromere regulator for centrosome-associated protein A recruitment12 and has been reported as a mitotic regulator of HJURP (holliday junction recognition protein) interaction.41 In our study, we verified that OIP5 knockdown led to G2/M cell cycle arrest (Fig. 2D). However, a recent study found that OIP5 knockdown could induce breast cancer cell apoptosis,39 which disagrees with our finding that OIP5 depletion had no obvious effect on apoptosis (Supplementary Fig. S3C). Reasons for this disagreement include that OIP5 may function differently in different types of cancer or that the effects of OIP5 are pathway dependent, as the epidermal growth factor receptor pathway is highly activated in glioblastoma but not in breast cancer.
Importantly, our study elucidated the mechanism of how OIP5 regulates E2F1 signaling in glioblastoma. E2F1 expression impaired patient survival and was positively correlated with tumor stage, suggesting a tumor-supporting role of E2F1 in glioblastoma (Fig. 6F and G). The correlation among OIP5, E2F1, and E2F1-regulated genes strongly suggests a deeper connection between OIP5 and E2F1 signaling (Fig. 6D and E). We hypothesized and proved that E2F1 transcriptionally regulates OIP5 expression, as E2F1 is a transcription factor (Fig. 6B and C). We also detected the binding of OIP5 to the E2F1 promoter, but this result was negative (data not shown). Therefore, the inhibition of E2F1 signaling caused by OIP5 depletion may regulate the protein stability of E2F1. While we demonstrated that OIP5 interacts with E2F1 and stabilizes E2F1 signaling in glioblastoma cells (Fig. 5 and Supplementary Fig. S5), the interaction between OIP5 and E2F1 may be indirect, as other proteins may be involved in this interaction. Mechanistically, our studies demonstrate a novel feedback loop in which E2F1 activates the expression of OIP5 to ultimately stabilize E2F1 signaling, persistently augment pathway activity, and promote the E2F1-regulated gene expression required for aggressive tumor biology.
In summary, our research shows the tremendous potential of using cancer-testis specific genes for anticancer therapy. Our study not only identified OIP5 as a tumor promoter in glioblastoma progression and metastasis but also illuminated its underlying mechanisms, suggesting that OIP5 is a potential target for cancer treatment.
Supplementary material
Supplementary material is available at Neuro-Oncology online.
Funding
This study was supported by the National Natural Science Foundation of China (81672502, 81602479), the National Key Research and Development Program of China (2016YFC1302204), and Chongqing University Innovation Team Building Program funded projects (CXTDX201601010).
Conflict of interest statement. No conflicts of interest exist.
Supplementary Material
References
- 1. Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014;54:251–272. [DOI] [PubMed] [Google Scholar]
- 2. van der Bruggen P, Traversari C, Chomez P, et al. . A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. [DOI] [PubMed] [Google Scholar]
- 3. Boël P, Wildmann C, Sensi ML, et al. . BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2(2):167–175. [DOI] [PubMed] [Google Scholar]
- 4. De Backer O, Arden KC, Boretti M, et al. . Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res. 1999;59(13):3157–3165. [PubMed] [Google Scholar]
- 5. Türeci O, Sahin U, Schobert I, et al. . The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res. 1996;56(20):4766–4772. [PubMed] [Google Scholar]
- 6. Türeci Ö, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M. Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc Natl Acad Sci U S A. 1998;95(9):5211–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Y-T, Scanlan MJ, Sahin U, et al. . A testicular antigen aberrantly expressed in human cancers detected by autologous antibodyscreening. Proc Natl Acad Sci U S A. 1997;94(5):1914–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sahin U, Türeci O, Schmitt H, et al. . Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A. 1995;92(25):11810–11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Almeida LG, Sakabe NJ, deOliveira AR, et al. . CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37(Database issue):D816–D819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hofmann O, Caballero OL, Stevenson BJ, et al. . Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105(51):20422–20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naetar N, Hutter S, Dorner D, et al. . LAP2alpha-binding protein LINT-25 is a novel chromatin-associated protein involved in cell cycle exit. J Cell Sci. 2007;120(Pt 5):737–747. [DOI] [PubMed] [Google Scholar]
- 12. Fujita Y, Hayashi T, Kiyomitsu T, et al. . Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12(1):17–30. [DOI] [PubMed] [Google Scholar]
- 13. Williams JM, Chen GC, Zhu L, Rest RF. Using the yeast two-hybrid system to identify human epithelial cell proteins that bind gonococcal Opa proteins: intracellular gonococci bind pyruvate kinase via their Opa proteins and require host pyruvate for growth. Mol Microbiol. 1998;27(1):171–186. [DOI] [PubMed] [Google Scholar]
- 14. Chun HK, Chung KS, Kim HC, et al. . OIP5 is a highly expressed potential therapeutic target for colorectal and gastric cancers. BMB Rep. 2010;43(5):349–354. [DOI] [PubMed] [Google Scholar]
- 15. Koinuma J, Akiyama H, Fujita M, et al. . Characterization of an Opa interacting protein 5 involved in lung and esophageal carcinogenesis. Cancer Sci. 2012;103(3):577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gong M, Xu Y, Dong W, et al. . Expression of Opa interacting protein 5 (OIP5) is associated with tumor stage and prognosis of clear cell renal cell carcinoma. Acta Histochem. 2013;115(8):810–815. [DOI] [PubMed] [Google Scholar]
- 17. Mobasheri MB, Shirkoohi R, Modarressi MH. Cancer/testis OIP5 and TAF7L genes are up-regulated in breast cancer. Asian Pac J Cancer Prev. 2015;16(11):4623–4628. [DOI] [PubMed] [Google Scholar]
- 18. Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alla V, Engelmann D, Niemetz A, et al. . E2F1 in melanoma progression and metastasis. J Natl Cancer Inst. 2010;102(2):127–133. [DOI] [PubMed] [Google Scholar]
- 20. Lee JS, Leem SH, Lee SY, et al. . Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28(16):2660–2667. [DOI] [PubMed] [Google Scholar]
- 21. Hu F, Gartenhaus RB, Zhao XF, et al. . c-Myc and E2F1 drive PBK/TOPK expression in high-grade malignant lymphomas. Leuk Res. 2013;37(4):447–454. [DOI] [PubMed] [Google Scholar]
- 22. Ma X, Gao Y, Fan Y, et al. . Overexpression of E2F1 promotes tumor malignancy and correlates with TNM stages in clear cell renal cell carcinoma. PLoS One. 2013;8(9):e73436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan Y, Wei X, Zhang W, et al. . Resveratrol enhances the radiosensitivity of nasopharyngeal carcinoma cells by downregulating E2F1. Oncol Rep. 2017;37(3):1833–1841. [DOI] [PubMed] [Google Scholar]
- 24. Yan LH, Wei WY, Cao WL, Zhang XS, Xie YB, Xiao Q. Overexpression of E2F1 in human gastric carcinoma is involved in anti-cancer drug resistance. BMC Cancer. 2014;14:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yan LH, Wang XT, Yang J, et al. . Reversal of multidrug resistance in gastric cancer cells by E2F-1 downregulation in vitro and in vivo. J Cell Biochem. 2014;115(1):34–41. [DOI] [PubMed] [Google Scholar]
- 26. Pützer BM, Engelmann D. E2F1 apoptosis counterattacked: evil strikes back. Trends Mol Med. 2013;19(2):89–98. [DOI] [PubMed] [Google Scholar]
- 27. Pierce AM, Gimenez-Conti IB, Schneider-Broussard R, Martinez LA, Conti CJ, Johnson DG. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci U S A. 1998;95(15):8858–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20(1):273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moon NS, Di Stefano L, Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol. 2006;26(20):7601–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng C, Ren Z, Wang H, et al. . E2F1 Induces tumor cell survival via nuclear factor-kappaB-dependent induction of EGR1 transcription in prostate cancer cells. Cancer Res. 2009;69(6):2324–2331. [DOI] [PubMed] [Google Scholar]
- 32. Engelmann D, Mayoli-Nüssle D, Mayrhofer C, et al. . E2F1 promotes angiogenesis through the VEGF-C/VEGFR-3 axis in a feedback loop for cooperative induction of PDGF-B. J Mol Cell Biol. 2013;5(6):391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Knoll S, Fürst K, Kowtharapu B, et al. . E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep. 2014;15(12):1315–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang B, Ma A, Zhang L, et al. . POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun. 2015;6:8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xuan F, Huang M, Liu W, Ding H, Yang L, Cui H. Homeobox C9 suppresses Beclin1-mediated autophagy in glioblastoma by directly inhibiting the transcription of death-associated protein kinase 1. Neuro Oncol. 2016;18(6):819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang R, Wu Y, Wang M, et al. . HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation. Oncotarget. 2015;6(10):7644–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19(1):139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brat DJ, Verhaak RG, Aldape KD, et al. . Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li HC, Chen YF, Feng W, et al. . Loss of the Opa interacting protein 5 inhibits breast cancer proliferation through miR-139-5p/NOTCH1 pathway. Gene. 2017;603:1–8. [DOI] [PubMed] [Google Scholar]
- 40. Li H, Zhang J, Lee MJ, Yu GR, Han X, Kim DG. OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways. Oncotarget. 2017;8(11):18129–18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J, Liu X, Dou Z, et al. . Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP). J Biol Chem. 2014;289(12):8326–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.