Abstract
Mycobacterium africanum consists of Lineages L5 and L6 of the Mycobacterium tuberculosis complex (MTBC) and causes human tuberculosis in specific regions of Western Africa, but is generally not transmitted in other parts of the world. Since M. africanum is evolutionarily closely placed between the globally dispersed Mycobacterium tuberculosis and animal-adapted MTBC-members, these lineages provide valuable insight into M. tuberculosis evolution. Here, we have collected 15 M. africanum L5 strains isolated in France over 4 decades. Illumina sequencing and phylogenomic analysis revealed a previously underappreciated diversity within L5, which consists of distinct sublineages. L5 strains caused relatively high levels of extrapulmonary tuberculosis and included multi- and extensively drug-resistant isolates, especially in the newly defined sublineage L5.2. The specific L5 sublineages also exhibit distinct phenotypic characteristics related to in vitro growth, protein secretion and in vivo immunogenicity. In particular, we identified a PE_PGRS and PPE-MPTR secretion defect specific for sublineage L5.2, which was independent of PPE38. Furthermore, L5 isolates were able to efficiently secrete and induce immune responses against ESX-1 substrates contrary to previous predictions. These phenotypes of Type VII protein secretion and immunogenicity provide valuable information to better link genome sequences to phenotypic traits and thereby understand the evolution of the MTBC.
Keywords: Mycobacterium africanum, Lineage 5, Tuberculosis, PPE38, PhoPR, Antibiotic resistance
Introduction
Tuberculosis remains the most deadly infectious disease of humankind (WHO 2017). Mycobacterium tuberculosis is the main causative agent of human tuberculosis, but closely related species of the M. tuberculosis complex (MTBC), such as Mycobacterium bovis, are able to infect a wide range of domestic and wild animals (Malone and Gordon 2017). The majority of the more than 10 million annual human cases are caused by M. tuberculosis strains of Lineages (L)1–4, but besides M. tuberculosis, the so-called Mycobacterium africanum strains are also known as causative agents of human tuberculosis in West Africa. M. africanum strains were first described in 1968 from tuberculosis patients in Senegal (Castets et al. 1968), and based on phenotypic differentiation criteria these strains showed characteristics that were intermediate between M. tuberculosis and M. bovis (David et al. 1978; Thorel 1980). However, the use of primarily phenotypic criteria resulted in the situation that the exact phylogenomic position of M. africanum strains remained rather vague, as within the strains defined as M. africanum, some groups of strains also closely resembled M. tuberculosis (Haas et al. 1997; Frothingham et al. 1999; Mostowy et al. 2002; Niemann et al. 2002; Sola et al. 2003). Finally, it was the use of comparative genomics, which allowed a clearer phylogenic positioning of M. africanum strains to be established (Brosch et al. 2002; Gagneux et al. 2006). According to this classification, M. africanum strains share a common ancestor that has undergone deletion of the region of difference RD9 and are subdivided into Lineages L5 (also known as M. africanum 1) and L6 (also known as M. africanum 2) (Brosch et al. 2002; Gagneux et al. 2006). In addition to the absence of region RD9, M. africanum L6 strains also lack RD7, RD8, and RD10, similar to the animal adapted strains of the MTBC. These latter three regions have been retained in the genomes of M. africanum L5 strains, indicating that L5 strains have branched from the common RD9-deleted ancestor before the deletion of regions RD7, RD8, and RD10 occurred. This grouping was confirmed by single nucleotide polymorphism (SNP) analyses of whole genome sequences from a few selected strains, which suggest that M. africanum L5 strains show about 2,100 SNPs (filtered) in their genomes relative to the M. tuberculosis H37Rv reference sequence (Comas et al. 2010; Coscolla et al. 2013). Mycobacterium africanum L5 strains thus occupy an ancestral and particularly interesting place in the phylogeny of the tubercle bacilli, which encouraged us to investigate the M. africanum L5 strains in more detail.
Up to now, only a small number of L5 strain genomes have been analyzed and published (Comas et al. 2010; Winglee et al. 2016; Zhu et al. 2016) and phenotypic traits of these strains that might affect their diagnosis, transmission, or virulence characteristics are rarely investigated (de Jong et al. 2010; Gehre, Otu et al. 2013). M. africanum L5 strains are endemic in a number of countries around the Gulf of Guinea, such as Cameroon, Nigeria, Benin, Ghana, Ivory coast, and Sierra Leone, where they cause between 6% and 39% of all tuberculosis cases (de Jong et al. 2010). Some rare M. africanum L5 cases are recorded outside of this region, which are in the large majority associated with patients that were born in this region (de Jong et al. 2010; Sharma et al. 2016). Thus, the geographic restriction of M. africanum is an interesting characteristic, investigation of which may lead to novel insights into the global dispersal of related MTBC members, such as M. tuberculosis sensu stricto and the animal adapted M. bovis (Brites and Gagneux 2015; Stucki et al. 2016).
In this study, we have investigated a panel of 15 L5 strains comprising a large part of the collection of both the current and former French national reference centers for tuberculosis during the last four decades. We sequenced the genomes of these isolates and performed phylogenomic analysis and comparative genomics. The obtained data were combined with phenotypic assessment of drug susceptibility, protein secretion, immunogenicity, and growth characteristics and allow identification of genomic sublineages with a surprising level of genomic and phenotypic variation.
Materials and Methods
Growth Conditions
All newly analyzed isolates of M. africanum (supplementary table 1, Supplementary Material online) were isolated in different locations in France and subsequently sent to the national reference laboratory. Strains were grown on solid culture media of Löwenstein–Jensen (Beckton–Dickinson) and Coletsos.
Cultivation of all isolates (supplementary table 2, Supplementary Material online) was performed on 7H11 solid medium with OADC supplement (Difco), or in liquid 7H9 medium supplemented with ADC (Difco), 0.05% Tween 80, and 0.2% w/v pyruvate (Keating et al. 2005). Kanamycin was added at a concentration of 25 mg/l where appropriate.
Plasmids and Primers
All primers that were used for initial antibiotic resistance testing have been described (Brossier et al. 2017). Other primers used in previous studies (Brosch et al. 2002; Abdallah et al. 2006; McEvoy et al. 2009; Otchere et al. 2016) can be found in supplementary table 4, Supplementary Material online. The pMV::mt2419-22 (ppe38-71) plasmid was previously published (Ates, Dippenaar et al. 2018).
Phenotypic Drug Susceptibility Testing
In vitro drug susceptibility testing was performed on Löwenstein–Jensen medium with the standard proportions method, as previously described (Canetti et al. 1963), with the following concentrations: 40 mg/l rifampicin, 0.2 (Low) and 1 (High) mg/l isoniazid, 2 mg/l ethambutol, 4 mg/l streptomycin, 2 mg/l ofloxacin, 20 mg/l amikacin, 30 mg/l kanamycin, 40 mg/l capreomycin, 40 mg/l ethionamide, 30 mg/l cycloserin, 1.0 mg/l para-aminosalicylic acid, 1.0 mg/l linezolid (Barrera et al. 2008; WHO 2014).
DNA Sequencing of Genes Associated with Drug Resistance
Genomic DNA, used for genomic susceptibility testing, was isolated from bacteria grown on Löwenstein–Jensen medium. A loop of culture was suspended in water (500 µl) and heated at 95 °C for 15 min. The DNA used for polymerase chain reaction (PCR) amplification was obtained by heat shock extraction (1 min at 95 °C and 1 min on ice, repeated five times). Five microliters were submitted to PCR amplification using the oligonucleotide primers described (Brossier et al. 2017). After amplification, unincorporated nucleotides and primers were removed by filtration with Microcon 100 microconcentrators (Amicon, Inc., Beverly, MA), and the amplicons were sequenced by using the BigDye Terminator cycle sequencing-ready kit according to the manufacturer’s instructions.
Determination of Phylogenomic Lineage
The phylogenomic lineages of our MTBC isolates were determined by using the mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) molecular typing method for all NRC-isolates. Standard 24-locus-based MIRU-VNTR typing was performed as described previously (Supply et al. 2006), with the MIRU-VNTR typing kit from GenoScreen. The amplified fragments were analyzed on a 16-capillary Applied Biosystems 3130 genetic analyzer. To determine the lineages of the isolates, the 24 numerical values generated by MIRU-VNTR analysis were compared with those in the MIRU-VNTRplus database (http://www.miru-vntrplus.org/).
DNA Isolation for Genome Sequencing
Mycobacterial genomic DNA was isolated by bead-beating, boiling, and ethanol precipitation as described previously (Howard 2006; Pawlik et al. 2013). Libraries were prepared from the mycobacterial DNA using the Nextera DNA Library Preparation Kit (Illumina, San Diego, CA) following the manufacturer’s recommendations for all strains except for strain 01, 72, 65, 66, 68, and 77, which were prepared with the NextFlex PCR-Free Library preparation. Specific indexes were introduced in both library adapters (double indexing) to allow multiplexed paired-end sequencing. Quality and quantity for each library were checked with the Fragment Analyzer (Advanced Analytical Technologies, Inc., Ames, IA). A total of 108 bp paired-end runs were performed on an Illumina HiSeq 2500 platform for all previously published strains (supplementary table 2, Supplementary Material online) and newly described isolates were sequenced by an Illumina MiSeq using a paired-end (2× 255 bp) approach.
Preprocessing, Alignment, and Variant Detection
Illumina paired-end sequencing data for all isolates published in the current study (N = 16) and 49 genomes, representative of the members of the MTBC, published previously or available in public databases (supplementary table 3, Supplementary Material online) were analyzed with a pipeline composed of open source software as described previously (Dippenaar et al. 2017). Briefly, trimming of adapters and low-quality bases (Phred quality score of less than 20) using a sliding window approach, and filtering for a minimum read length of 36 was done with Trimmomatic (Bolger et al. 2014). Reads were aligned to the M. tuberculosis H37Rv (Genbank: AL123456) and M. tuberculosis CDC1551 genome (Genbank: AE000516) using three different alignment algorithms: The Burrows–Wheeler Alignment (BWA) Tool (Li and Durbin 2009), Novoalign, and SMALT (Ponstingl and Ning 2010). For all sequenced isolates, over 97% of the reference genome was covered by at least one read and an average depth of coverage of 189× (minimum 89, maximum 328) was achieved, when considering the average depth of coverage for each isolate aligned to M. tuberculosis H37Rv with BWA, Novoalign and SMALT. The alignment files were subjected to local realignment and deduplication using the Genome Analysis Toolkit (GATK) (McKenna et al. 2010) and Picard (Winglee et al. 2016). Variants [SNPs and insertion/deletions (indels)] in coding and noncoding regions were identified from each alignment file using GATK (McKenna et al. 2010), and the variants identified in all three alignments were used for further analysis. Variants were annotated using annotation data from TubercuList (Lew et al. 2011), and drug resistance was inferred from a drug resistance mutation library (Coll et al. 2015). SpolPred and SpoTyping were used to predict the spoligotype patterns of all the sequenced M. africanum isolates (Coll et al. 2012; Xia et al. 2016). Genotypic drug resistance prediction based on WGS was done using TBprofiler (Coll et al. 2015).
Phylogenomic Analysis
The raw sequencing data generated for the purpose of this study and 49 selected mycobacterial genomes published previously or available in public databases (Comas et al. 2010; Biek et al. 2012; Blouin et al. 2012, 2014; Coscolla et al. 2013; Bradley et al. 2015; Alexander et al. 2016; Winglee et al. 2016; Zhu et al. 2016), depicted in supplementary table 3, Supplementary Material online, were analyzed. Variants in repetitive regions, such as pe/ppe and pe_pgrs gene families, were excluded for phylogenomic analysis and only variants supported by at least 90% of reads at a given position with a minimum depth coverage of ten reads were considered. Concatenated sequences containing high-confidence variable sites (coding and noncoding SNPs) with respect to the M. tuberculosis H37Rv reference genome were used to infer the phylogenomic relationship between the various strains analyzed as previously described (Dippenaar et al. 2017). The general time reversal model of nucleotide substitution was applied to generate a maximum likelihood phylogeny of the isolates included in this analysis with Randomized Axelerated Maximum Likelihood (RaxML) with 1,000 bootstrap pseudoreplicates (Stamatakis 2015). Positions containing gaps or missing data were not considered for the analysis.
De Novo Assembly and Annotation
De novo assembly of the sequenced M. africanum isolates was done with SPAdes using one Illumina paired-end library for each isolate (Bankevich et al. 2012). The M. tuberculosis CDC1551 genome annotation was transferred to the assembled scaffolds of each isolate using RATT (Otto et al. 2011). Contigs containing the regions of interest were further studied in Artemis (Rutherford et al. 2000), Artemis comparison tool (Carver et al. 2005) and/or Integrative genomics viewer (Robinson et al. 2011).
Growth Assessment
Growth in liquid culture was performed as previously described (Le Chevalier et al. 2015). In short, precultures were grown until exponential phase, washed one time in fresh growth medium and inoculated in 5 ml 7H9 containing ADC, Pyruvate, and Tween 80 in 25 ml glass tube. OD600 was measured at indicated time-points. Regular aeriation of the tubes was undertaken by opening tubes under sterile conditions to ensure the presence of oxygen for optimal growth. The experiment was performed in biological triplicates of technical duplicates. Please note that OD600 was only measured during weekdays and therefore not every data point represents three biological replicates.
Growth on solid medium was assessed by diluting 200 µl of the liquid culture at Day 1 of the growth curve in 10-fold dilutions. A 20 µl of each dilution was spotted on 7H11 + OADC solid growth medium and incubated for 18 days, after which colonies were counted and photographed, colonies were recounted 7 days later.
Secretion Analysis
Secretion analysis was performed broadly as described previously, with some modifications (Houben et al. 2012; Ates et al. 2015). Strains were precultured as described above until all strains reached the exponential phase (OD600 between 0.6 and 1.2), after which they were washed with 7H9 medium without ADC, but supplemented with 0.2% dextrose, 0.2% pyruvate, and 0.025% Tween 80. Washed cells were inoculated in the same medium at a starting OD600 of 0.5 for M. africanum strains or 0.4 OD600 for M. tuberculosis H37Rv and were incubated at 37 °C under shaking conditions over two nights. Cultures were centrifuged and supernatants were filtered (0.22 μm filters, Millipore) and precipitated with Trichloroacetic acid (TCA). Cells were washed with PBS and suspended in solubilization/denaturation buffer, heat inactivated for 2–4 h at 80 °C, sonicated to disrupt cells and boiled for 10 min at 95 °C.
SDS-PAGE gels were loaded with an equivalent of 0.225 OD-units for whole cell lysates and 0.6 OD-units for culture filtrates. Western blots were stained with antibodies: Anti-PGRS 7C4.1F7 (antibody-producing clone kind gift by MJ Brennan, Aeras, Rockville, MD) (Abdallah et al. 2009), anti-SigA (Kind gift from Ida Rosenkrands, Statens Serum Institut, Copenhagen, Denmark), anti-GroEL2 antibody CS-44 (Kind gift from J. Belisle, Colorado State University, Fort Collins, CO) Rabbit polyclonal anti-EsxN (rMTb9.9A) (Alderson et al. 2000), and monoclonal ESAT-6 hyb 76-8 (Harboe et al. 1998).
Immunogenicity of M. africanum Sublineages
Six-week-old female C57BL/6 x CBA F1 (H-2b/k) mice (n = 2 mice/group) received subcutaneous injection of 1 ×106 CFU/mouse of diverse M. africanum, or M. tuberculosis H37Rv strains. Three weeks postimmunization, total splenocytes were cultured in 96-well flat-bottom plates (TPP, Denmark) at 5×105 cells per well in HL-1 medium (Biowhittaker, Lonza, France), complemented with 2 mM GlutaMax (Invitrogen, Life Technologies, France), 5×10−5 M β-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich, France) in the presence of 10 μg/ml of synthetic peptides bearing MHC-II or -I-restricted mycobacterial epitopes. Concanavalin A and PPD were used as positive controls for cell viabilities and successful immunization, respectively, while MalE: 100–114 peptide was used as negative control as previously described (Sayes et al. 2016; Ates, Sayes et al. 2018).
After 3 days of incubation at 37 °C and 5% CO2, IFN-γ produced in the culture supernatant was quantified by sandwich ELISA (clone AN-18 for coating and clone R46A2 for detection, BD Biosciences, San Jose, CA). Data were analyzed by using Prism software (GraphPad Software, La Jolla, CA).
Results
Strain Isolation and Clinical Characterization
The French National Reference Centre for Mycobacteria (NRC MyRMA, http://cnrmyctb.free.fr) routinely receives approximately 1,000 clinical samples and mycobacterial strains per year that are isolated in France. The isolated MTBC strains are subjected to phenotypic and genotypic drug susceptibility testing (WHO 2014). The phylogenetic diversity is evaluated by determining mycobacterial interspersed repetitive unit variable-number tandem repeat (MIRU-VNTR) codes of the strains (Supply et al. 2006). In this capacity, a total of seven M. africanum L5 strains were isolated between 2007 and 2015 (supplementary table 1, Supplementary Material online).
Although strains were isolated from patients living in different locations in France, the places of birth of these patients are located in Western or Central Africa in all cases where this information was available (table 1). Average patient age was 41.7 years [standard deviation (SD) = 0.7]. Three patients were reported as HIV positive (average age 32.7, SD = 6.1) and three patients were HIV negative (average age 45.7, SD = 6.5), while the HIV status of one patient was unknown. Bacterial isolates were derived from different patient samples and remarkably, were isolated from extrapulmonary sites in five out of the seven cases, which was not strictly dependent on HIV status (table 1). The high proportion of extrapulmonary tuberculosis caused by these strains suggests that L5 strains might show a different ability to cause pulmonary disease than M. tuberculosis senso stricto strains. The low number of M. africanum L5 strains isolated over 9 years and the lack of detected transmission episodes are in agreement with an absence of transmission of M. africanum L5 within France. Phenotypic and genotypic drug susceptibility testing classified isolates NRC6 and NRC7 as multidrug resistant (MDR), because they were resistant to both isoniazid and rifampicin. In addition, NRC7 was classified as pre-extensively drug-resistant (pre-XDR: MDR also resistant to the injectable aminoglycoside kanamycin). Three strains were susceptible to first-line drugs (table 2). Corresponding to the described association between HIV and drug resistance described for M. tuberculosis sensu stricto (Mesfin et al. 2014), the pan-susceptible strains were all isolated from HIV negative patients. The patient infected with NRC3 was previously treated for tuberculosis, while no previous treatment information was available for the patient infected with NRC6. In contrast, the five other patients were not documented to have previously received tuberculosis treatment, suggesting transmission of drug-resistant isolates instead of de novo in-patient acquisition of drug resistance.
Table 1.
Patient Characteristics Corresponding to Newly Isolated Strains
| Strain name | Gender | Age | Country of birth | Patient material | Isolated in location, year | HIV-status | Ziehl-Neelsen |
|---|---|---|---|---|---|---|---|
| NRC1 | Female | 39 | Benin | Bronchial aspirate | Besançon, 2007 | Negative | Negative |
| NRC2 | Male | 38 | Cameroon | Lymph node | Paris, 2008 | Positive | Negative |
| NRC3 | Female | 26 | Unknown | Blood | Lyon, 2008 | Positive | Negative |
| NRC4 | Male | 52 | Unknown | Vertebral biopsy | Caen, 2008 | Negative | Positive |
| NRC5 | Male | 46 | Ivory Coast | Lymph node | Poitiers, 2010 | Negative | Positive |
| NRC6 | Male | 57 | DR Congo | Lymph node | Angers, 2011 | Unknown | Unknown |
| NRC7 | Female | 34 | DR Congo | Sputum | Lille, 2015 | Positive | Negative |
Information is reported as provided by physicians sending the isolates to the NRC.
DR, Democratic Republic of
Table 2.
Phenotypic and Genotypic Antibiotic Susceptibility Profiles of Newly Isolated Strains
| Genotypic Resistance mutations |
|||||||
|---|---|---|---|---|---|---|---|
| Name | Previously treated | Phenotypic resistance | rpoB | katG | PinhA | pncA | Other |
| NRC1 | No | Susceptible | wt | wt | wt | wt | |
| NRC2 | No | Isoniazid (Low), streptomycin | wt | wt | c−15t | wt | gyrA/gyrB: wt |
| NRC3 | Yes | Streptomycin | wt | wt | wt | wt | gyrA/gyrB: wt |
| NRC4 | No | Susceptible | wt | wt | wt | wt | |
| NRC5 | No | Susceptible | wt | wt | wt | wt | |
| NRC6 | Unknown | Isoniazid (High), rifampicin, ethionamide, streptomycin, para-aminosalicylic acid | S450(531)L | S315T | wt | wt |
|
| NRC7 | No | Isoniazid (High), rifampicin, pyrazinamide, ethambutol, ethionamide, streptomycin, kanamycin, para-aminosalicylic acid | S450(531)L | S315T | wt | S104R |
|
Genotypic antibiotic susceptibility was tested for first-line drugs isoniazid (katG, PinhA, promoter of inhA), rifampicin (rpoB), pyrazinamide (pncA) (Barrera et al. 2008; WHO 2014), by different PCRs and Sanger-sequencing as described previously (Brossier et al. 2017). When an isolate was resistant to any of these antituberculosis drugs, second-line drugs ethionamide (ethA, ethR); aminoglycosides (*, rrs region 1400, eis promoter), fluoroquinolones (gyrA/gyrB) and para-aminosalicylic acid were tested. Genotypic susceptibility is indicated as “wt” (wild-type), or the resulting amino acid (upper case), or base (lower case) changes. First letter depicts amino acid in reference sequence; second letter depicts detected amino acid/base; rpoB positions between brackets represent positions in older nomenclature system; Position is depicted as number in subscript. N.P., not performed.
Phylogeny of M. africanum Lineage 5 Reveals Distinct Sublineages
To gain more insight into the diversity of M. africanum L5, genomic DNA was isolated from all L5 strains described above. This panel of strains was supplemented with eight additional L5 and two L6 strains from the former National Reference Laboratory for Mycobacteria (at the Institut Pasteur, Paris, France). This second collection of strains was isolated between 1965 and 1998 and these strains were previously genotyped by spoligotyping to belong to their respective lineages (Viana-Niero et al. 2001; Brudey et al. 2004) (supplementary table 2, Supplementary Material online).
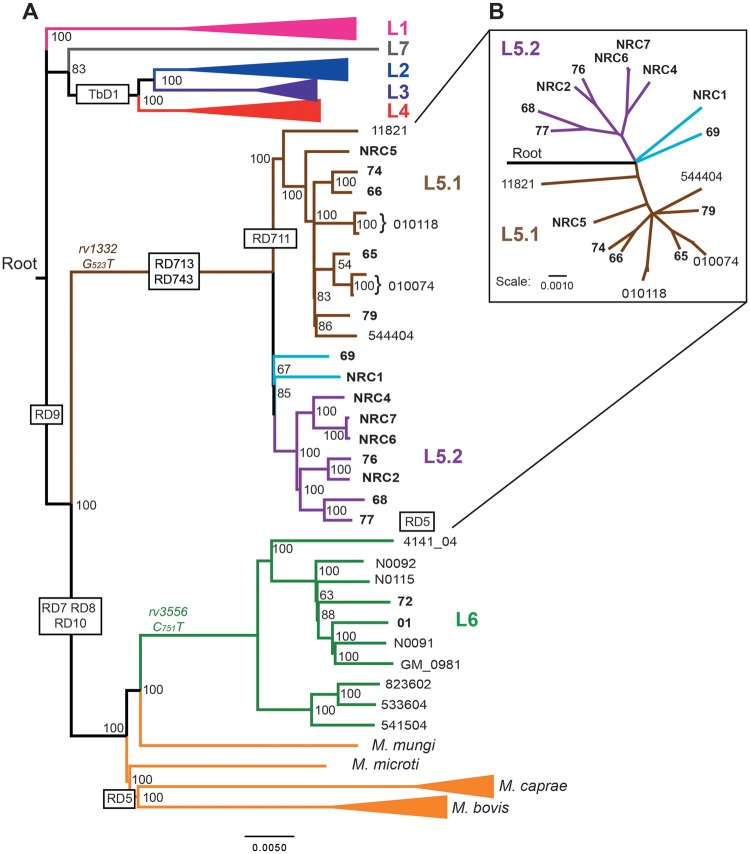
Illumina sequencing of the genomic DNA resulted in an average depth of coverage of between 84- and 328-fold and between 97% and 99% of reads were mapped to the Lineage 4 reference genome H37Rv (supplementary file 1, sheet 1, Supplementary Material online). Based on these data, a comprehensive table of SNPs and insertions/deletions (indels) was constructed to compare isolates (supplementary file 2, sheet 2, Supplementary Material online). More stringent filtering criteria (Materials and Methods) were used to define a panel of SNPs with high confidence that was used to define the phylogenomic relationship between the sequenced M. africanum isolates and a panel of previously published genome-sequences (supplementary table 3, Supplementary Material online) by maximum likelihood analysis. The constructed bootstrap consensus tree fully supported the previous classification of all strains to L5, or L6 (fig. 1A). L5 strains were clearly separated from L6 and were more distantly related to the animal adapted strains of the MTBC, consistent with previous reports (Brosch et al. 2002; Hershberg et al. 2008). Strikingly, a comparison of only the L5 strains among each other, revealed at least two separated subgroups in the phylogeny, which we propose to name M. africanum sublineage 5.1 and 5.2, respectively (figs. 1A and 1B).
Fig. 1.
—Phylogenomic analysis of sequenced and previously published isolates reveals three sublineages within M. africanum L5. (A) Bootstrap consensus tree based on 32,510 variable positions inferred from 1,000 bootstrap replicates is taken to represent the evolutionary history of the isolates (Felsenstein 1989). Previously published genomes and associated accession numbers are listed in supplementary table 3, Supplementary Material online. The phylogenomic analysis was performed with RaxML and is based on variable positions identified with respect to M. tuberculosis H37Rv ( Stamatakis 2006, 2014). Lineages 1, 2, 3, and 4 as well as M. bovis and M. caprae strains are depicted as collapsed branches. A strain of the nonclonal, smooth tubercle bacilli (Blouin et al. 2014; Boritsch et al. 2014) known as Mycobacterium canettii, was used to establish the root of the tree, but is not depicted. Large sequence polymorphisms specific for the (sub)lineages are depicted within black boxes. SNPs specific for either L5 or L6 are depicted in brown or green, respectively. (B) Radial tree of L5 strains depicts the clear separation of L5.1 and L5.2 and the intermediate position of the orphan L5 isolates (light blue). Isolates sequenced in this study are in bold typeset. Newly proposed sublineages are color coded in the tree as: L5.1 (brown) and L5.2 (Lilac). Scale bars depict SNPs/1,000 base pairs.
The identified L5 sublineages correspond well to the classification proposed by Brudey et al., which was based on extended spoligotyping profiles, obtained from a selection of these strains (supplementary table 2, Supplementary Material online) (Brudey et al. 2004). Sublineage 5.1 (L5.1) corresponds to the previously identified Subgroup A2 described by Viana-Niero et al. (2001) or Subgroup A2-1 described by Brudey et al. (2004). A second cluster of seven strains from our collection, which we propose to name L5.2, corresponds to the previously described Subtypes A2-3 (Brudey et al. 2004). Two strains (69 and NRC1) diverged close to the root of L5, in between the two larger sublineages. Strain 69 was previously classified as subtype A2-2 (Brudey et al. 2004) and based on phenotypic characteristics these isolates could be seen as an independent sublineage. However, it should be noted that we were not able to identify sublineage-specific polymorphisms shared between these two strains and the phylogenomic distance from each other and the other sublineages suggest that they are orphan representatives of independent sublineages. Notably, all previously published L5 genomes, clustered inside L5.1 (Comas et al. 2010; Winglee et al. 2016; Zhu et al. 2016), suggesting that the genomic diversity within L5 may have been underestimated in previous whole-genome sequencing studies (Comas et al. 2010; Copin et al. 2014).
Together, these results largely confirm the previous classification of these strains based on spoligotyping and PCR-based methods, but show a markedly different phylogenomic relationship between the isolates than previously suggested (Viana-Niero et al. 2001; Brudey et al. 2004). Our analysis suggests that the orphan representatives NRC1 and isolate 69 diverged from the most-recent common ancestor at a similar timescale as the sublineages L5.1 and L5.2 and these may represent independent sublineages, although more insight might be obtained as more sequences become available.
Previously Proposed Phylogenomic Markers Underestimate L5 Diversity
Different molecular methods have been previously proposed to distinguish M. africanum L5 from other MTBC lineages. Since the phylogenomic analysis revealed that all published L5 genomes clustered with the L5.1 strains, we investigated whether these genetic methods were able to classify all L5 strains accordingly. Vasconcellos et al. (2010) previously described two SNPs that were used to uniquely identify L5 and L6 strains of M. africanum. Indeed, all L5 strains in our collection harbored the rv1332G-523->T polymorphism that was absent from the L6 strains. Conversely, both L6 strains harbored the nat(rv3566c)C-751->T polymorphism, which was absent from all L5 strains (fig. 1A;supplementary file 1, Supplementary Material online). These results confirm these SNPs as reliable genotyping markers for these lineages. However, single-SNP analysis carries risks of creating false positives, compared with the analysis of large sequence polymorphisms or regions of difference (RD). PCR-based analysis of RDs has allowed reliable classification of the different lineages of the MTBC in a cost- and labor-effective way (Gordon et al. 1999; Brosch et al. 2002; Tsolaki et al. 2004). Mycobacterium africanum L5 has in this way been defined as a TbD1-intact lineage having a deletion of the RD9 region, carrying intact RD7, RD8, RD10, and RD702 loci, whereas M. africanum L6 and animal adapted strains additionally have deleted RD7, RD8, RD10, and RD702 regions (fig. 1A) (Brosch et al. 2002; Hershberg et al. 2008). We utilized the obtained Illumina sequence data to catalog these large deletions by mapping paired-end reads to the M. tuberculosis H37Rv reference genome, taking also into account the genome-wide depth of coverage (supplementary file 3, Supplementary Material online) and visual inspection of the alignment of suspected deleted regions (supplementary fig. 1, Supplementary Material online).
The thereby obtained lineage differentiation was found to be robust for all strains tested (fig. 1; supplementary files 1 and 3, Supplementary Material online). However, it would be preferable to have a single unique and lineage-specific marker for L5. The RD711 deletion in genes rv1333-36, was first proposed by Mostowy et al. and is often suggested as such a specific marker for L5 strains (Mostowy et al. 2004; Gagneux et al. 2006; de Jong et al. 2010). Strikingly, our mapping and de novo assembly analyses revealed that this region was indeed deleted in all L5.1 strains, but was intact in all other L5 strains (fig. 1A). These results show that RD711 cannot be used as a universal marker of L5 strains, which was previously suggested (Huard et al. 2006), although it may still be useful to define L5.1 strains.
Three other RD described by Mostowy et al. were here found to be shared between all L5 strains: that is, RD713 affecting rv1977-rv1979c, RD743 affecting rv1992c-1996, and RD715 affecting rv2479c-rv2480c (supplementary fig. 1, Supplementary Material online). Therefore, we propose that these RD have the potential to pose as more reliable markers specific for L5 isolates. However, the flanking region of RD713 partially overlaps with that of RD7 and therefore may fail to produce a PCR product in certain strains (supplementary fig. 1, panel 2, Supplementary Material online) (Mostowy et al. 2004; Huard et al. 2006; Vasconcellos et al. 2010). To our knowledge RD715 and RD743 have not been widely used for genotyping of M. africanum and should first be tested rigorously for specificity, before being implemented in further studies. This is especially true for RD715, which is corresponding to an IS6110 element and may therefore be variable and/or unspecific. Unfortunately, we were unable to identify any large deletions specific for sublineage 5.2 and therefore propose that this sublineage is better identified by analyzing combinations of L5.2-specific SNPs (supplementary file 2, sheet 3, Supplementary Material online). We did not identify deletions or SNPs specific for the orphan representatives, and therefore these isolates were not classified as an independent sublineage.
Bioinformatic Prediction of Drug Resistance is Applicable to M. africanum L5 and Uncovers a 33-kb Deletion Responsible for Isoniazid Resistance and a Putative Association Between L5.2 and Drug Resistance
Genome sequencing is used with increasing frequency in a clinical setting to predict phenotypic antibiotic resistance of M. tuberculosis. Development and validation of these methods are usually based on isolates that are most prevalent in the local clinical setting. However, it is not clear whether the same prediction parameters also apply to more distantly related isolates, such as M. africanum L5 strains. The availability of phenotypic drug-resistance profiles and whole genome sequences of the isolated L5 strains, prompted us to compare the bioinformatic, PCR-based and culture-based drug-resistance profiles. The predicted drug-resistance profiles of first-line antibiotics isoniazid, rifampicin, and pyrazinamide fully corresponded to the observed culture-based antibiotic profiles (table 2; supplementary file 1, sheet 2, Supplementary Material online). All the L5.1 strains included in this study were susceptible to all the antituberculosis drugs tested (table 2; supplementary file 1, sheet 2, Supplementary Material online). Orphan strain NRC1 isolated in 2007, was also pan-susceptible and orphan isolate 69 harbored a L533P mutation conferring only low level resistance to rifampin. In contrast, four out of the seven L5.2 isolates were resistant to multiple anti-TB drugs (supplementary file 1, sheet 2, Supplementary Material online). Isolate NRC2 was resistant to isoniazid because of a nucleotide substitution c-15t in the inhA promoter. Isolates NRC6 and NRC7 were both MDR (Rifampicin-R plus Isoniazid-R) and displayed S450(531)L in RpoB and S315T in KatG, which together represent the most commonly encountered mutations in MDR MTBC clinical isolates. For isolate NRC7, which is pre-extensively drug-resistant (MDR also resistant to the injectable aminoglycoside kanamycin), resistance to kanamycin was correctly predicted by the mutation in the eis promoter (g-37t). In addition, NRC7 was also resistant to pyrazinamide (mutation S103R in PncA) and to ethambutol (mutation M306I in EmbB). The comparison of the 24 loci-MIRU-VNTR profiles of the NRC isolates clearly indicates that NRC6 and NRC7, isolated in 2011 and 2015, respectively, from 2 patients born in the Democratic Republic of the Congo, are closely related (supplementary table 1, Supplementary Material online). They differ only by 1/24 loci, at the level of locus 2163b/QUB11B and are separated by nineteen high-confidence SNPs (defined as a position read coverage >10 and a frequency of >90%; supplementary file 2, Supplementary Material online), suggesting that the two patients belonged to a chain of transmission of a clone in circulation during the 2011–2015 period in this country. However, direct transmission thresholds are usually set at 5–10 SNPs and these cases are therefore likely separated by other cases, which is supported by the finding that both isolates had unique SNPs (i.e. NRC6: 12 SNPs; NRC7: 7 SNPs). Considering the extended resistance profile of NRC7 compared with NRC6, it is likely that the 2 isolates are epidemiologically linked to a common L5.2 MDR epidemic clone that has evolved to gain additional resistance to kanamycin, pyrazinamide and ethambutol, to yield the pre-XDR NRC7 strain isolated in 2015. Finally, isolate 77 was rifampin-resistant with an RpoB N435(516)V mutation. Interestingly, isolate 77 was also classified as a MDR isolate, since it was predicted to be resistant to isoniazid due to deletion of katG. Certain mutations in katG (rv1908) are known to confer isoniazid resistance, through the role of KatG as a catalase/peroxidase that transforms the prodrug isoniazid to its active form (Zhang et al. 1992; Johnsson et al. 1997). Complete deletion of katG as a mechanism of isoniazid resistance has been described before, but is rare and such katG deletion events are commonly associated with a decreased virulence, as determined in animal models (Middlebrook and Cohn 1953; Li et al. 1998; Pym et al. 2002; Tsolaki et al. 2004). However, the ahpC promotor polymorphism at position −48 likely functions as a compensatory mutation for the negative effect of this katG-deletion (supplementary file 1, sheet 2, Supplementary Material online). PCR amplification confirmed the absence of katG in isolate 77 and revealed its presence in all other isolates tested. Culture-based phenotypic resistance showed that isolate 77 was indeed highly resistant (MIC > 1.0 µg/ml) to isoniazid, whereas the closely related isolate 68 was phenotypically susceptible (MIC < 0.2 µg/ml). Closer inspection of read-coverage in the genetic region containing katG, revealed a very large deletion affecting genes corresponding to rv1879 to rv1917c (supplementary fig. 1, panel 2, Supplementary Material online). The deletion concerns a 33.8 kb genomic region corresponding to positions 2,129,643 to 2,163,457 in the M. tuberculosis H37Rv genome (Cole et al. 1998; Kapopoulou et al. 2011). This is to our knowledge the largest-known genomic deletion in the MTBC and includes 38 open reading frames (supplementary file 3, sheets 2 and 16, Supplementary Material online).
Sublineage-Dependent In Vitro Growth Characteristics
M. africanum is known to exhibit specific in vitro growth characteristics compared with M. tuberculosis sensu stricto (Castets et al. 1968; Gehre, Otu et al. 2013). Most notable are a relatively slow growth, dependence on pyruvate in liquid media and small colonies on solid media, which can lead to underdiagnosis in a clinical setting (Castets et al. 1968; Keating et al. 2005; Gehre, Otu et al. 2013). Since early biochemical studies did not discern between different lineages of M. africanum (Castets et al. 1968) and to our knowledge the only systematic evaluation of M. africanum growth rate was performed in L6 strains (Bold et al. 2012; Gehre, Otu et al. 2013), we investigated in vitro growth behavior of our L5 strain collection.
When handling the strains, we noticed a trend of slower growth on solid media for L5.1 strains and therefore performed in vitro growth curves in our standard M. africanum culture medium. M. tuberculosis H37Rv clearly grew faster than all M. africanum isolates (fig. 2). During exponential growth, no lineage-, or sublineage-specific growth characteristics could be observed. However, a sublineage-dependent trend of different maximum optical densities was seen. While L6 and L5.1 strains all reached a plateau of optical density between OD600 1.35 and 1.50, all L5.2 strains reached a lower plateau between OD600 1.06–1.28. The two L5 orphan strains seemed to exhibit an intermediate phenotype. In parallel, we performed assessment of growth on solid 7H11 medium, by serial dilution and spotting of 20 µl of liquid cultures at Day 1 of the growth curves. This confirmed a previous observation by Bold et al. that a culture of M. africanum L6 of a given OD600 contains fewer CFU than that of the L4 isolate H37Rv, possibly because of larger bacterial size (Bold et al. 2012) and extends this observation to all M. africanum sublineages tested here (supplementary fig. 2, Supplementary Material online). In contrast to the growth curve, this analysis confirmed the observed slower growth phenotype of L5.1 compared with all other (sub)lineages (microcolonies appearing after >20 days of culture). L6 and other L5 isolates grew at similar rates (microcolonies appearing after 15 days culture) while H37Rv clearly outgrew all M. africanum isolates (microcolonies after 12 days). This observation was confirmed and quantified by counting the CFUs of all isolates after 17 and 24 days of culture (supplementary fig. 2, Supplementary Material online). These results show that different sublineages of M. africanum L5 have different growth characteristics in vitro depending on the used culture medium.
Fig. 2.
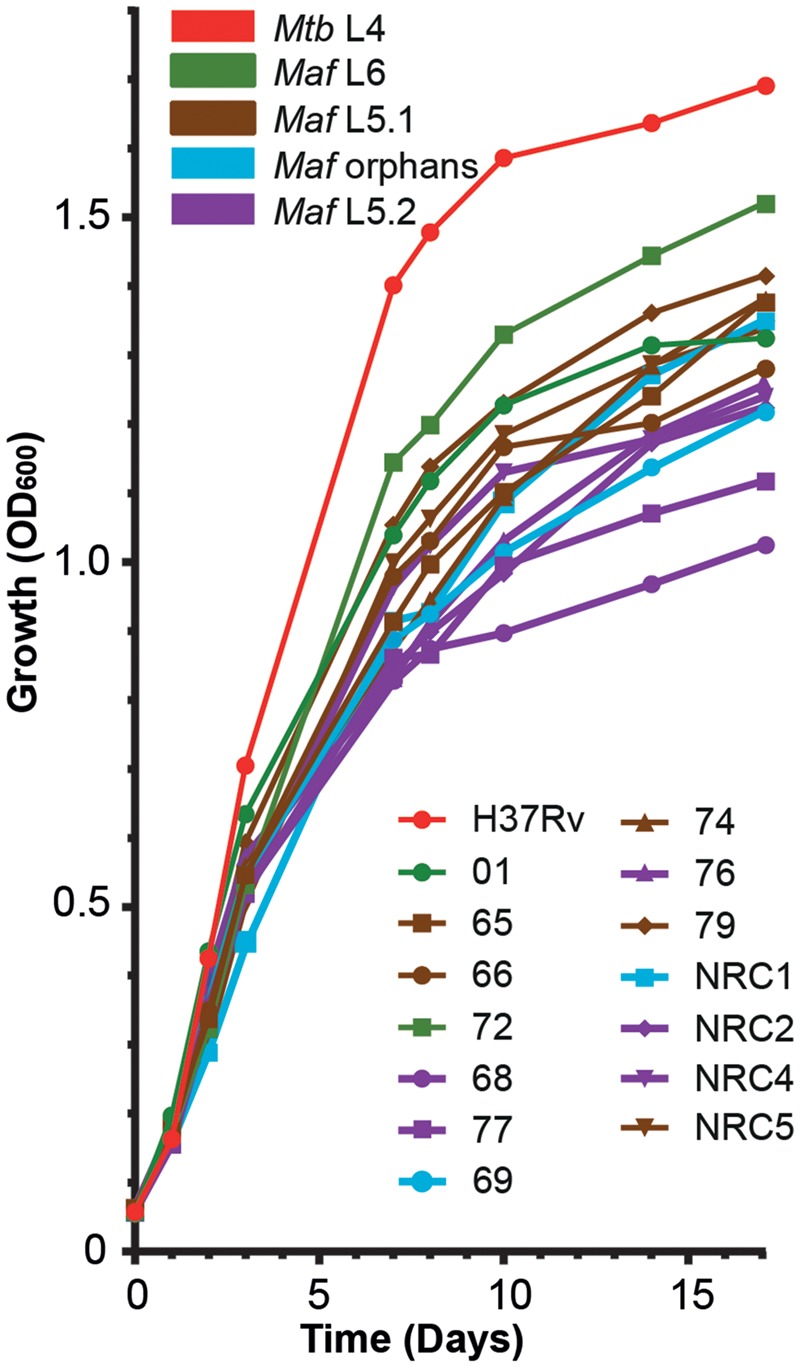
—Sublineage-specific growth characteristics of M. africanum sublineages. Precultures of the indicated strains were grown until midlogarithmic phase and were diluted to 0.05 OD600/ml in glass tubes containing 5 ml of liquid 7H9 medium supplemented with 0.05% Tween 80, ADC supplement and 0.2% pyruvate. Optical density was measured at the indicated time points (symbols). Data points are averages of three individual experiments performed in technical duplicates. Colors of line indicate the specific (sub)lineage of M. tuberculosis (Mtb) or M. africanum (Maf) on the legend (top left). Individual isolates can be identified by combination of colors and symbols used (bottom right).
Secretion analysis of M. africanum Unexpectedly Reveals Fully Functional ESX-1 Secretion in L5
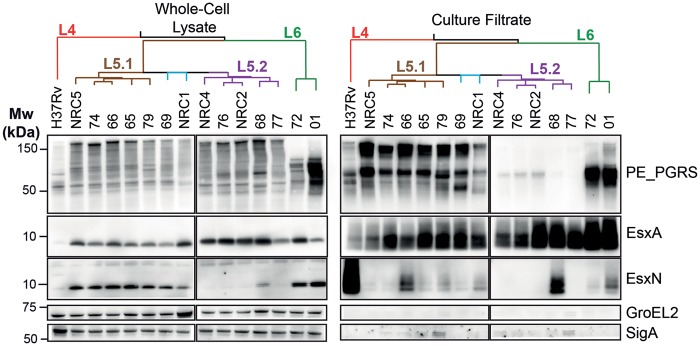
Type VII protein secretion by the secretion systems ESX-1 and ESX-5 are important virulence determinants for M. tuberculosis isolates, which can be affected by relatively small polymorphisms (Gonzalo-Asensio et al. 2014; Ates, Dippenaar et al. 2018). Recently, polymorphisms in the PhoPR two component regulatory system were found to impair virulence, by impacting espACD-mediated secretion of ESX-1 proteins (Blasco et al. 2012; Gonzalo-Asensio et al. 2014; Lou et al. 2017). Since some of these mutations were predicted to be present in M. africanum strains (Gonzalo-Asensio et al. 2014; Malone and Gordon 2017) (supplementary file 2, Supplementary Material online), we analyzed their occurrence in combination with a detailed phenotypic protein expression and secretion analysis. Our analysis showed that the PhoR V71I polymorphism was present in all M. africanum strains of our collection (supplementary file 2, Supplementary Material online), while the deletion of region RD8 (Orgeur and Brosch 2018) was only detected in the L6 strains (supplementary fig. 1, Supplementary Material online). Secretion of EsxA (also known as the 6 kDa early secretory antigenic target ESAT-6) was detected in all M. africanum L5 isolates and occurred at comparable levels to M. tuberculosis H37Rv, while the two M. africanum L6-isolates secreted elevated levels of EsxA (fig. 3). Interestingly, EsxA was detected efficiently in the whole-cell lysates of all L5 and L6 isolates, but only scarcely in H37Rv, probably because H37Rv is a relatively weak EsxA producing strain due to the specific mutation in the promoter region of whiB6 regulatory gene (Solans et al. 2014). In conclusion, we found similar EsxA secretion in M. tuberculosis H37Rv and M. africanum L5, and relatively elevated levels in L6 strains, whereby the latter phenomenon is likely linked to the RD8 deletion.
Fig. 3.
—Secretion analysis of different M. africanum sublineages reveals functional EsxA secretion in L5 isolates and a PE_PGRS secretion defect in L5.2. The phylogenomic relationship of the indicated isolates is depicted by the tree on top of the figure where red indicates L4, brown L5.1, blue orphan L5 isolates, purple L5.2 and green L6. Whole-cell lysates (Left) reveal similar expression levels of PE_PGRS proteins although the detected pattern is different in L6 isolates. EsxN is intracellularly accumulated in all M. africanum isolates except Lineage 5.2. Culture filtrates (right) reveal the L5.2 specific PE_PGRS secretion defect and overall functional EsxA secretion, while EsxN secretion is lower in all M. africanum isolates compared with H37Rv. GroEL2 and SigA staining were used as loading and lysis controls.
A Sublineage Specific PE_PGRS Secretion Defect Independent of PPE38
We previously showed that the ESX-5 substrate PPE38 is required for the secretion of two large subgroups of other ESX-5 substrates called the PE_PGRS and PPE-MPTR proteins (Ates, Dippenaar et al. 2018). Deletions of the ppe38-locus and concurring secretion defects increase virulence of M. tuberculosis during late stage infection. The locus encoding ppe38 and its highly similar copy ppe71, is highly variable in different M. tuberculosis isolates due to frequent recombination/deletion events and/or introduction of an IS6110 insertion element and is part of the RD5 locus that is deleted in M. bovis, M. bovis BCG, and several other animal adapted isolates (Brodin et al. 2002; Garnier et al. 2003; McEvoy et al. 2009; Ates, Dippenaar et al. 2018; Ates, Sayes et al. 2018). Conflicting results about the deletion of RD5 in M. africanum have been reported varying between 0% and 45.4% of M. africanum strains (Viana-Niero et al. 2001; Brosch et al. 2002; Gaudrat et al. 2006). Genome analysis and PCR amplification of the RD5-associated gene plcA (Brosch et al. 2002) and the ppe38-71 locus revealed that only one L5 strain (77, L5.2) had RD5 deleted. Interestingly, all other L5 strains were found to have only one copy of ppe38, while the L6 strains and H37Rv possessed the more common four-gene ppe38-71 locus (McEvoy et al. 2009; Ates, Dippenaar et al. 2018; Ates, Sayes et al. 2018).
Based on the finding that only one strain was a natural ppe38-deletion mutant, we hypothesized that only this strain would be deficient in PE_PGRS secretion. Contrary to this hypothesis, all lineage 5.2 strains exhibited a clear deficiency in PE_PGRS secretion, while all other M. africanum strains secreted these proteins (fig. 3). It should be noted that three L5.2 isolates (76, NRC2, and NRC4), reproducibly secreted residual amounts of PE_PGRS, which is unlike the phenotype of M. tuberculosis ppe38-deleted strains (Ates, Dippenaar et al. 2018). The pattern of both expressed (whole-cell lysate) and secreted PE_PGRS proteins of L6 strains were clearly distinguishable from the other strains (fig. 3).
Furthermore, all M. africanum isolates secreted relatively low levels of the ESX-5 substrate EsxN (fig. 3). This reduced secretion coincided with accumulation of EsxN in the whole-cell lysate of L6 strains as well as L5.1 and L5 orphan isolates, but not in L5.2, where EsxN seemed to be expressed and secreted only in isolate 68 (fig. 3).
Together, these data reveal a L5.2-specific PE_PGRS secretion defect that is seemingly independent of the genetic presence of ppe38. Therefore, we investigated whether this defect might be caused by a loss of transcription or translation of ppe38. Closer investigation of the ppe38 gene locus in all affected isolates (except the RD5-deleted isolate 77) revealed a c → a synonymous SNP corresponding at position 1026 of the ppe38 gene in the L5.2 strains, but not in other M. africanum isolates (supplementary file 2, sheet 2, Supplementary Material online). This SNP results in a codon that is approximately four times less commonly utilized in M. tuberculosis (codon occurrence per 1,000 in M. tuberculosis H37Rv coding sequences: gcg =48.7 → gcu 10.9. Source: http://www.kazusa.or.jp/codon/). To test if reduced ppe38 transcription or translation was a possible explanation for the PE_PGRS secretion defect, we introduced an integrative plasmid that constitutively expresses the ppe38-71 locus (mt2419-22) and successfully complements PPE38-dependent secretion in L2, L4, or BCG isolates, in two L5.2 strains, that is, isolate 68, with functional EsxN secretion and in the RD5-deleted isolate 77 (Ates, Dippenaar et al. 2018; Ates, Sayes et al. 2018). Three PCR-confirmed plasmid-containing clones were tested for PE_PGRS secretion. Low levels of PE_PGRS secretion could be observed in all complemented colonies, showing that L5.2 strains have a PE_PGRS secretion defect, which seems to be independent of ppe38 and may therefore help to uncover novel factors required for the secretion of these proteins.
We set out to uncover such a novel factor by comparison of our genome sequence of L5 orphan representatives and L5.2 and identifying genomic variants specific for L5.2, which may contribute to this PE_PGRS secretion defect. A comparison of large sequence polymorphisms uncovered only one deletion shared by all L5.2 strains, but none of the other M. africanum strains. This polymorphism was a putative in-frame deletion of 1,728 bp within the ppe-mptr gene ppe54. Unfortunately, we were unable to confirm this deletion by PCR, because of the highly repetitive nature and large size of the gene. In parallel, 43 nonsynonymous SNPs or indels were found to be shared between all L5.2 strains (isolate 77 was excluded for this analysis), but with no other M. africanum strains (supplementary file 2, sheet 3, Supplementary Material online). None of these L5.2 specific variants were located in the esx5 locus or could be directly linked to a PE_PGRS secretion defect, although a number of variants were found in ESX-5 substrates, which could be of future interest.
Together, these results confirm that ESX-5 secretion can be markedly different in closely related strains of M. africanum. Since EsxN secretion is clearly functional in strain 68, the PE_PGRS secretion defect in L5.2 strains is no general ESX-5 secretion defect and occurs in spite of a genetically intact ppe38. Therefore additional mechanisms may be required for the secretion of PE_PGRS/PPE-MPTR proteins.
T-Cell Responses Against M. africanum Correlate With In Vitro Secretion Analysis and Include Strong Responses Against ESX-1-Dependent Substrates
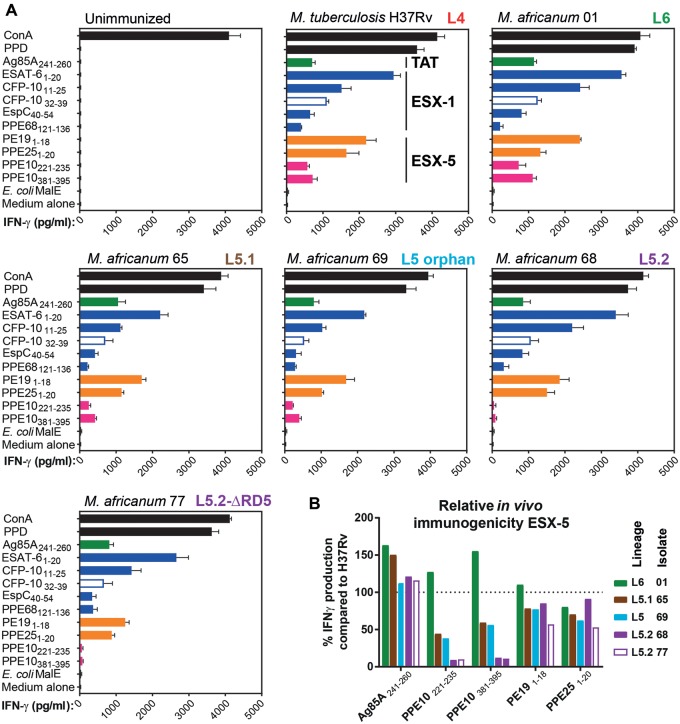
Secretion of ESX Type VII secretion substrates is required for the efficient induction of CD4+ T-cell responses (Majlessi et al. 2005; Brodin et al. 2006; Sayes et al. 2012; Ates, Sayes et al. 2018; Sayes et al. 2018). Since we were surprised by the observed in vitro secretion phenotypes of L5 isolates, we set out to assess how these characteristics affect the cellular immune responses induced by strains belonging to different M. africanum sublineages. We infected C57BL/6 ×CBA F1 mice with M. tuberculosis H37Rv, or with M. africanum (sub)lineage representatives of L6 (strain 01), L5.1 (strain 65), an L5 orphan isolate (strain 69), or Lineage 5.2 with (strain 77) or without (strains 68) an RD5-like deletion. Three weeks postinfection, mice were killed and splenocytes were stimulated with previously identified immunogenic peptides. Interferon-γ (IFNγ) production was measured as a readout of induced T-cell-mediated immunogenicity (Sayes et al. 2012, 2018; Ates, Sayes et al. 2018). Splenocytes from infected mice produced similar patterns of IFN-γ upon stimulation with immunogenic peptides derived from the twin-arginine transporter substrate FbpA (Ag85A241–260), ESX-1 substrates EsxA (EsxA1:20), EsxB (both MHC-II-restricted EsxB11–25 and MHC-I-restricted EsxB32–39), EspC (EspC40–54) and PPE68 (PPE68121–136), or ESX-5 substrates PE19 (PE191–18, an epitope highly specific to the esx-5 region) and PPE25 (PPE251–20, a shared epitope by other PPE proteins encoded outside esx-5 region) (Sayes et al. 2012, 2016) (fig. 4A;supplementary fig. 4, Supplementary Material online). Only the splenocyte responses to the two immunogenic peptides PPE10221–235 and PPE10381–395 derived from the ESX-5 dependent PPE-MPTR protein PPE10 differed markedly between sublineages, with an approximately 90% reduction of IFN-γ production in splenocytes from mice immunized with L5.2 strains, compared with L4 reference H37Rv (fig. 4B;supplementary fig. 4, Supplementary Material online). We previously showed that not only in vitro secretion, but also in vivo induction of T-cell immunity against PPE10 is dependent on PPE38 (Ates, Sayes et al. 2018). However, similar to the observed in vitro PE_PGRS secretion phenotype, both L5.2 isolates were unable to induce immune responses to PPE10 even though isolate 68 does contain one copy of the ppe38 gene. This result suggests that the presumed PPE38-independent PE_PGRS secretion defect of L5.2 isolate can also be extended to the PPE-MPTR proteins. Sublineage L5.1 and L5 orphan representatives induced moderately reduced IFN-γ production in response to PPE10-peptides in comparison to L4 and L6 isolates, possibly because these latter isolates possess a full-length ppe38-locus, while L5 strains only have one copy of ppe38 and no esxXY genes.
Fig. 4.
—Sublineage-specific differences in immunogenicity of M. africanum. (A) Two 6-week-old female C57BL/6 x CBA F1 (H-2b/k) mice per group subcutaneously received 1×106 CFU/mouse of the indicated mycobacterial strains. Three weeks later, mouse splenocytes were stimulated with peptides containing known immunogenic MHC-II (closed bars) or MHC-I (open bars) restricted epitopes (listed on the y axes). IFN-γ production of stimulated splenocytes was measured as a readout of T-cell stimulation (x axes). The (sub)lineage of the tested strains is indicated in bold typeset in the top-right of each panel. Colors indicate the nature of the tested immunogenic peptides. Green: twin-arginine transported substrate; blue: ESX-1 substrate; orange/pink ESX-5 substrate; pink putative PPE38-dependent substrate; black: positive and negative controls. (B) Relative immunogenicity of ESX-5-secreted substrate PPE10 in M. africanum compared with M. tuberculosis H37Rv (dotted line at 100%), reveals PPE-MPTR secretion defect in L5.2 isolates 68 and 77 (purple bars), while other ESX-5 dependent substrates PE19 and PPE25, or the TAT-secreted substrate Ag85A were not significantly affected.
Moreover, we found that M. africanum L5 and L6 isolates were able to induce T-cell responses against selected ESX-1 dependent substrates, confirming the above reported results on EsxA secretion under in vitro conditions. These results also suggest that the previously reported mutation in phoR in M. africanum might not have a strong impact on EsxA secretion in the genomic background of L5 strains. Alternatively, L5 strains might carry yet unknown compensatory mutations that limit the impact of the phoR mutation in these strains.
Discussion
In this work, we systematically compared genomic phylogeny and phenotypic traits of M. africanum L5 isolated over multiple decades in France. In contrast to the high incidence of these strains in their endemic region (de Jong et al. 2010), L5-isolates are only rarely detected in France and are strongly associated with patients born in countries endemic for L5 (Viana-Niero et al. 2001; de Jong et al. 2010; Sharma et al. 2016). This is emphasized by the finding that only seven L5 isolates were submitted to the French National Reference Centre for tuberculosis between 2007 and 2016, while approximately 1,000 samples of M. tuberculosis/MTBC strains are received each year. Furthermore, all these strains were isolated from patients in different French cities and therefore fully support the hypothesis that there is no direct transmission of M. africanum L5 in France and the transmission events probably occurred in the countries of birth of the patients. However, the finding that five out of the seven newly described strains were isolated from extrapulmonary tissues opens the question whether L5 strains are reduced in their pathogenic potential once infection has been established. In fact, it would be an interesting subject of further work to assess whether this high rate of extrapulmonary tuberculosis is truly associated with M. africanum L5 and if this may contribute to the low levels of transmission. Although three of the seven patients carrying L5 isolates were HIV positive, three others were tested to be HIV-negative, suggesting that L5 strains can also multiply in immunocompetent hosts. An important clinical finding is the isolation of pre-XDR L5-isolate NRC7, which was isolated from a patient who did not report previous antituberculosis treatment, suggesting that this strain was transmitted in its current pre-XDR form. MDR isolate NRC6 and pre-XDR isolate NRC7, which share identical spoligotypes and very similar MIRU-VNTR profiles, were the two most-recently isolated L5-isolates submitted to the NRC and highlight that development of drug-resistance is not limited to M. tuberculosis L1–L4, but may also be increasing for M. africanum L5 isolates. Finally, it is particularly intriguing that all the multidrug-resistant strains belonged to sublineage L5.2, suggesting these strains may transmit more efficiently or develop drug-resistance more readily than their closely related counterparts.
Although the isolation of these strains from France is an interesting observation, these strains do not significantly contribute to the French tuberculosis burden. However, the strong geographical restriction of M. africanum and lack of transmission within France, in spite of relatively severe symptoms associated with infection, is of significant fundamental scientific interest to identify the factors that have contributed to the global success of the so-called generalist (sub)lineages of M. tuberculosis (Brites and Gagneux 2015; Stucki et al. 2016). Furthermore, the recent increase in availability of mycobacterial genomes allows for valuable meta-analyses about general variation and adaptation of the MTBC (Comas et al. 2010; Copin et al. 2014; Brites and Gagneux 2015). However, for such analyses to reach maximum potential it is important to include strains reflecting the true diversity of the MTBC. In this light, it is important to note that all previously published L5 genomes were found to belong to sublineage 5.1 and that therefore the genomic diversity of L5 has been underestimated. We would like to suggest future phylogenomic and meta-genomic studies to include at least one representative of sublineage 5.2 and the genomes of orphan isolates 69 and NRC1. The latter two strains were classified as orphan L5 representatives, despite of their previous classification as A2–2 by Brudey et al. (2004), because of their reciprocal genomic distances from L5.1 as well as L5.2 strains. These isolates may represent orphan members of two different sublineages. The genomes of all these isolates are publicly available (Accession number: PRJEB25506) and we have provided extensive supplementary analyses that may aid biologists in further pursuing the questions that have been raised by our work (supplementary files 1–3, Supplementary Material online).
A direct effect of the low amount of diversity in previously analyzed L5 isolates is the use of RD711 as a genomic marker to identify L5 isolates (Mostowy et al. 2004). It was suggested previously, that this marker may not identify all L5 isolates (Huard et al. 2006) and this is now confirmed by our finding that this region is deleted in all L5.1 isolates, but not in other L5 sublineages. Therefore, the SNP-polymorphisms proposed by Vasconcellos et al., RD713, or RD743 may be better suited as lineage-specific markers for L5 (Mostowy et al. 2004; Huard et al. 2006; Vasconcellos et al. 2010). Hopefully, the continuing increase in power and decrease in cost of sequencing techniques will make such typing techniques obsolete, but they are currently still of importance for epidemiological studies in the countries where M. africanum is prevalent and resources and facilities are relatively scarce. This is also true for spoligotyping and MIRU-VNTR, methods which are more informative from an epidemiological perspective than lineage-specific polymorphisms. Spoligotyping has been the predominant technique in the few studies examining the phylogeny of L5 isolates (Viana-Niero et al. 2001; Brudey et al. 2004; Gehre, Antonio et al. 2013). Of course, the resolution of spoligotyping is inferior to that of the whole genome sequencing applied here. However, the combination of whole-genome sequences with spoligotypes and MIRU-VNTR patterns provided here (supplementary table 1, sheet 3, Supplementary Material online) or previously may increase the resolution of newly performed analysis by providing a robust phylogenetic framework (Viana-Niero et al. 2001; Brudey et al. 2004). Retrospective analysis of the data sets from Viana-Niero et al. and our study showed that of the eleven L5.2 isolates, seven were associated with patients from Cameroon and two with patients from the Democratic Republic of Congo (the 2 remaining being of unknown origin), but not with countries such as Ivory Coast, Benin, Ghana and Nigeria, where L5 is also prevalent (Viana-Niero et al. 2001; Brudey et al. 2004). Therefore, L5.2 may be even more geographically restricted to the Central-African part of the L5-endemic area, compared with the Western African countries, which could explain the scarcity of these isolates in previous publications.
Having the genomes of the different sublineages available, allows hypothesis-formation with respect to specific mutations associated with predicted or observed phenotypic traits. One such hypothesis was that L5 strains may secrete lower levels of EsxA in comparison with L6 or L1–4 strains. This was based on the previous finding that L5 strains share the phoR71 (GTT → ATT) mutation that was shown to reduce ESX-1 secretion, but do not have the RD8 deletion, which compensates for this reduction by constitutive expression of the EspACD proteins (Gonzalo-Asensio et al. 2014; Ates and Brosch 2017). However, we were unable to provide evidence for this hypothesis, since all L5 isolates secreted EsxA at levels that were at least comparable to H37Rv and induced comparable T-cell responses to different ESX-1-dependent secreted antigens in vivo. It has to be mentioned, however, that M. tuberculosis H37Rv expresses and secretes lower amounts of EsxA than other M. tuberculosis strains (Solans et al. 2014). Another possible explanation for this observation could be that the L5 isolates have another yet unknown secondary mutation to compensate for the reduction in EsxA secretion caused by the phoR71 (GTT → ATT) polymorphism, which is not present in the M. tuberculosis genomic background. The two L6 isolates secreted slightly, but reproducibly higher levels of EsxA, which are likely to be caused by the deregulation of the espACD locus linked to the RD8 deletion in these strains. Finally, it should be mentioned that EsxA is only one of several ESX-1 substrates and might not be solely responsible for the PhoPR/ESX-1-associated virulence phenotype of tubercle bacilli (Conrad et al. 2017), which also involves a wide range of virulence lipids (Gonzalo-Asensio et al. 2014; Augenstreich et al. 2017). Further work on the variation of the PhoPR system and the associated secretion and virulence phenotypes in the different lineages of tubercle bacilli will be of great interest.
Perhaps one of the most striking phenotypic differences between the different L5 sublineage was the PPE38-independent PE_PGRS secretion defect of sublineage 5.2. Since deletion of ppe38 and the associated PE_PGRS/PPE-MPTR secretion defect were found to increase virulence of M. tuberculosis L2 and L4 isolates, L5.2 isolates may have a similar increased potential of transmission compared with the other L5 isolates. This hypothesis would account for the finding that (i) 67% (4/6) of the L5 isolates received at the French National Reference Center between 2007 and 2015 belong to sublineage 5.2, and (ii) all the resistant NRC isolates, in particular one MDR and one pre-XDR, are L5.2 isolates. However, a larger-scale epidemiological study in local West-African setting would be required to verify this hypothesis. Even though previously published work suggested that approximately half of all M. africanum strains may harbor RD5-deletions (Viana-Niero et al. 2001), we only identified one strain with such a deletion. Intriguingly, all other L5.2 strains also had a clear PE_PGRS secretion deficiency even though they contain an intact ppe38 gene. Constitutive expression of ppe38 by introduction of the vector that was previously successfully used to complement L2, L4 and BCG strains (Ates, Dippenaar et al. 2018; Ates, Sayes et al. 2018), did not restore the PE_PGRS secretion defect, suggesting that this defect is PPE38-independent. Unfortunately, we were unable to identify the genomic cause of this phenotype. However, these results provide the basis to uncover such additional factors in future work with the help of the here reported genomes. Such insight might aid in understanding the molecular mechanism by which PPE38 is required for the secretion of PE_PGRS and PPE-MPTR proteins. Since deletion of ppe38 and the associated PE_PGRS/PPE-MPTR secretion defect were found to cause increase virulence in M. tuberculosis L2 and L4 isolates, it is tempting to speculate that the PE_PGRS/PPE-MPTR secretion defect in L5.2 isolates is balanced by another mutation that limits the increase in virulence, since these isolates are also not transmitted outside their endemic area. In this perspective it should be emphasized that many of the animal-adapted strains of the MTBC have independent deletions of the RD5 region, leading to a similar secretion defect (Ates, Sayes et al. 2018), which could be suggestive of convergent evolution in these branches of the MTBC.
Taken together, by sequencing 15 M. africanum L5 strains isolated in France during the previous decades, we have uncovered a surprisingly high level of genomic and phenotypic diversity. Mycobacterium africanum L5 caused pulmonary and/or extrapulmonary tuberculosis in both HIV-positive and negative patients and we report a pre-XDR isolate from a patient that was not previously treated. The reported genome sequences of these isolates and their associated phenotypic traits with respect to growth, secretion, and immunogenicity may help to increase our understanding of the interesting L5 isolates of M. africanum, which are highly geographically restricted. Understanding this geographic restriction may uncover the factors that have allowed other M. tuberculosis lineages to spread worldwide.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Ethics
Experiments on animals were performed according to the institutional and national guidelines for the care and use of laboratory animals, in strict accordance with European and French regulations (Directive 86/609/CEE and Decree 8-848 of 19 October 1987). All protocols were reviewed and approved by the Institut Pasteur Safety and Animal Care and Use Committee (Protocol 11.245) and the local ethical committee CETEA (Comité d'Ethique en Expérimentation Animale), (approved protocols CETEA 2012-0005 and CETEA 2013-0036).
Supplementary Material
Acknowledgments
We are grateful for help by Gérald Millot, Wafa Frigui, Merel Damen, and Eva Boritsch. We are also grateful for initial discussions and suggestions for Data Analysis by Mickael Orgeur and members of the Bioinformatics and Biostatistics HUB at the Institut Pasteur. We acknowledge the support from the Centre de Recherche Translationnelle (CRT) of the Institut Pasteur. This work was supported in part by the French National Research Council ANR (ANR-14-JAMR-001-02, ANR-10-LABX-62-IBEID, ANR-16-CE35-0009, ANR-16-CE15-0003), the Fondation pour la Recherche Médicale (DEQ20130326471) and the European Union’s Horizon 2020 Research and Innovation Program (643381). LSA and JWJvH are supported by the Netherlands Organisation for Scientific Research (Vidi grant 91717305 to JWJvH). The Genomics Platform of the Institut Pasteur is a member of “France Génomique” consortium (ANR10-INBS-09-08). This study was supported in part by the South African Medical Research Council. The content is the solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council.
Literature cited
- Abdallah AM, et al. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 62:667–679. [DOI] [PubMed] [Google Scholar]
- Abdallah AM, et al. 2009. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 73:329–340. [DOI] [PubMed] [Google Scholar]
- Alderson MR, et al. 2000. Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using human CD4(+) T cells. J Exp Med. 191:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander KA, Larsen MH, Robbe-Austerman S, Stuber TP, Camp PM.. 2016. Draft Genome Sequence of the Mycobacterium tuberculosis Complex Pathogen M. mungi, Identified in a Banded Mongoose (Mungos mungo) in Northern Botswana. Genome Announc. 4:e00471–e00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates LS, et al. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic Mycobacteria. PLoS Genet. 11(5):e1005270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates LS, Dippenaar A, et al. 2018. Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat Microbiol. 3(2):181–188. [DOI] [PubMed] [Google Scholar]
- Ates LS, Sayes F, et al. 2018. RD5-mediated lack of PE_PGRS and PPE-MPTR export in BCG vaccine strains results in strong reduction of antigenic repertoire but little impact on protection. PLoS Pathog. 14(6):e1007139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates LS, Brosch R.. 2017. Discovery of the type VII ESX-1 secretion needle? Mol Microbiol. 103(1):7–12. [DOI] [PubMed] [Google Scholar]
- Augenstreich J, et al. 2017. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol 2017; 19(7):e12726. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera L, et al. 2008. Policy Guidance on Drug-Susceptibility Testing (DST) of Second-Line Antituberculosis Drugs. Geneva: World Health Organization. [PubMed] [Google Scholar]
- Biek R, et al. 2012. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog. 8(11):e1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco B, et al. 2012. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 8(3):e1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin Y, et al. 2014. Progenitor ‘Mycobacterium canettii’ clone responsible for lymph node Tuberculosis epidemic, Djibouti. Emerg Infect Dis. 20(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin Y, et al. 2012. Significance of the identification in the Horn of Africa of an exceptionally deep branching Mycobacterium tuberculosis clade. PLoS One 7(12):e52841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bold TD, et al. 2012. Impaired fitness of Mycobacterium africanum despite secretion of ESAT-6. J Infect Dis. 205(6):984–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritsch EC, et al. 2014. A glimpse into the past and predictions for the future: the molecular evolution of the tuberculosis agent. Mol Microbiol. 93(5):835–852. [DOI] [PubMed] [Google Scholar]
- Bradley P, et al. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun 6(1):10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites D, Gagneux S.. 2015. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev. 264(1):6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, et al. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect Immun. 70(10):5568–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, et al. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 74(1):88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 99(6):3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brossier F, et al. 2017. Molecular detection methods of resistance to antituberculosis drugs in Mycobacterium tuberculosis. Med Mal Infect., doi:10.1016/j.medmal.2017.04.008 [DOI] [PubMed] [Google Scholar]
- Brudey K, et al. 2004. Mycobacterium africanum genotyping using novel spacer oligonucleotides in the direct repeat locus. J Clin Microbiol. 42(11):5053–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canetti G, Rist N, Grosset J.. 1963. [Measurement of sensitivity of the tuberculous bacillus to antibacillary drugs by the method of proportions. Methodology, resistance criteria, results and interpretation]. Rev Tuberc Pneumol (Paris) 27:217–272. [PubMed] [Google Scholar]
- Carver TJ, et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21(16):3422–3423. [DOI] [PubMed] [Google Scholar]
- Castets M, Boisvert H, Grumbach F, Brunel M, Rist N.. 1968. [Tuberculosis bacilli of the African type: preliminary note]. Rev Tuberc Pneumol (Paris) 32(2):179–184. [PubMed] [Google Scholar]
- Le Chevalier F, et al. 2015. Revisiting the role of phospholipases C in virulence and the lifecycle of Mycobacterium tuberculosis. Sci Rep. 5(1):16918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(6685):537–544. [DOI] [PubMed] [Google Scholar]
- Coll F, et al. 2015. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 7(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll F, et al. 2012. SpolPred: rapid and accurate prediction of Mycobacterium tuberculosis spoligotypes from short genomic sequences. Bioinformatics 28(22):2991–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. 2010. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 42(6):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad WH, et al. 2017. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci U S A. 114(6):1371–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin R, et al. 2014. Sequence diversity in the pe_pgrs genes of Mycobacterium tuberculosis is independent of human T cell recognition. MBio 5(1):e00960–e009613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscolla M, et al. 2013. Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerg Infect Dis. 19(6):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David HL, Jahan M-T, Jumin A, Grandry J, Lehman EH.. 1978. Numerical taxonomy analysis of Mycobacterium africanum. Int J Syst Bacteriol. 28(4):464–472. [Google Scholar]
- Dippenaar A, et al. 2017. Progenitor strain introduction of Mycobacterium bovis at the wildlife-livestock interface can lead to clonal expansion of the disease in a single ecosystem. Infect Genet Evol. 51:235–238. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- Frothingham R, et al. 1999. Phenotypic and genotypic characterization of Mycobacterium africanum isolates from West Africa. J Clin Microbiol. 37(6):1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, et al. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 103(8):2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T, et al. 2003. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A. 100(13):7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudrat F, et al. 2006. [Molecular identification of species within the Mycobacterium tuberculosis complex using regions of difference]. Ann Biol Clin (Paris) 64:61–66. [PubMed] [Google Scholar]
- Gehre F, Antonio M, et al. 2013. The first phylogeographic population structure and analysis of transmission dynamics of M. africanum West African 1—Combining molecular data from Benin, Nigeria and Sierra Leone. PLoS One 8(10):e77000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehre F, Otu J, et al. 2013. Deciphering the growth behaviour of Mycobacterium africanum. PLoS Negl Trop Dis. 7(5):e2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo-Asensio J, et al. 2014. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator. Proc Natl Acad Sci U S A. 111(31):11491–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SV, et al. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 32(3):643–655. [DOI] [PubMed] [Google Scholar]
- Haas WH, et al. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob Agents Chemother. 41(7):1601–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harboe M, et al. 1998. B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect Immun. 66(2):717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, et al. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography Blaser, MJ, editor. PLoS Biol. 6(12):e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben ENG, et al. 2012. Composition of the type VII secretion system membrane complex. Mol Microbiol. 86(2):472–484. [DOI] [PubMed] [Google Scholar]
- Howard ST. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152(6):1581–1590. [DOI] [PubMed] [Google Scholar]
- Huard RC, et al. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol. 188(12):4271–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson K, Froland WA, Schultz PG.. 1997. Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J Biol Chem. 272(5):2834–2840. [DOI] [PubMed] [Google Scholar]
- de Jong BC, Antonio M, Gagneux S.. 2010. Mycobacterium africanum—Review of an important cause of human tuberculosis in West Africa Picardeau, M, editor. PLoS Negl Trop Dis. 4(9):e744.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapopoulou A, Lew JM, Cole ST.. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91(1):8–13. [DOI] [PubMed] [Google Scholar]
- Keating LA, et al. 2005. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol Microbiol. 56(1):163–174. [DOI] [PubMed] [Google Scholar]
- Lew JM, Kapopoulou A, Jones LM, Cole ST.. 2011. TubercuList – 10 years after. Tuberculosis 91(1):1–7. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kelley C, Collins F, Rouse D, Morris S.. 1998. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J Infect Dis. 177(4):1030–1035. [DOI] [PubMed] [Google Scholar]
- Lou Y, Rybniker J, Sala C, Cole ST.. 2017. EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol Microbiol. 103(1):26–38. [DOI] [PubMed] [Google Scholar]
- Majlessi L, et al. 2005. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J Immunol. 174(6):3570–3579. [DOI] [PubMed] [Google Scholar]
- Malone KM, Gordon SV.. 2017. Mycobacterium tuberculosis complex members adapted to wild and domestic animals. Adv Exp Med Biol. 1019:135–154. [DOI] [PubMed]
- McEvoy CRE, van Helden PD, Warren RM, Gey van Pittius NC.. 2009. Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol Biol. 9(1):237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesfin YM, Hailemariam D, Biadglign S, Kibret KT, Kibret KT.. 2014. Association between HIV/AIDS and multi-drug resistance tuberculosis: a systematic review and meta-analysis. PLoS One 9(1):e82235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middlebrook G, Cohn ML.. 1953. Some observations on the pathogenicity of isoniazid-resistant variants of tubercle bacilli. Science 118(3063):297–299. [DOI] [PubMed] [Google Scholar]
- Mostowy S, et al. 2004. Genomic analysis distinguishes Mycobacterium africanum. J Clin Microbiol. 42(8):3594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA.. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis. 186(1):74–80. [DOI] [PubMed] [Google Scholar]
- Niemann S, et al. 2002. Mycobacterium africanum subtype II is associated with two distinct genotypes and is a major cause of human tuberculosis in Kampala, Uganda. J Clin Microbiol. 40(9):3398–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgeur M, Brosch R.. 2018. Evolution of virulence in the Mycobacterium tuberculosis complex. Curr Opin Microbiol 41:68–75. [DOI] [PubMed] [Google Scholar]
- Otchere ID, et al. 2016. Detection and characterization of drug-resistant conferring genes in Mycobacterium tuberculosis complex strains: a prospective study in two distant regions of Ghana. Tuberculosis 99:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Dillon GP, Degrave WS, Berriman M.. 2011. RATT: rapid Annotation Transfer Tool. Nucleic Acids Res. 39(9):e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik A, et al. 2013. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol. 90(3):612–629. [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Ning Z.. 2010. SMALT – A new mapper for DNA sequencing reads. F1000Research 1, doi: 10.7490/f1000research.327.1 [Google Scholar]
- Pym AS, Saint-Joanis B, Cole ST.. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun. 70(9):4955–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945. [DOI] [PubMed] [Google Scholar]
- Sayes F, et al. 2016. CD4+ T cells recognizing PE/PPE antigens directly or via cross reactivity are protective against pulmonary Mycobacterium tuberculosis infection. PLOS Pathog. 12(7):e1005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayes F, et al. 2018. Multiplexed quantitation of intraphagocyte Mycobacterium tuberculosis secreted protein effectors. Cell Rep. 23(4):1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayes F, et al. 2012. Strong immunogenicity and cross-reactivity of Mycobacterium tuberculosis ESX-5 type VII secretion: encoded PE-PPE proteins predicts vaccine potential. Cell Host Microbe 11(4):352–363. [DOI] [PubMed] [Google Scholar]
- Sharma A, Bloss E, Heilig CM, Click ES.. 2016. Tuberculosis caused by Mycobacterium africanum, United States, 2004-2013. Emerg Infect Dis. 22(3):396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola C, et al. 2003. Is Mycobacterium africanum subtype II (Uganda I and Uganda II) a genetically well-defined subspecies of the Mycobacterium tuberculosis complex? J Clin Microbiol. 41(3):1345–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solans L, et al. 2014. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WhiB6 as a novel ESX-1 component. Infect Immun. 82(8):3446–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2015. Using RAxML to infer phylogenies. Curr. Protoc. Bioinform. 1:6.14.1–6.14.14. [DOI] [PubMed] [Google Scholar]
- Stucki D, et al. 2016. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. 48(12):1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 44(12):4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel MF. 1980. Isolation of Mycobacterium africanum from monkeys. Tubercle 61(2):101–104. [DOI] [PubMed] [Google Scholar]
- Tsolaki AG, et al. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 101(14):4865–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcellos SEG, et al. 2010. Distinct genotypic profiles of the two major clades of Mycobacterium africanum. BMC Infect Dis. 10(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana-Niero C, et al. 2001. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 39(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2014. Companion Handbook to the WHO Guidelines for the Programmatic Management of Drug-Resistant Tuberculosis. Geneva: World Health Organization. [PubMed] [Google Scholar]
- WHO. 2017. Global Tuberculosis Report 2017. Geneva: WHO; Available from: http://www.who.int/tb/publications/global_report/en/ (accessed November 13, 2017). [Google Scholar]
- Winglee K, et al. 2016. Whole genome sequencing of Mycobacterium africanum strains from Mali provides insights into the mechanisms of geographic restriction. PLoS Negl Trop Dis. 10(1):e0004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia E, Teo Y-Y, Ong RT-H.. 2016. SpoTyping: fast and accurate in silico Mycobacterium spoligotyping from sequence reads. Genome Med. 8(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heym B, Allen B, Young D, Cole S.. 1992. The catalase—peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358(6387):591–593. [DOI] [PubMed] [Google Scholar]
- Zhu L, et al. 2016. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 44(2):730–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.