Abstract
MMSET/WHSC1 is a histone methyltransferase (HMT) overexpressed in t(4;14)+ multiple myeloma (MM) patients, believed to be the driving factor in the pathogenesis of this MM subtype. MMSET overexpression in MM leads to an increase in histone 3 lysine 36 dimethylation (H3K36me2), and a decrease in histone 3 lysine 27 trimethylation (H3K27me3), as well as changes in proliferation, gene expression, and chromatin accessibility. Prior work linked methylation of histones to the ability of cells to undergo DNA damage repair. In addition, t(4;14)+ patients frequently relapse after regimens that include DNA damage-inducing agents, suggesting that MMSET may play a role in DNA damage repair and response. In U2OS cells, we found that MMSET is required for efficient non-homologous end joining as well as homologous recombination. Loss of MMSET led to loss of expression of several DNA repair proteins, as well as decreased recruitment of DNA repair proteins to sites of DNA double strand breaks (DSBs). Using genetically matched MM cell lines that had either high (pathological) or low (physiological) expression of MMSET, we found that MMSET high cells had increased damage at baseline. Upon addition of a DNA damaging agent, MMSET high cells repaired DNA damage at an enhanced rate and continued to proliferate, whereas MMSET low cells accumulated DNA damage and entered cell cycle arrest. In a murine xenograft model using t(4;14)+ KMS11 MM cells harboring an inducible MMSET shRNA, depletion of MMSET enhanced the efficacy of chemotherapy, inhibiting tumor growth and extending survival. These findings help explain the poorer prognosis of t(4;14) MM and further validate MMSET as a potential therapeutic target in MM and other cancers.
Keywords: MMSET/WHSC1/NSD2, histone methylation, DNA damage, DNA repair, NHEJ, HR
Introduction
Multiple myeloma (MM), which accounts for 20% of deaths from hematologic malignancies 1, is a tumor of post-germinal center B-cells that have undergone somatic hypermutation, antigen selection and immunoglobulin heavy chain class switching 2. MM is often characterized by chromosomal translocations that link an oncogene with a strong immunoglobulin promoter/enhancer. Among these, the t(4;14) translocation, occurring in 15–20% of patients, leads to the overexpression of MMSET/WHSC1/NSD2 3. These patients have a poorer prognosis and response to therapy compared to other MM subtypes 4. MMSET was first identified in Wolf-Hirschorn Syndrome, characterized by growth deficiency, craniofacial abnormalities and developmental delays 5. MMSET is one of three NSD family members, all of which possess a SET domain and lysine methyltransferase activity. Overexpression of MMSET is also observed in neuroblastoma, prostate and breast cancer, and is associated with a poorer prognosis 6, 7.
We showed previously that elevated levels of MMSET in t(4;14)+ MM lead to a global increase in histone 3 lysine 36 dimethylation (H3K36me2) and a concomitant decrease in histone 3 lysine 27 trimethylation (H3K27me3) 8–10. H3K36me2 is a chromatin modification associated with active transcription, whereas H3K27me3 is a repressive mark. MMSET overexpression leads to increased proliferation, clonogenicity and chromatin accessibility, as well as changes in gene expression (including DNA repair genes) 8, 11, 12. In vivo targeting of MMSET reverses histone methylation changes and leads to decreased tumor burden in athymic mice, indicating that MMSET is required for MM proliferation and represents a therapeutic target 10.
Treatment of MM has improved but most patients will eventually relapse, especially individuals with adverse cytogenetics such as t(4;14). Melphalan, an alkylating agent that generates either monoadducts or DNA interstrand crosslinks 13, is used at high doses with autologous stem cell transplant in MM patients, resulting in improved survival 14. In the pre-bortezomib and lenalidomide era, t(4;14)+ patients responded well to induction melphalan initially, but then experienced quicker disease progression 15. Including drugs like bortezomib improves patient response rates 16. Furthermore, in relapsed patients, melphalan plus salvage transplant resulted in poorer outcomes in t(4;14)+ patients 15, 17, suggesting that MMSET may influence chemotherapy response. There is still a need for more targeted MM therapies and a clearer understanding of the molecular mechanism of drug resistance and response.
Recently, MMSET was implicated in the DNA damage response (DDR), although differing data exist regarding its function. MMSET was identified in a screen for genes involved in DNA replication stress and its depletion sensitized cells to genotoxic agents 18. Another group reported that MMSET accumulates at DSBs, leading to recruitment of 53BP1, a critical regulator of the DDR 18, 19. MMSET also plays a role in class switch recombination (CSR) during B-cell development, accumulates at immunoglobulin gene switch regions with H3K36me2 and γH2AX, and its depletion led to defects in CSR 20.
H3K36 methylation, created by MMSET and other SET domain proteins in vivo, can also influence DNA repair pathway choice. The SET-domain protein SETMAR/Metnase deposits H3K36me2 at DSBs, enhancing association of non-homologous end joining (NHEJ) repair components at those sites 21. In yeast, loss of Set2/H3K36 methylation led to decreased DNA damage signaling and more open chromatin around DSBs 22, 23. Set2 promoted Ku recruitment to damaged DNA 23 and inhibited homologous recombination (HR) 22, increasing NHEJ. In human cells, loss of the H3K36me3 methyltransferase SETD2 led to reduced DSB end resection and decreased recruitment of HR repair proteins such as RAD51 24–26. Collectively, this background suggests that MMSET may modulate DNA repair and chemotherapy response in t(4;14)+ MM.
Here, we demonstrate that MMSET affects DNA repair when expressed at physiological and pathological levels. MMSET is necessary for both efficient NHEJ and HR repair in U2OS cells, and loss of MMSET leads to decreased expression of DNA repair genes, as well as decreased recruitment of DNA repair proteins to the site of damage. Using genetically matched MM cell lines differing only in their levels of MMSET expression, we found that MMSET promoted the survival and proliferation of cells in the face of DNA damage due to an enhanced rate of DNA repair. Loss of MMSET enhanced the efficacy of chemotherapy in a murine xenograft model, further substantiating MMSET as a therapeutic target.
Results
MMSET participates in multiple pathways of DNA repair
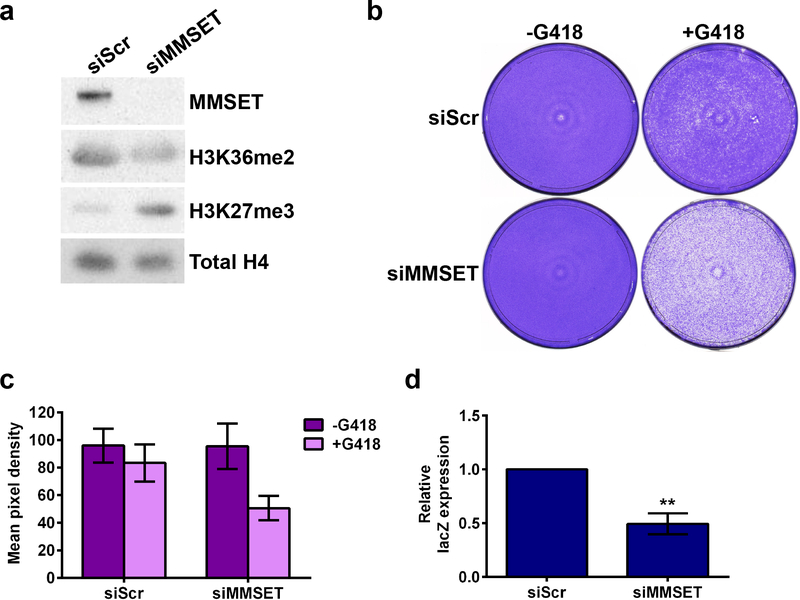
To investigate the role of MMSET in NHEJ, U2OS cells were transfected with a linearized vector harboring a neomycin-resistant gene 27 and an siRNA targeting MMSET. In the presence of neomycin, only cells that can integrate the plasmid via NHEJ survive. As expected 8, 28, 10, MMSET depletion led to decreased levels of H3K36 dimethylation and increased levels of H3K27 trimethylation (Figure 1a). Furthermore, knockdown of MMSET led to decreased formation of drug-resistant colonies (Figure 1b and 1c, Supplemental Figure 1a), suggesting that MMSET is important in NHEJ. In parallel, siRNA-mediated depletion of Ligase IV, which is required to form the NHEJ complex, led to an expected decrease in cell survival (Supplemental Figure 1b) 27. To assess if MMSET had a role in HR, cells were transfected with two mutant β-galactosidase (lacZ) plasmids. Only cells that repair the plasmids via HR express lacZ. Again, the efficiency of HR decreased when MMSET was depleted (Figure 1d and Supplemental Figure 1c).
Figure 1. Loss of MMSET in U2OS cells leads to decreased efficiency of NHEJ and HR.
(a) Immunoblot for MMSET, H3K36me2 and H3K27me3 upon MMSET knockdown in U2OS cells. 5 μg of nuclear protein was loaded per sample. (b) NHEJ assay upon MMSET knockdown in the presence of G418. One representative experiment is shown out of three performed. (c) Quantification of NHEJ assay shown in (b) and Supplemental Figure 1a. The average ± SEM is shown. (d) HR assay measuring relative lacZ expression by qPCR in cells with MMSET knockdown. The average ± SEM is shown for 3 independent experiments. ** p<0.007 by Student’s t-test. A pooled MMSET siRNA was used for all experiments shown.
Using a qPCR-based array, we found that knockdown of MMSET in U2OS cells led to decreased expression of many genes implicated in DNA repair pathways (Supplemental Figure 2a). We used two siRNAs directed against MMSET, one that was a pool of siRNAs (Supplemental Figure 2a, top) and one that was directed toward the C-terminal region of MMSET (Supplemental Figure 2a, bottom). Both siRNA reagents led to downregulation of many of the same genes, including DDIT3, PRKDC, MSH2, MSH3, XRCC2, BARD1 and BLM. We confirmed that MMSET knockdown did not affect cell cycle progression in U2OS cells (Supplemental Figure 2b) and therefore the changes in DNA repair were not simply related to changes in cell proliferation.
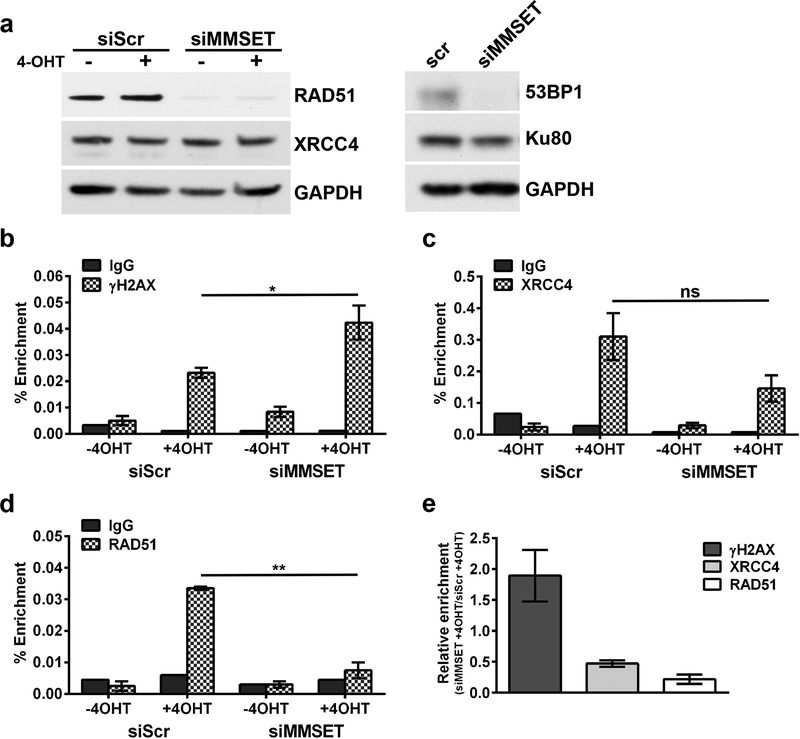
The U2OS cells were engineered to express the AsiSI enzyme fused to an estrogen receptor hormone-binding domain 29. Upon 4-hydroxytamoxifen (4-OHT) treatment, the enzyme translocates into the nucleus to induce DSBs at AsiSI sites throughout the genome. We confirmed an increase in γH2AX levels after addition of 4-OHT (Supplemental Figure 2c). Upon MMSET depletion there was decreased expression of RAD51 and 53BP1 (Figure 2a), and this depletion was not altered by DSB induction. We also observed loss of CtIP expression (data not shown). By contrast, no loss of expression of XRCC4 and Ku80 was observed (Figure 2a). RAD51 binds the ends of single-stranded DNA during HR 30, whereas 53BP1 is a regulator of the DSB response 31. XRCC4 and Ku80 complex with Ligase IV to promote end joining in NHEJ 32.
Figure 2. Loss of MMSET in U2OS cells leads to loss of expression and recruitment of some DNA repair proteins.
(a) Left, immunoblot for RAD51 and XRCC4 upon siRNA knockdown of MMSET. Cells were treated with or without 4-hydroxytamoxifen (4-OHT). 25 μg of total protein was loaded per sample. Right, immunoblot for 53BP1 and Ku80 after siRNA knockdown of MMSET. ChIP-qPCR for (b) γH2AX (n=3 experiments, * p<0.05 by Student’s t-test), (c) XRCC4 (n=3 experiments, not significant), and (d) RAD51 (n=2 experiments, ** p<0.01) occupancy at a DSB site on chromosome 1. For (b-d), the average ± SEM is shown. A pooled MMSET siRNA was used for all experiments shown. (e) Average relative enrichment ± SEM for γH2AX, XRCC4 and RAD41 in siMMSET + 4-OHT relative to siScr + 4-OHT.
We performed chromatin immunoprecipitation (ChIP) and monitored a specific AsiSI-induced DSB site for recruitment of DNA repair proteins. After MMSET knockdown, we observed increased levels of γH2AX, a well-established indicator of DNA damage (Figure 2b and 2e). Simultaneously, XRCC4 recruitment was decreased (Figure 2c and 2e) even though its protein expression was unchanged. ChIP showed that RAD51 was recruited to this DSB but this failed to occur with MMSET knockdown (Figure 2d and 2e), likely due to the striking loss of RAD51 protein expression. These findings imply that MMSET is important for regulating expression of certain DNA repair proteins in both major repair pathways, and may facilitate recruitment of DDR proteins to DSBs.
MMSET extends a proliferative advantage in MM cells treated with a DNA damaging agent
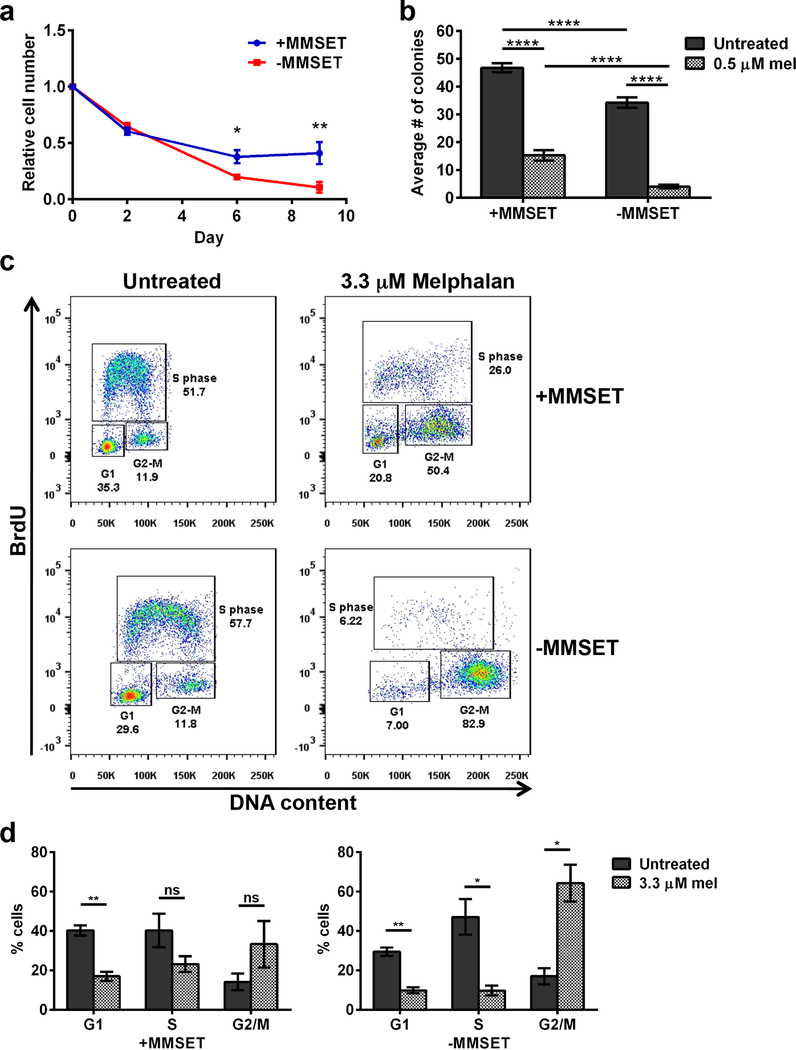
To study the effects of MMSET on the DDR in MM, we used two cell lines derived from the t(4;14)+ KMS11 myeloma cell line and manipulated by HR-mediated gene disruption 11. NTKO, or MMSET high cells, express only the t(4;14) allele, and TKO, or MMSET low cells, express only the wild-type allele (Supplemental Figure 3a). When treated with a modest dose of the DNA crosslinking agent melphalan (0.5 μM) (Figure 3a and 3b) or the DSB inducer bleomycin 33 (Supplemental Figure 3b and 3c), MMSET high cells had increased proliferation and formed more colonies. Based on the differences in proliferation, we next determined if a checkpoint response and cell cycle arrest was occurring. At baseline, both cell lines showed a similar cell cycle profile (Figure 3c, left and 3d). MMSET low cells treated with melphalan had a significant decrease of cells in S phase and an accumulation of cells in G2/M (Figure 3c and 3d, right), in accordance with prior findings 34. However, even when treated for an extended time, MMSET high cells continued to progress through the cell cycle. Treatment of these cells with bleomycin and monitoring response by immunoblot for ATM, DNA-PK and Chk1 showed that both cell lines activated DDR pathways in a similar manner (Supplemental Figure 3d), suggesting that both cell types were sensing and responding to DNA damage. Nevertheless, cell cycle arrest only occurred in MMSET low cells.
Figure 3. MMSET high cells continue to proliferate and form colonies after a DNA damaging insult, whereas MMSET low cells enter cell cycle arrest.
(a) Proliferation of MMSET high (blue) and low (red) cells grown in the presence of 0.5 μM melphalan for 9 days. The average ± SEM is shown for 3 independent experiments. * p<0.02 at day 6, ** p<0.001 at day 10 by Student’s t-test. (b) Colony-forming assay of MMSET high and low cells grown in the presence of 0.5 μM melphalan. Two individual experiments were performed in duplicate. A minimum of 4 fields/well were counted and the average number of colonies is shown ± SEM. **** p<2.3 x 10−5 by Student’s t-test. (c) Cell cycle analysis of MMSET high and low cells treated with or without 3.3 μM melphalan for 5 days. The percentage of cells in each cell cycle phase is shown on each FACS plot. One representative experiment is shown. (d) Quantification of percentage of cells in each phase of the cell cycle in MMSET high (left) or MMSET low (right) cells. The average ± SEM is shown for 3 independent experiments. * p<0.02, ** p<0.002 by Student’s t-test. ns, not significant.
MMSET high cells have increased DNA damage at baseline and enhanced repair
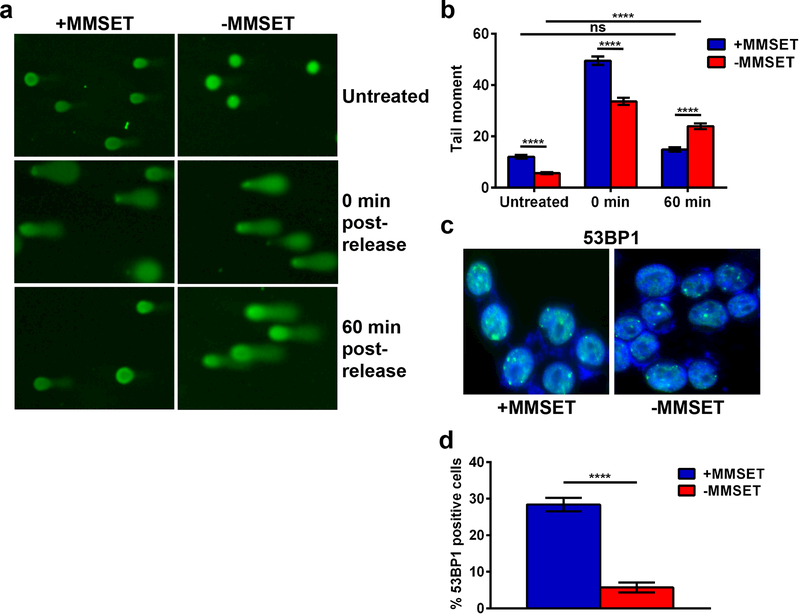
Since DDR signaling appeared intact in MMSET high and low cells, we determined whether the differential response of the cells could be due to differences in the handling of DNA damage. MMSET high and low cell lines were treated with bleomycin for one hour, collected either immediately after treatment or washed, fed with drug-free media and collected one hour later. Intriguingly, at baseline before treatment, MMSET high cells demonstrated higher levels of DNA damage, as measured by the alkaline comet assay (Figure 4a, top and 4b) 35. This was corroborated by immunostaining the MMSET high and low cells for 53BP1, which binds to damaged chromatin 31 (Figure 4c and 4d). After one hour of bleomycin treatment, both cell types had increased levels DNA damage as assayed by tail moment compared to untreated (Figure 4a, middle), but one hour after drug release, MMSET high cells displayed a much shorter tail moment, indicating a significantly greater extent of DNA repair (Figure 4a, bottom and Figure 4b). MMSET low cells still had a significant increase in tail length after drug release. Similar results were found when these cells were treated with melphalan (Supplemental Figure 3e).
Figure 4. MMSET high cells display enhanced DNA damage repair and increased survival.
(a) Images obtained after performing the comet tail assay in MMSET high (left) and low (right) cells. Cells were left untreated (top), treated with 0.33 μg/ml bleomycin for 1 h, washed and then collected immediately after (middle) or 1 h later (bottom). (b) Quantification of the comet tail assay shown in (a). Blue, MMSET high; red, MMSET low. The average ± SEM is shown for 3 independent experiments. **** p<0.0001 by Mann-Whitney U test. ns, not significant. (c) Immunofluorescence for 53BP1 in untreated MMSET high and low cells. Green, 53BP1. Blue, DAPI. (d) Quantification of 53BP1 positive cells shown in (c). The average ± SEM is shown. **** p<0.0006 by Student’s t-test.
Next, MMSET low (TKO) cells were repleted using retroviruses harboring wild-type (wt) MMSET or an HMT-inactive form (Y1118A) of MMSET 8. TKO cells overexpressing wt MMSET had higher levels of baseline DNA damage than cells infected with the control vector. Importantly, the HMT inactive mutant did not induce increased DNA damage (Supplemental Figure 4a and 4b). When cells were treated with a pulse of melphalan for one hour and collected at 0 h and 24 h post-release, we again observed that cells with wt MMSET repaired DNA damage more rapidly (Supplemental Figure 4b). TKO cells overexpressing wt MMSET displayed increased survival when continuously treated with melphalan than did cells containing the vector or HMT-inactive mutant (Supplemental Figure 4c). These data suggest that the increased survival of MMSET overexpressing cells after DNA damaging agents is linked to accelerated DNA repair and that HMT activity of MMSET is critical for its role in DDR in MM.
MMSET high cells have increased rates of DNA damage and repair
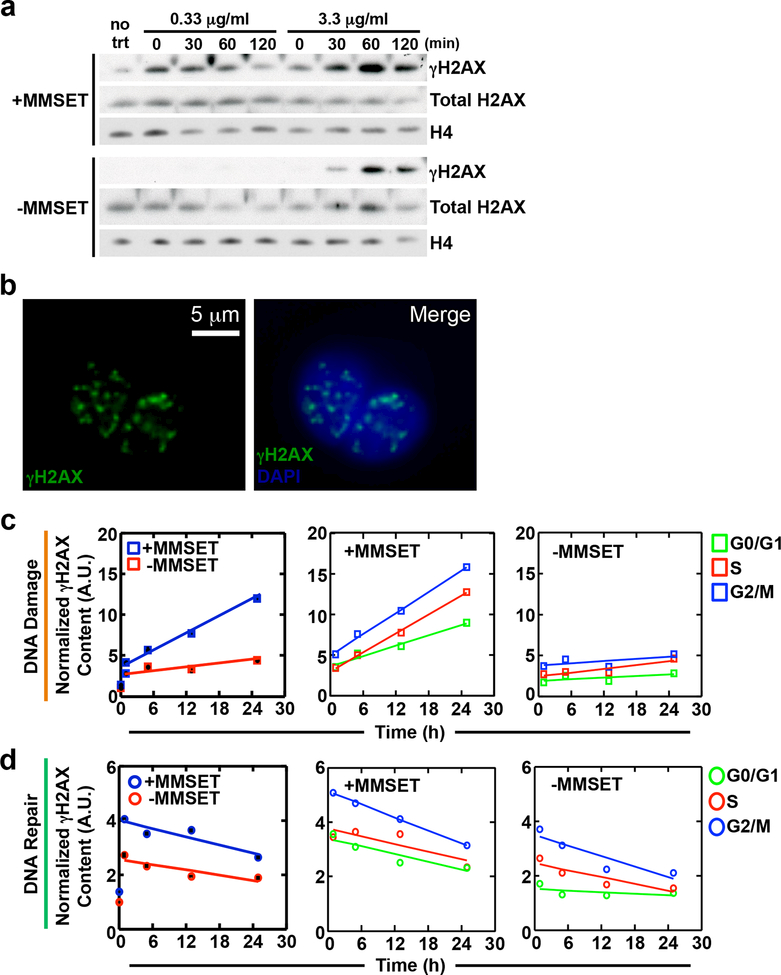
To further examine the induction and resolution of DNA damage in MM cells, we performed kinetic experiments (Figure 5) in which MMSET high and low cells were treated with bleomycin and assayed for γH2AX protein expression over time. In MMSET high cells, a dramatic increase in γH2AX expression was seen immediately following bleomycin release, which returned to baseline levels after 2 hours (Figure 5a). This effect was amplified with higher concentrations of bleomycin. However, in MMSET low cells, no γH2AX expression was observed at the lower concentration of bleomycin. Even at the higher concentration, γH2AX expression was not seen until 30 minutes following drug release and continued to increase over time (Figure 5a). These data extend the results of the comet assay and suggest that MMSET high cells have a higher baseline level of DNA damage, and accumulate more DNA damage after a genotoxic insult.
Figure 5. MMSET high cells have higher levels of DNA damage and an increased rate of DNA repair.
(a) Immunoblot showing γH2AX levels in MMSET high and low cells. Cells were either untreated or treated for 1 h with a low (0.33 μg/ml) or high (3.3 μg/ml) concentration of bleomycin, washed and then collected at 0, 30, 60 and 120 m after drug washout. Total H2AX is a control for γH2AX and H4 is used as a loading control. (b-d) High-throughput cell phenotyping was performed on MMSET high and low cells using γH2AX content to determine rates of DNA damage and repair. Cells were treated continuously with 0.33 μg/ml bleomycin for up to 24 h and γH2AX levels were assessed in each phase of the cell cycle. (b) Left panel, representative image of single cells showing fluorescent staining of γH2AX foci (green). Right panel, merged image with DAPI (blue) for DNA content. (c) DNA damage rates and (d) DNA repair rates in MMSET high and low cells. In (c) and (d), the left graph shows the overall rate of DNA damage or repair for MMSET high (blue) and low (red) cells. The middle (MMSET high) and right (MMSET low) graphs show the rate of damage or repair in each phase of the cell cycle. G0/G1, green; S-phase, red; G2/M, blue.
We further attempted to define differences in DNA damage and repair kinetics using a high throughput single cell phenotyping (htCP) platform 36–38 to elucidate cell cycle-dependent contributions on the DDR. DNA damage and repair kinetics were evaluated by the total γH2AX content present within the nucleus (per single cell) at baseline and after bleomycin treatment (Figure 5b-d) and by using linear regression of the change in γH2AX content as a function of time, we quantified the rates of DNA damage and repair. MMSET high cells displayed an increased rate of DNA damage as seen by a 5-fold increase in γH2AX content relative to MMSET low cells (Figure 5c, left and Supplemental Figure 5a and 5b). The rate of accumulation of γH2AX content/time was significantly increased for all phases of the cell cycle in MMSET high cells (Figure 5c, middle panel) versus MMSET low cells (Figure 5c, right panel, and, Supplemental Figure 5a and 5b). Furthermore, actively cycling cells in G2/M showed higher rates of damage relative to cells in G0/G1 and S phases in both MMSET high and low cells, which may be attributed to differences in the chromatin state as a function of cell cycle progression 36, 39. DNA repair rates were evaluated by the loss of γH2AX signal per single nuclei following bleomycin removal. When cells across all phases of the cell cycle were examined there was a trend towards higher repair rates in MMSET high cells (Figure 5d, left). Upon closer examination of cells in each phase the cell cycle, there was an elevated rate of repair in MMSET high cells in G0/G1 (when NHEJ is active) and G2/M (when HR may occur) as measured by the rate of loss of γH2AX expression, but not S phase (Figure 5d, compare middle and right panels, Supplemental Figure 5a and 5b). Together, these experiments confirm that MMSET high cells have increased levels of DNA damage at baseline, demonstrate that MMSET high cells can tolerate higher levels of DNA damage and accumulate DNA damage at a higher rate, and repair damage faster than MMSET low cells.
MMSET low cells have delayed restoration of chromatin architecture after induced DNA damage
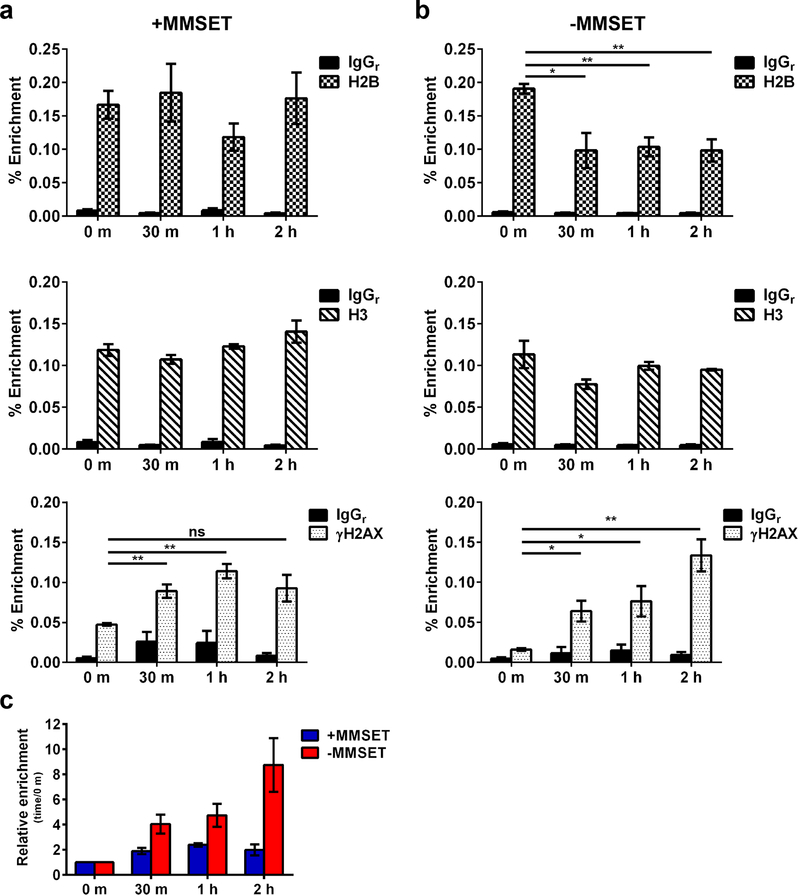
For the DDR to ensue, chromatin is reorganized according to the “access, repair, restore” model 40, 41. DNA damage is recognized, chromatin remodeling occurs to allow DNA repair proteins to access the damage and finally, the original chromatin architecture is restored. Histone dynamics, including histone variants like γH2AX, are important in the DDR and histone chaperones and other chromatin remodelers are required to disassemble and reassemble chromatin during these phases. We hypothesized that MMSET may affect nucleosome disruption caused by DSBs which can be monitored by loss and regain of the H2A/H2B histone dimer 42, 43 at Alu sequences 44. These repetitive elements position nucleosomes with the central and 3’ flanking regions being nucleosome-free 45, thus showing increased sensitivity to DNA damage 45–47. MMSET high and low cells were treated with a pulse of bleomycin and cells were collected at various timepoints after drug release for ChIP followed by amplification of the Alu repeats 48. MMSET high cells exhibited a depletion of H2B, representing a partial loss of nucleosomes one hour after bleomycin treatment, but regained H2B occupancy after 2 hours (Figure 6a, top). By contrast, while MMSET low cells also exhibited eviction of H2B, they regained full nucleosomes more slowly (Figure 6b, top). As expected 43, histone H3 levels over the Alu sequences stayed relatively constant over time (Figure 6a and 6b, middle). We also observed a dynamic change in γH2AX occupancy, similar to Figure 5a (Figure 6a and 6b, bottom). MMSET high cells had increased γH2AX occupancy immediately following drug release at 0 m, whereas MMSET low cells still had low levels of γH2AX (Figure 6a and 6b, bottom). MMSET high cells showed an initial increase of γH2AX and then a plateau of accumulation by 2 hours, but MMSET low cells demonstrated a steady increase of γH2AX over time, again suggesting that MMSET high cells may be capable of limiting DNA damage through accelerated repair.
Figure 6. MMSET low cells have delayed histone restoration at Alu sequences after DNA damage.
MMSET high and low cells were treated with a pulse of bleomycin (3.3 μg/ml) for 1 h, washed and collected at 0 m, 30 m, 1h and 2h after release. ChIP-qPCR was performed over Alu repeat sequences to determine occupancy of H2B, H3 and γH2AX in (a) MMSET high and (b) MMSET low cells. The average ± SEM is shown for 3 independent experiments. * p<0.03, ** p<0.008 by Student’s t-test. (c) Average relative enrichment ± SEM of γH2AX at each timepoint relative to 0 m after drug release in MMSET high and low cells.
Loss of MMSET combined with chemotherapy in mice leads to decreased tumor size and increased survival
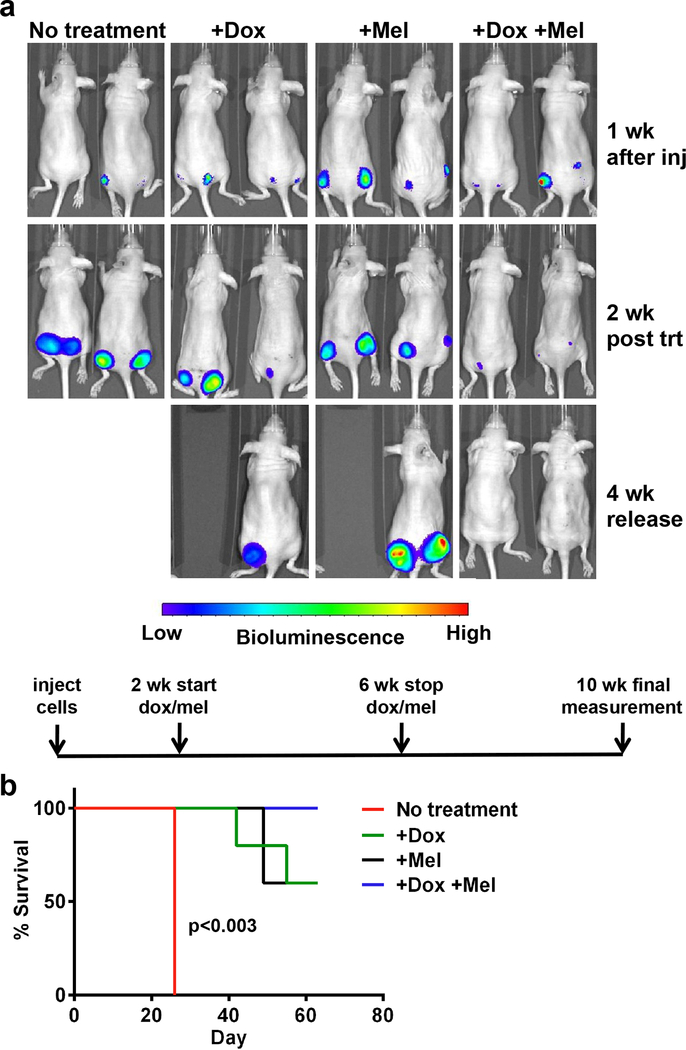
The increased repair capacity and survival of MMSET high cells after chemotherapy represents a barrier to effective therapy for t(4;14)+ MM. Increased DNA repair by MMSET requires its HMT activity. Accordingly, loss of MMSET expression or the application of a potential MMSET enzyme inhibitor would be predicted to enhance chemotherapy efficacy. To test this idea, we injected nude mice with t(4;14)+ KMS11 cells tagged with luciferase and expressing a doxycycline-inducible shRNA targeting MMSET 8, 10 (Figure 7). Tumors formed over two weeks, after which mice were left untreated, administered doxycycline to down-regulate MMSET expression, administered melphalan chemotherapy, or were given both treatments. Treatments were administered for four weeks, and mice were monitored non-invasively for an additional four weeks (Figure 7a, lower schematic). Untreated mice had rapidly growing tumors and were sacrificed due to tumor burden (Figure 7a and 7b). Knockdown of MMSET slowed tumor progression and melphalan chemotherapy had a similar effect on survival (Figure 7b). Strikingly, the combination of MMSET knockdown and melphalan led to decreased tumor size (Figure 7a) and increased survival (Figure 7b), with several mice experiencing complete tumor regression. This suggests that MMSET inhibition might synergize with other therapies in MM patients.
Figure 7. MMSET knockdown and melphalan treatment eliminates tumors in a mouse xenograft model.
(a) Mice were divided into four treatment groups (n=5) and imaged using a fluorescent imaging system to measure tumor formation over time. Two mice from each group are represented. Mice received no treatment (first column), doxycycline alone (second column), melphalan alone (third column), or both doxycycline and melphalan together (fourth column). Images were obtained one week after injection of cells (first row), two weeks after starting doxycycline and/or melphalan treatment (second row), and four weeks after stopping all treatment (bottom row). An experimental timeline is shown below the pictures. Wk, week. Inj, injection. Trt, treatment. (b) Kaplan-Meier survival curve. Red, no treatment. Green, doxycycline alone. Black, melphalan alone. Blue, doxycycline and melphalan in combination. p<0.003 for no treatment vs. +Dox+Mel by Mantel-Cox test.
Discussion
Our work and that of others provide growing evidence that MMSET and histone methylation are important for DNA repair. In U2OS cells, loss of MMSET leads to less efficient NHEJ and HR (Figure 1), correlating with decreased expression of specific DNA repair genes and decreased recruitment of particular DNA repair proteins to DSBs (Figure 2). This suggests that MMSET may act as a transcriptional co-factor to assure the transcription of key DDR components. This does not exclude others modes of action and how MMSET may affect recruitment of factors to sites of DNA damage is not yet understood. Along these lines, when we isolated MMSET partner proteins in 293 cells, we identified KAP1 49, which has been implicated in the DDR 50, suggesting that MMSET may assist in recruitment of some DDR components to chromatin. Recent work has shown that MMSET is recruited to DSBs as part of a larger protein complex, which results in chromatin remodeling and recruitment of RAD51 to the DSB site 51. Alternatively, the loss of expression of key constituents of the DDR pathways, such as CtIP, may lead to inefficient recruitment of other DDR proteins, like XRCC4, to DNA lesions. For example, loss of the histone chaperone nucleolin led to decreased recruitment of XRCC4, abrogating repair at DSB sites 43.
The role of MMSET in the normal DDR may not be reflective of effects of pathological overexpression of MMSET in MM. High levels of MMSET lead to altered gene expression, including expression of DNA repair genes 8, but whether the modest increase in DDR genes we observed in t(4;14) cells can explain the increased rates of DNA damage at baseline and after genotoxic insult is uncertain. Furthermore, there are still conflicting data regarding MMSET and the role of the specific histone marks it makes in relation to DNA repair. While some groups showed that MMSET might accumulate at DSBs and induce H4K20 methylation at those sites, resulting in recruitment of 53BP1 18, 19, two independent groups showed that MMSET had no effect on H4K20 methylation or 53BP1 formation 52, 53. We, and others, have not observed MMSET-induced H4K20 methylation in vivo and instead find that H3K36 is the main target of MMSET 8, 10, 54. A number of studies have shown that the H3K36 mark helps determine DNA repair pathway choice, favoring NHEJ 21–26. We showed that the global increase of H3K36me2 and decrease of H3K27me3 across the genome 10 due to MMSET overexpression was associated with a more open chromatin state characterized by increased chromatin susceptibility to micrococcal nuclease 8. The open chromatin of MMSET high cells may allow DNA to be more accessible to damaging agents, including the genotoxic stresses experienced by cells in culture. In support of this idea, single cell analysis showed that MMSET high cells had increased baseline levels of γH2AX and accumulated more of this modification, reflecting increased DNA damage, when treated with bleomycin (Figure 5). The increased accessibility of chromatin in t(4;14) cells would also allow the DNA repair machinery to more rapidly access lesions, thus facilitating repair of the breaks and restoration of normal chromatin 40, 41 to an intact, undamaged state. Alu repeat sequence analysis supported this idea with MMSET high cells showing more rapid restoration of chromatin (Figure 6). Given that DDR signaling pathways appeared intact in both MMSET high and low cells, we propose that the rapid DNA repair of MMSET high cells underlies their relative insensitivity to genotoxic chemotherapy.
There is precedent for the physical status of chromatin altering the efficiency of DNA repair. Embryonic stem (ES) cells have a more open chromatin structure with more chromatin remodeling occurring at any given time 55, 56. Accordingly, murine ES cells demonstrate a high level of DNA single strand breaks and γH2AX accumulation, which was attributed to global chromatin decondensation 57. Increased levels of histone acetylation characterize transcriptionally active euchromatin and use of histone deacetylase inhibitors (HDACi) leads to a more relaxed chromatin state and increased gene expression. Treating leukemia cells with the HDACi trichostatin A led to DNA damage in regions of DNA containing H4 acetylation and stimulated apoptosis as a result of that damage in leukemic cells 58. Treating cancer cells with a different HDACi, vorinostat, also led to increased γH2AX levels 59. Normal cells could repair DSBs upon HDACi removal, but cancer cells could not, which was attributed to decreased expression of DNA repair proteins 59. These examples all support the idea that altered chromatin structure can lead to increased DNA damage and altered DNA repair in cancer cells.
Melphalan resistance in MM has been associated with enhanced DNA repair. The melphalan-resistant cell line RPMI8226-LR5 demonstrated upregulated expression of NHEJ proteins, including XRCC4 60. Melphalan-resistant cells had an increased number of cells with γH2AX foci compared to -sensitive cells, and the resistant cells showed a greater decrease in the number of γH2AX foci over time 60. A connection between chemotherapy resistance and enhanced DNA repair has also been documented in other malignancies. In a mouse lung cancer model, prolonged treatment with cisplatin led to drug resistance, elevated expression of DNA damage repair genes and enhanced DNA repair 61. The endonuclease Ape1/Ref-1, a key component for base excision repair, can be elevated in human gliomas, contributing to alkylating agent chemotherapy resistance 62. These findings support our data showing that loss of MMSET leads to a decrease in repair protein expression (Figure 2) and that MMSET high cells can repair DNA damage more quickly than MMSET low cells (Figure 4 and 5).
In addition to MM, neuroblastoma, prostate, breast and ovarian cancers also have high levels of MMSET 6, 7. Our findings show that MMSET enhances multiple pathways of DNA repair and provides a rational target for therapy. The search for an MMSET inhibitor is already underway and could prove fruitful for MM and other cancers.
Materials and Methods
See also Supplementary Materials and Methods.
Cell culture
U2OS cells harboring the pBabe-AsiSI-ER vector 29 were grown in DMEM with 10% heat-inactivated fetal bovine serum, 25 mM HEPES and antibiotics. Cells were treated with 300 nM 4-hydroxytamoxifen for 6–8 h to induce DSBs. All MM cell lines were cultured in RPMI as described 8 and treated with varying concentrations of melphalan or bleomycin.
NHEJ assay
U2OS cells were transfected with siScramble, siMMSET pool or siLigase IV. 24 h after transfection, cells were transfected with BamHI-XhoI linearized pEGFP-C1 (Clontech, Mountain View, CA, USA). The next day, cells were trypsinized, counted and plated. Cells with and without G418 (0.5 mg/ml) were incubated for 14 days at 37°C and colonies visualized with 0.05% crystal violet, 1% formaldehyde, 1% methanol. Quantification was performed using ImageJ.
HR assay
A Homologous Recombination Assay (Norgen Biotek, Thorold, ON, Canada) was performed in U2OS cells after transfection with siScramble, siMMSET pool or siMMSET C-terminal. On day 5 of knockdown, cells were trypsinized and 1x105 cells were replated. On day 6, cells were transfected using TurboFect (ThermoScientific, Pittsburgh, PA, USA) with 2.5 μg of Positive Control Plasmid or 2.5 μg each of dl-1 and dl-2 plasmids. 24 h after transfection DNA was isolated using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA). qPCR (Primer sequences in Supplementary Table 1) was performed on a Roche LightCycler 480 II using the Assay Primers from the kit and LightCycler 480 SYBR Green I Master (Roche Applied Science, Indianapolis, IN, USA). GAPDH was used for normalization.
Comet assays
Cells were processed using the CometAssay® Kit (Trevigen, Gaithersburg, MD, USA). Images were obtained using a Leica DFC320 microscope camera with Leica Application Suite V4.4 software. (Leica Microsystems, Wetzlar, Germany). At least 100 cells were analyzed per sample using CometScore (TriTek Corp., Sumerduck, VA, USA).
High-throughput cell phenotyping
MM cells were seeded at a density of 2x105 cells into 6-well plates and treated with 0.33 μg/ml bleomycin for 24 h followed by drug washout and incubation in drug-free media. After treatment, cells were washed and incubated with 1 μg/ml LIVE/DEAD® Fixable Far-Red Dead Cell Stain (L10120, Molecular Probes, Eugene, OR, USA). Cells were fixed, permeabilized 36 and stained for γH2AX (EMD Millipore, Billerica, MA, USA), F-actin (phalloidin, Molecular Probes), and DNA (Hoechst 33342). Fluorescently-labeled suspension cells were injected into a custom microscopy imaging chamber and an 8x8 square grid covering a contiguous area of approximately 30 mm2 was scanned for each sample at low magnification (20x). Four fluorescence channels for Hoescht 33342 (DNA), Alexa Fluor 488 (γH2AX), Alexa Fluor 568 (F-Actin), and 647 live/dead cell stain were recorded to obtain cell and nuclear information at single cell resolution. Image correction, calibration, segmentation and signal quantification was performed as described 36–38.
Xenograft experiments
5x106 KMS11 cells harboring a doxycycline-inducible MMSET shRNA 8 were resuspended in PBS, mixed with CultreX PathClear BME (Trevigen) and injected into six-week-old female athymic nude mice (Crl:NU(NCr)-Foxn1nu, Charles River Laboratories, Wilmington, MA, USA). One week later, mice were divided into groups of 5. Group 1 (control) was administered regular drinking water; group 2, water with doxycycline (2 mg/ml); group 3, regular water and melphalan by intraperitoneal (ip) injection; and group 4, water with doxycycline and melphalan by ip injection. Groups 1 and 2 were given PBS by ip injection as control. Beginning two weeks after treatment initiation, images were acquired using an IVISR Spectrum (Caliper Life Sciences, Inc., Hopkinton, MA, USA) every other week. 150 mg/kg firefly luciferin was injected by ip injection and images were taken 10 min after luciferin injection. Bioluminescence was quantified using the Living Images software (Caliper Life Sciences, Inc.). Kaplan-Meier survival analysis was calculated using GraphPad Prism software.
Supplementary Material
Acknowledgements
NIH grant R01CA123204, Leukemia & Lymphoma Society Specialized Center for Research Award, Physical Sciences Oncology Center U54CA143869, Celgene and the Samuel Waxman Cancer Research Foundation (J.D.L); NIH grants R01CA174388 and U54CA143868 (D.W.); and Canadian Institute of Health and Research operating grant (287300) (N.J.B). We thank Dr. Gaëlle Legube for the AsiSI-U2OS cells and members of the Licht lab for their helpful comments.
Footnotes
Conflict of interest: J.D.L. receives funding from Celgene.
References
- 1.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 2005; 23: 6333–6338. [DOI] [PubMed] [Google Scholar]
- 2.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med 2011; 364: 1046–1060. [DOI] [PubMed] [Google Scholar]
- 3.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998; 92: 3025–3034. [PubMed] [Google Scholar]
- 4.Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood 2003; 101: 1520–1529. [DOI] [PubMed] [Google Scholar]
- 5.Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 1998; 7: 1071–1082. [DOI] [PubMed] [Google Scholar]
- 6.Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiaer E, Rossing HH, Johansen JV et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 2011; 17: 2919–2933. [DOI] [PubMed] [Google Scholar]
- 7.Hudlebusch HR, Skotte J, Santoni-Rugiu E, Zimling ZG, Lees MJ, Simon R et al. MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer research 2011; 71: 4226–4235. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011; 117: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, Sweet SM, Popovic R, Martinez-Garcia E, Tipton JD, Thomas PM et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proceedings of the National Academy of Sciences of the United States of America 2012; 109: 13549–13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Ezponda T, Shah MY et al. Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation. PLoS Genet 2014; 10: e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood 2008; 111: 856–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 2011; 44: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gourzones-Dmitriev C, Kassambara A, Sahota S, Reme T, Moreaux J, Bourquard P et al. DNA repair pathways in human multiple myeloma: role in oncogenesis and potential targets for treatment. Cell Cycle 2013; 12: 2760–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palumbo A, Cavallo F, Gay F, Di Raimondo F, Ben Yehuda D, Petrucci MT et al. Autologous transplantation and maintenance therapy in multiple myeloma. N Engl J Med 2014; 371: 895–905. [DOI] [PubMed] [Google Scholar]
- 15.Jaksic W, Trudel S, Chang H, Trieu Y, Qi X, Mikhael J et al. Clinical outcomes in t(4;14) multiple myeloma: a chemotherapy-sensitive disease characterized by rapid relapse and alkylating agent resistance. J Clin Oncol 2005; 23: 7069–7073. [DOI] [PubMed] [Google Scholar]
- 16.Roussel M, Moreau P, Huynh A, Mary JY, Danho C, Caillot D et al. Bortezomib and high-dose melphalan as conditioning regimen before autologous stem cell transplantation in patients with de novo multiple myeloma: a phase 2 study of the Intergroupe Francophone du Myelome (IFM). Blood 2010; 115: 32–37. [DOI] [PubMed] [Google Scholar]
- 17.Cook G, Williams C, Brown JM, Cairns DA, Cavenagh J, Snowden JA et al. High-dose chemotherapy plus autologous stem-cell transplantation as consolidation therapy in patients with relapsed multiple myeloma after previous autologous stem-cell transplantation (NCRI Myeloma X Relapse [Intensive trial]): a randomised, open-label, phase 3 trial. Lancet Oncol 2014; 15: 874–885. [DOI] [PubMed] [Google Scholar]
- 18.Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America 2011; 108: 13130–13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011; 470: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pei H, Wu X, Liu T, Yu K, Jelinek DF, Lou Z. The histone methyltransferase MMSET regulates class switch recombination. J Immunol 2013; 190: 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proceedings of the National Academy of Sciences of the United States of America 2011; 108: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jha DK, Strahl BD. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun 2014; 5: 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pai CC, Deegan RS, Subramanian L, Gal C, Sarkar S, Blaikley EJ et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 2014; 5: 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 2014; 21: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 2014; 3: e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell reports 2014; 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010; 17: 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene 2013; 32: 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 2010; 29: 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 2012; 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 2014; 15: 7–18. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Robert C, Rassool FV. The Role of Error-Prone Alternative Non-Homologous End-Joining in Genomic Instability in Cancer In: Vengrova S (ed). DNA Repair and Human Health, 2011. [Google Scholar]
- 33.Povirk LF, Han YH, Steighner RJ. Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5' extensions. Biochemistry 1989; 28: 5808–5814. [DOI] [PubMed] [Google Scholar]
- 34.Lupi M, Cappella P, Matera G, Natoli C, Ubezio P. Interpreting cell cycle effects of drugs: the case of melphalan. Cancer chemotherapy and pharmacology 2006; 57: 443–457. [DOI] [PubMed] [Google Scholar]
- 35.Liao W, McNutt MA, Zhu WG. The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods 2009; 48: 46–53. [DOI] [PubMed] [Google Scholar]
- 36.Chambliss AB, Wu PH, Chen WC, Sun SX, Wirtz D. Simultaneously defining cell phenotypes, cell cycle, and chromatin modifications at single-cell resolution. FASEB J 2013; 27: 2667–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Q, Phillip JM, Majumdar S, Wu PH, Chen J, Calderon-Colon X et al. Modulation of keratocyte phenotype by collagen fibril nanoarchitecture in membranes for corneal repair. Biomaterials 2013; 34: 9365–9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WC, Wu PH, Phillip JM, Khatau SB, Choi JM, Dallas MR et al. Functional interplay between the cell cycle and cell phenotypes. Integrative biology : quantitative biosciences from nano to macro 2013; 5: 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulli G, Di Micco R, d'Adda di Fagagna F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 2012; 12: 709–720. [DOI] [PubMed] [Google Scholar]
- 40.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell 2010; 140: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 2012; 46: 722–734. [DOI] [PubMed] [Google Scholar]
- 42.Berkovich E, Monnat RJ Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 2007; 9: 683–690. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America 2013; 110: 16874–16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deininger P Alu elements: know the SINEs. Genome Biol 2011; 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Englander EW, Howard BH. Nucleosome positioning by human Alu elements in chromatin. J Biol Chem 1995; 270: 10091–10096. [DOI] [PubMed] [Google Scholar]
- 46.Englander EW, Wolffe AP, Howard BH. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J Biol Chem 1993; 268: 19565–19573. [PubMed] [Google Scholar]
- 47.Englander EW, Howard BH. Alu-mediated detection of DNA damage in the human genome. Mutat Res 1997; 385: 31–39. [DOI] [PubMed] [Google Scholar]
- 48.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011; 332: 1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Min DJ, Ezponda T, Kim MK, Will CM, Martinez-Garcia E, Popovic R et al. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia 2013; 27: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cann KL, Dellaire G. Heterochromatin and the DNA damage response: the need to relax. Biochemistry and cell biology = Biochimie et biologie cellulaire 2011; 89: 45–60. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q, Goldstein M. Small RNAs recruit chromatin modifying enzymes MMSET and Tip60 to reconfigure damaged DNA upon double-strain break and facilitate repair. Cancer Research 2016. [DOI] [PubMed] [Google Scholar]
- 52.Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol 2013; 5: 157–165. [DOI] [PubMed] [Google Scholar]
- 53.Tuzon CT, Spektor T, Kong X, Congdon LM, Wu S, Schotta G et al. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell reports 2014; 8: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 2009; 284: 34283–34295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developmental cell 2006; 10: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C et al. Global transcription in pluripotent embryonic stem cells. Cell stem cell 2008; 2: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banath JP, Banuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res 2009; 315: 1505–1520. [DOI] [PubMed] [Google Scholar]
- 58.Gaymes TJ, Padua RA, Pla M, Orr S, Omidvar N, Chomienne C et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Molecular cancer research : MCR 2006; 4: 563–573. [DOI] [PubMed] [Google Scholar]
- 59.Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proceedings of the National Academy of Sciences of the United States of America 2010; 107: 14639–14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sousa MM, Zub KA, Aas PA, Hanssen-Bauer A, Demirovic A, Sarno A et al. An inverse switch in DNA base excision and strand break repair contributes to melphalan resistance in multiple myeloma cells. PLoS One 2013; 8: e55493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes & development 2010; 24: 837–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB et al. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clinical cancer research : an official journal of the American Association for Cancer Research 2002; 8: 3008–3018. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.